Abstract
The β-subunit of DNA polymerase III is located as one or two condensed clusters within the nucleoid-occupied space in exponentially growing cells of Escherichia coli. When chromosome replication is terminated after incubation at nonpermissive temperature in a temperature-sensitive dnaC mutant, the β-subunit is located in the cytosolic spaces of the cell poles.
The dnaN gene in Escherichia coli encodes the β-subunit of DNA polymerase III. A dimer of the β-subunit forms the DNA sliding clamp. The dimer is loaded to DNA in an ATP-dependent manner catalyzed by the γ complex (a subassembly of DNA polymerase III) to form the preinitiation complex. In the next step, the DNA polymerase III core that catalyzes DNA synthesis associates with the preinitiation complex to form the initiation complex. The β-subunit exists as 300 to 5,000 dimers per cell (for reviews, see references 1, 6, 7, and 9). In this work, we have analyzed the subcellular localization of the β-subunit in exponentially growing wild-type cells and temperature-sensitive dnaC mutant cells synchronized for initiation of chromosome replication.
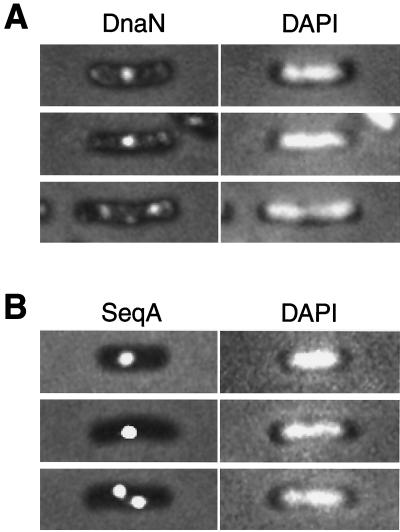
Subcellular localization of the β-subunit of DNA polymerase III and SeqA in exponentially growing cells.
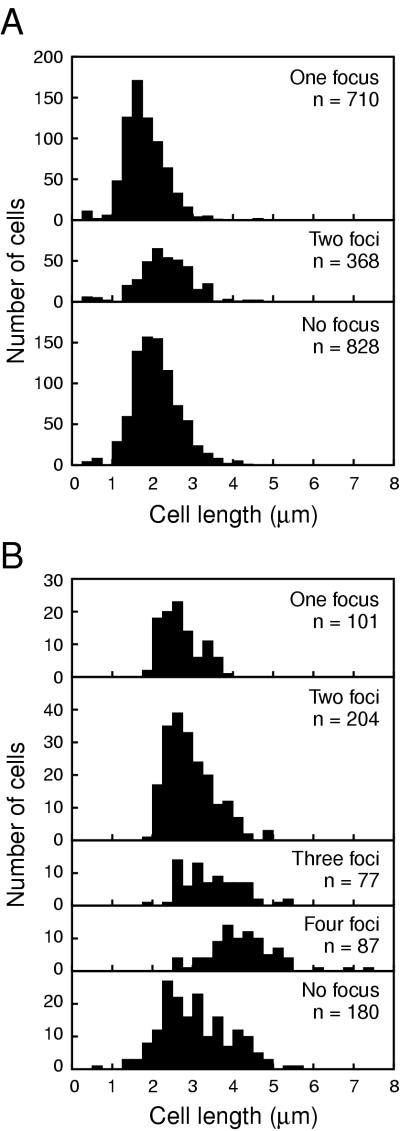
We analyzed the subcellular localization of the β-subunit of DNA polymerase III by indirect immunofluorescence microscopy (4) using rabbit anti-β-subunit polyclonal antibody. In exponentially growing cells of strain YK1100 (a tryptophan-deficient mutant derived from W3110 [11]) at 37°C in M9 glucose medium supplemented with l-tryptophan (50 μg/ml), the β-subunit formed one or two distinct condensed clusters within the nucleoid. Besides clear clusters, a portion of β-subunit molecules was distributed in polar cytosolic spaces in some cells (Fig. 1A and Table 1). The average length of cells with two β-subunit clusters was longer than that of cells with one β-subunit cluster (Table 1 and Fig. 2A). A portion (45.8%) of cells did not have clear clusters of the β-subunit, which was dispersed throughout the whole cell. The average length (2.09 ± 0.58 μm [mean ± standard deviation]) of cells without clear clusters was similar to that (1.98 ± 0.62 μm) of total cells with one and two clusters (Table 1 and Fig. 2A). This result eliminates the possibility that this type of cells without clusters was in a specific stage of the cell cycle, for example, the D period (the nonreplication period under the slow-growth conditions at a 55- to 60-min doubling time). Presumably, it is difficult to detect faint clusters, because of a high background of β-subunit dispersed throughout the whole cell.
FIG. 1.
Subcellular localization of the β-subunit of DNA polymerase III and the SeqA protein in exponentially growing cells. Cells of strain YK1100 (11) growing in M9 medium supplemented with 0.5% glucose and 50 μg of l-tryptophan per ml at 37°C were fixed and analyzed by indirect immunofluorescence microscopy (4) with rabbit anti β-subunit of DNA polymerase III polyclonal antibodies and rabbit anti-SeqA polyclonal antibodies. (A) Subcellular localization of the β-subunit of DNA polymerase III (left). Pictures on the right show 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells. (B) Subcellular localization of the SeqA protein (left). Pictures on the right show DAPI-stained cells.
TABLE 1.
Statistical analysis of β-subunit clusters in cells growing in different media
| Medium | Doubling time at 37°C (min) | Total no. of cells | No. of foci | No. of cells | Ratio (%) | Avg cell length (μm)b |
|---|---|---|---|---|---|---|
| M9-Gluc-Trp | 60 | 1,906 | 1 | 710 | 37.3 | 1.81 ± 0.51 |
| 2 | 368 | 19.3 | 2.32 ± 0.67 | |||
| 0a | 828 | 45.8 | 2.09 ± 0.58 | |||
| L | 30 | 647 | 1 | 101 | 15.6 | 2.67 ± 0.47 |
| 2 | 204 | 31.5 | 2.94 ± 0.60 | |||
| 3 | 77 | 10.9 | 3.47 ± 0.72 | |||
| 4 | 87 | 13.4 | 4.23 ± 0.84 | |||
| 0a | 180 | 27.8 | 3.06 ± 0.86 |
β-Subunits were distributed in the whole cells without clear clusters.
Means and standard deviations were measured in pixels and converted to micrometers.
FIG. 2.
Histogram of cell length. Cells of YK1100 growing at 37°C in M9 medium supplemented with 0.5% of glucose and 50 μg of l-tryptophan per ml (A) or in L medium (B) were fixed and analyzed by immunofluorescence microscopy.
In a rich medium (L medium), cells had one, two, three, and four clusters of the β-subunit (Table 1). The doubling time of the strain was approximately 30 min at 37°C in L medium, suggesting that multiforked replication occurred. Note that the average length of the cells with three β-subunit clusters was between the average length of cells with two clusters and that of cells with four clusters. This indicates that cells with three clusters were in an intermediate stage between cells with two clusters and cells with four clusters. A portion (27.8%) of cells did not have clear clusters of the β-subunit, which was dispersed throughout the whole cell (Table 1). The average length (3.06 ± 0.86 μm) of cells without clear clusters was similar to that (3.21 ± 0.85 μm) of total cells with one, two, three, and four clusters (Table 1 and Fig. 2B). It is unlikely that this type of cells without clusters was in a specific stage of the cell cycle.
We previously reported that SeqA is localized as discrete foci in growing YK1100 cells. SeqA forms clusters with hemimethylated nascent DNA segments behind replication forks (4, 10; for a review, see reference 3). We therefore analyzed the subcellular localization of the SeqA protein as a landmark of hemimethylated nascent DNA segments in the cell cycle. As shown in Fig. 1B, one or two clear fluorescent foci of SeqA were observed in exponentially growing cells in M9 glucose medium supplemented with l-tryptophan (50 μg/ml). A single SeqA focus was localized at the middle position of the cell within a nucleoid. Two SeqA foci were localized at the one-fourth and three-fourths positions of the cell. SeqA foci were never localized at the cell poles or the septal site. In L medium, cells had one, two, three, and four SeqA foci. These results for SeqA foci were consistent with previous observations (4).
Comparison between the localization of β-subunit of DNA polymerase III and that of SeqA protein in cells synchronized for initiation of chromosome replication.
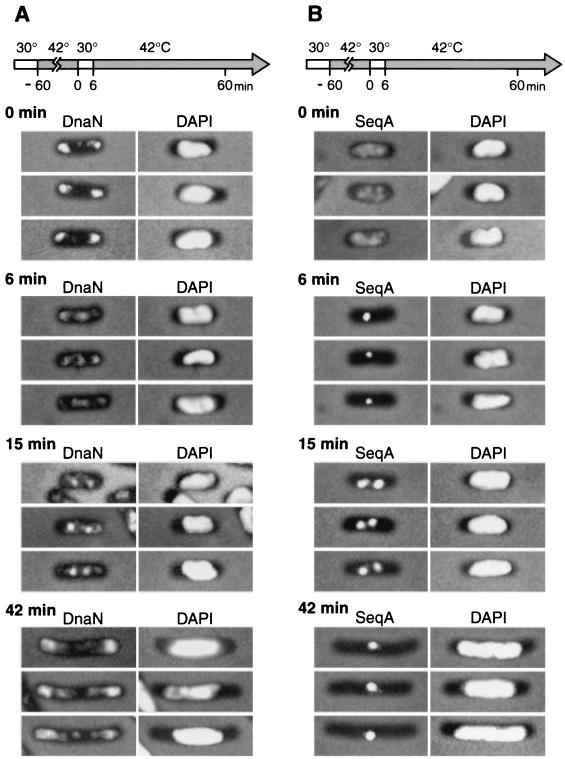
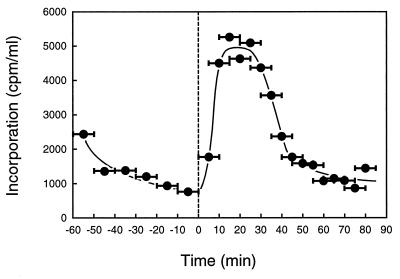
To examine whether the formation of condensed clusters of β-subunit depends on ongoing replication, we analyzed the localization of clusters in cells synchronized for initiation of chromosome replication. To synchronize the initiation of chromosome replication, we used a temperature-sensitive dnaC mutant strain, PC2 (2, 5). The dnaC gene codes for the DnaC protein, which loads the DnaB helicase onto DNA (for a review, see reference 9). The thermosensitive DnaC mutant protein of strain PC2 is inactive at 42°C, but its activity is immediately recovered after transfer to 30°C (5). Cells of PC2 growing exponentially at the permissive temperature of 30°C in L medium supplemented with thymine (50 μg/ml) were transferred to the nonpermissive temperature of 42°C for 60 min to complete ongoing rounds of replication without initiation. After the incubation at 42°C, the β-subunit was localized in cytosolic spaces at the cell poles (Fig. 3A). Subsequently, the cells were incubated at 30°C for 6 min to synchronously initiate chromosome replication from the replication origin oriC, and then they were transferred again to 42°C to inhibit further initiation of chromosome replication. During incubation at 30°C for 6 min, chromosome replication was initiated in more than 90% of the total cells (our unpublished data). Under these conditions, only one round of chromosome replication is presumed to occur. After 6 min at 30°C, β-subunit molecules were recruited from the polar cytosolic spaces to the nucleoid, resulting in a condensed cluster. A portion of β-subunit molecules still remained in the polar cytosolic spaces (Fig. 3A). At 15 min, β-subunit molecules were localized as two condensed clusters in the majority of cells or as one cluster in the minority of cells within the nucleoid (Fig. 3A). At 42 min, β-subunit molecules were either dispersed throughout the cells or localized again in the polar cytosolic spaces (Fig. 3A). DNA synthesis was analyzed as shown in Fig. 4. The incorporation rate of [3H]thymidine into the acid-insoluble fraction increased immediately after transfer to 30°C for 6 min. The incorporation rate was almost null after approximately 40 min as expected. We conclude that β-subunit clusters are formed within nucleoid-occupied spaces and that this mechanism depends on ongoing chromosome replication. Molecules of the β-subunit are certainly recruited at and near replication forks.
FIG. 3.
Comparison of subcellular localization between the β-subunit of DNA polymerase III and the SeqA protein in cells synchronized for initiation of chromosome replication. To synchronize the initiation of chromosome replication, cells of the temperature-sensitive dnaC mutant strain (strain PC2) were exponentially grown in L medium supplemented with thymine (50 μg/ml) at 30°C and then transferred sequentially to 42, 30, and 42°C as shown in arrows. Cells were removed, fixed, and analyzed by indirect immunofluorescence microscopy. (A) Subcellular localization of the β-subunit of DNA polymerase III (left). Pictures on the right show 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells. (B) Subcellular localization of the SeqA protein (left). Pictures on the right show DAPI-stained cells.
FIG. 4.
Rate of DNA synthesis in a synchronized culture. Strain KK488, which is a Thy+ transductant derived from PC2, was exponentially grown at 30°C in M9 medium containing 1% glucose, 0.5% Casamino Acids (Difco), and 100 μg of l-tryptophan (without thymine) per ml. The culture was synchronized for initiation of chromosome replication by triple temperature shifts as shown in Fig. 3. One milliliter of culture was removed and labeled with [methyl-3H]thymidine (3.1 TBq/mmol; 0.37 MBq/ml of culture) in the presence of deoxyadenosine (200 μg/ml of culture) for 10 min at 42°C. The labeling was stopped by the addition of 2 ml of ice-cooled 10% trichloroacetic acid (TCA). The acid-insoluble fraction was collected with a glass filter and washed six times with ice-cooled 5% TCA. Incorporation of [methyl-3H]thymidine into the acid-insoluble fraction was counted with a scintillation counter.
For the same synchronized culture of PC2, we also analyzed the subcellular localization of SeqA. SeqA was distributed throughout the whole nucleoid after the 60-min incubation at 42°C (Fig. 3B). During the incubation at 42°C, ongoing rounds of chromosome replication are completed and the chromosomal DNA becomes fully methylated. As a result, SeqA molecules, which bind preferentially to hemimethylated DNA, are distributed in the whole nucleoid. Six minutes after the temperature shift to 30°C, SeqA molecules had been recruited to a single focus located at the middle position. Fifteen minutes after the shift, the majority contained two SeqA foci localized at the one-fourth and three-fourths positions (Fig. 3B), while a minority contained a single SeqA focus in the midcell position. Forty-two minutes after the shift, most cells had elongated and had a single SeqA focus in the middle of the nucleoid (Fig. 3B). The single SeqA focus presumably corresponds to a cluster of SeqA-bound hemimethylated terminus DNA.
The present data suggest that in the absence of DNA replication, the unbound β-subunit molecules are localized in the polar cytosolic spaces. Following initiation of chromosome replication, a substantial fraction of β-subunit molecules are recruited into a single focus located in the middle of the nucleoid, sometimes with the rest of molecules in the polar cytosolic spaces. A translocating replication factories model was previously proposed (3, 5). In this model, paired replication apparatuses that mediate the bidirectional chromosome replication are first located close to each other at the middle position of the nucleoid. Subsequently, the replication apparatuses separate and migrate in opposite directions during replication fork progression. Each replication apparatus migrates together with a SeqA-hemimethylated DNA cluster that associates with the replication apparatus. The subcellular localization of β-subunit molecules shown in this work is consistent with this model. When temperature-sensitive initiation mutant cells are transferred back from nonpermissive to permissive temperature, β-subunit molecules may first be recruited to the oriC region, which is localized in the middle position of the cell (5), and then paired replication apparatuses may be formed in the oriC region for bidirectional replication of the chromosome. These replication apparatuses are located close to each other in the middle of the nucleoid for a substantial period. The closely linked replication apparatuses are observed as a single fluorescent cluster. Subsequently, the replication apparatuses separate and rapidly migrate in opposite directions up to the one-fourth and three-fourths cellular positions together with SeqA-bound hemimethylated DNA clusters. The present results for localization of the β-subunit of DNA polymerase III support the translocating replication factories model (5) in E. coli but not the fixed replication factories model (5, 8). In the latter, paired replication apparatuses of bidirectional replication would be localized at the midcell position throughout the one round of chromosome replication until termination, and new paired replication apparatuses of the second-round replication would be formed at the one-fourth and three-fourths positions (future division sites) and would exist at the same positions throughout the second-round replication.
Acknowledgments
We thank Chiyome Ichinose, Yuki Kawata, and Mizuho Yano for assistance in the laboratory.
This work was supported by grants from the Ministry of Education, Science, Sports, Culture and Technology of Japan, Japan Society for the Promotion of Science, and CREST.
REFERENCES
- 1.Baker, T. A., and S. P. Bell. 1998. Polymerases and the replisome: machines within machines. Cell 92:295–305. [DOI] [PubMed] [Google Scholar]
- 2.Carl, P. L. 1970. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol. Gen. Genet. 109:107–122. [DOI] [PubMed] [Google Scholar]
- 3.Hiraga, S. 2000. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 34:21–59. [DOI] [PubMed] [Google Scholar]
- 4.Hiraga, S., C. Ichinose, H. Niki, and M. Yamazoe. 1998. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol. Cell 1:381–387. [DOI] [PubMed] [Google Scholar]
- 5.Hiraga, S., C. Ichinose, T. Onogi, H. Niki, and M. Yamazoe. 2000. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells 5:327–341. [DOI] [PubMed] [Google Scholar]
- 6.Kawakami, H., T. Iwura, M. Takata, K. Sekimizu, S. Hiraga, and T. Katayama. 2001. Arrest of cell division and nucleoid partition by genetic alterations in the sliding clamp of the replicase and DnaA protein. Mol. Gen. Genet. 266:167–179. [DOI] [PubMed] [Google Scholar]
- 7.Kelman, Z., and M. O’Donnell. 1995. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64:171–200. [DOI] [PubMed] [Google Scholar]
- 8.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516–1519. [DOI] [PubMed] [Google Scholar]
- 9.Marians, K. J. 1996. Replication fork propagation, p.749–763. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 10.Onogi, T., H. Niki, M. Yamazoe, and S. Hiraga. 1999. The assembly and migration of SeqA-Gfp fusion in living cells of Escherichia coli. Mol. Microbiol. 31:1775–1782. [DOI] [PubMed] [Google Scholar]
- 11.Yamanaka, K., T. Ogura, H. Niki, and S. Hiraga. 1996. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250:241–251 [DOI] [PubMed] [Google Scholar]