Abstract
Sinorhizobium fredii USDA257 forms nitrogen-fixing nodules on soybean (Glycine max [L.] Merr.) in a cultivar-specific manner. This strain forms nodules on primitive soybean cultivars but fails to nodulate agronomically improved North American cultivars. Soybean cultivar specificity is regulated by the nolXWBTUV locus, which encodes part of a type III secretion system (TTSS). NolX, a soybean cultivar specificity protein, is secreted by TTSS and shows homology to HrpF of the plant pathogen Xanthomonas campestris pv. vesicatoria. It is not known whether NolX functions at the bacterium-plant interface or acts inside the host cell. Antibodies raised against S. fredii USDA257 NolX were used in immunocytochemical studies to investigate the subcellular localization of this protein. Immunostaining of paraffin-embedded sections of developing soybean and cowpea (Vigna unguiculata [L.] Walp) nodules revealed localization of NolX in the infection threads. Protein A-gold immunocytochemical localization studies utilizing affinity-purified NolX antibodies revealed specific deposition of gold particles in the fibrillar material inside infection threads. Similar immunogold localization studies failed to detect NolX in thin sections of mature soybean and cowpea nodules. The results from this study indicate that NolX is expressed in planta only during the early stages of nodule development.
Sinorhizobium fredii is a fast-growing, gram-negative bacterium that was originally isolated from Chinese soil (14). Even though it was initially identified as the soybean symbiont, this strain nodulates 79 diverse genera of legumes (25). Curiously, some strains of S. fredii are restricted in their ability to form nitrogen-fixing nodules on certain soybean cultivars (11, 14). For example, S. fredii USDA257 forms nodules on Peking, a primitive Chinese soybean cultivar, but fails to form nodules on the North American soybean cultivar McCall. S. fredii USDA257 is able to attach and penetrate cultivar McCall roots, but the infection process is aborted prior to the formation of infection threads (4). The inability of S. fredii USDA257 to nodulate cultivar McCall and other advanced soybean cultivars is regulated by a complex genetic locus, nolXWBTUV (2, 21). Inactivation of any of the genes in this locus allows the mutant to form nitrogen-fixing nodules on soybean cultivar McCall, indicating a functional role for this locus in soybean cultivar specificity. Since nolXWBTUV locus mutants form nitrogen-fixing nodules on their hosts, it has been suggested that this locus may not have a direct role in nitrogen fixation (21). The six genes present in the soybean cultivar specificity locus are organized into three transcriptional units, nolX, nolW, and nolBTUV (15).
Legume root exudates, especially flavonoids which are released into the rhizosphere, are potent inducers of nod, nol, and noe genes (24). These compounds interact with NodD, a member of the LysR family of transcriptional regulators, to activate the bacterial nodulation genes (30). In S. fredii, the activation of some of the nod genes results in the production of lipochitooligosaccharide Nod factors that are N-substituted with a C18:1 acyl chain on the nonreducing glucosamine and contain an l-fucose or 2-O-methyl-l-fucose on the reducing N-acetylglucosamine (1). The Nod factors provoke the deformation of soybean root hairs and the formation of nodule primordia. Interestingly, the flavonoid-dependent induction of some nod genes of S. fredii USDA257 also results in the secretion of proteins into the rhizosphere (16, 18). The secretion of these flavonoid-induced proteins is regulated by both nodD1 and nodD2 and is dependent in S. fredii on a functional nolXWBTUV locus (18).
Bacterial genome sequencing projects have identified a locus similar to the soybean cultivar specificity loci in Rhizobium sp. strain NGR234 (6), Bradyrhizobium japonicum USDA110 (9), and Mesorhizobium loti MAF303099 (13). Some of the genes located in or near these loci have significant homology to components of the type III secretion system (TTSS) of plant and animal pathogens (15, 20, 21, 36). The TTSS is employed by both the plant and mammalian pathogens to deliver effector proteins into host cells (8), suggesting that the TTSS may play an analogous role in symbiotic interactions. NolT, NolW, and NolX share substantial homology to some hrp (hypersensitive reaction and pathogenicity) genes of plant pathogens (21, 36). For example, NolX, with a molecular mass of 64 kDa (15), has 48% identity to HrpF of Xanthomonas campestris pv. vesicatoria (12). HrpF is secreted into the culture medium in an hrp-dependent manner and has been suggested to function as a translocator of effector proteins into the host cells (26). Similarly, the secretion of NolX is mediated by the TTSS and is nodD dependent (36). Even though this protein has been implicated in soybean cultivar specificity, the precise biochemical role of NolX is unknown. High-resolution transcriptional analysis of the TTSS locus of Rhizobium sp. strain NGR234 has indicated that some of the proteins encoded by the TTSS locus may function as determinants of nodule initiation (36). It has been suggested that effector proteins secreted by the TTSS could be involved in the release of bacteria from the infection thread (36).
Currently, there is no information available on the cellular location of proteins secreted by rhizobia inside the host plant. For example, we do not know if secreted proteins like NolX function at the bacterium-plant interface or act inside the host cytosol. Such information is crucial for understanding the role of the secreted proteins in nodulation. In this study, affinity-purified NolX polyclonal antibodies have been employed to investigate the cellular location of this secreted protein in soybean and cowpea nodules.
MATERIALS AND METHODS
Growth conditions.
S. fredii USDA257 and USDA257Mu78 (nolX mutant) were grown on a reciprocal shaker at 30°C in yeast extract mannitol (YEM) medium (35). Log-phase cultures were harvested by centrifugation at 4,000 × g for 10 min at room temperature. The cell pellet was washed in YEM medium and resuspended in YEM medium to a concentration of 108 cells/ml. Bacterial growth in liquid cultures was estimated turbidimetrically relative to a standard curve that had been validated by bacterial counts with a Petroff-Hausser counting chamber.
Extracellular protein preparation.
S. fredii USDA257 and USDA257Mu78 (nolX mutant) were inoculated in starter cultures of 5 ml of YEM medium and grown at 30°C on a rotary shaker at 160 cycles per min. Aliquots (100 μl) were removed and transferred to 125-ml Erlenmeyer flasks, each containing 25 ml of YEM medium. Cultures were induced with 2.5 μl of 10 mM genistein, which was dissolved in absolute ethanol to give a final concentration of 1 μM. Noninduced cultures were incubated with 2.5 μl of absolute ethanol only. The cultures were harvested after 36 h of growth by centrifugation at 11,200 × g for 15 min. The resulting supernatant was mixed with 4 volumes of ice-cold acetone and incubated overnight at −20°C. The precipitated proteins were recovered by centrifugation at 12,000 × g for 15 min. The protein pellet was air dried and dissolved in 500 μl of sodium dodecyl sulfate (SDS) sample buffer (60 mM Tris-HCl [pH 6.8], 2% SDS [wt/vol], 5% β-mercaptoethanol [vol/vol], 10% glycerol [vol/vol]).
Electrophoresis.
Extracellular proteins from S. fredii USDA257 and USDA257Mu78 (nolX mutant) were resolved by SDS-polyacrylamide gel electrophoresis with the discontinuous buffer system of Laemmli (19). The slab gel (10 by 8 by 0.75 cm) consisted of either a 15% separation gel or a 4% stacking gel. Electrophoresis was carried out at 20 mA of constant current at room temperature. Equal amounts of extracellular proteins (20 μg/lane) were loaded into all lanes. Resolved proteins were visualized by staining with Coomassie brilliant blue.
Purification of NolX antibodies.
The production of antibodies in rabbits to a NolX–glutathione S-transferase (GST) fusion protein has been described previously (15). Antibodies specific for the GST moiety of the fusion protein were removed by repeated adsorption of the serum with Escherichia coli extracts expressing GST (10). Western blot analysis indicated that this preadsorbed antiserum reacted strongly with NolX but also showed weak cross-reaction with a few other proteins (15). In order to eliminate this nonspecific cross-reaction, the adsorbed antiserum was purified further by chromatography on a NolX affinity column by using the AminoLink immobilization kit (Pierce, Rockford, Ill.).
Western blot analysis.
Proteins resolved by SDS-polyacrylamide gel electrophoresis were electrophoretically transferred to a nitrocellulose membrane and incubated with affinity-purified NolX antibodies that were diluted 1:500 in Tris-buffered saline (TBS; 10 mM Tris-HCl [pH 7.5], 500 mM NaCl) containing 5% (wt/vol) nonfat dried milk. Immunologically reactive proteins were identified by the horseradish peroxidase color development procedure recommended by the manufacturer (Bio-Rad Laboratories, Inc., Richmond, Calif.).
Plant tests.
Vigna unguiculata (cowpea) cv. Pink Eye Purple Hull and Glycine max (soybean) cv. Peking seeds were surface sterilized and germinated on 1% water agar at 30°C for 3 days. Three-day-old seedlings were dipped into bacterial suspensions (108 cells/ml) and transferred to autoclaved plastic growth pouches that had been premoistened with nitrogen-free nutrient solution. The location of the root tip of each seedling was marked on the surface of the plastic growth pouches. The pouches were incubated in a growth chamber at 400 μmol of photons/m2/s with a 12-h photoperiod.
Embedment of developing nodules in paraffin.
Soybean and cowpea primary root segments extending 10 mm above and 10 mm below the root tip mark were harvested 4, 6, 8, and 20 days after inoculation with S. fredii USDA257 and USDA257Mu78 (nolX mutant). The root segments harvested at 4, 6, and 8 days after inoculation were viewed under a dissection microscope to locate the positions of nodule initiation sites. Mature nodules and root segments that had the highest concentrations of initiation sites were trimmed into small segments and immediately fixed in 50% ethyl alcohol-5% glacial acetic acid-10% formaldehyde for 24 h at 4°C. The tissue fixed in this mixture was dehydrated in a graded ethanol series followed by a graded xylene series. The tissue was infiltrated with several changes of paraffin at 60°C over a 3-day time period and embedded in Paraplast Plus tissue embedding medium (Oxford Labware, St. Louis, Mo.). The Paraplast Plus-embedded tissues were sectioned (10 μm thick) with a microtome and collected on poly-l-lysine-coated slides. The paraffin from the sections was removed with xylene, and sections were stained with hematoxylin.
Immunostaining of paraffin sections.
Deparaffinized and rehydrated sections were treated with methanol-hydrogen peroxide to inactivate endogenous peroxidase activity. Sections were incubated for 1 h in 5% goat serum-1% bovine serum albumin (BSA)-0.03% Triton X-100 in phosphate-buffered saline and then were incubated for 20 min with S. fredii USDA257 NolX antibodies that had been diluted 1:500 in 2% goat serum-1% BSA-0.03% Triton X-100 in phosphate-buffered saline. The sections were treated sequentially with biotinylated link, streptavidin-conjugated horseradish peroxidase, and substrate-chromogen solution according to the manufacturer’s recommendation (DAKO Corporation, Carpinteria, Calif.). The sections were counterstained with hematoxylin and examined with bright-field optics.
Electron microscopy.
Soybean and cowpea roots containing nodule primordia were dissected into 2- to 3-mm-thick pieces with a razor blade and fixed immediately in 2.5% glutaraldehyde buffered at pH 7.2 with 50 mM sodium phosphate buffer. The samples were fixed at room temperature for 4 h and washed extensively with several changes of phosphate buffer. The tissue samples were then postfixed with 1% aqueous osmium tetroxide for 1 h. After several rinses in distilled water, the samples were dehydrated in a graded acetone series and infiltrated with Spurr’s resin essentially as described by Krishnan et al. (17). The resin was cured at 65°C for 24 h. Thick sections were cut with a glass knife, stained with 1% toluidine blue for 2 min, and examined with bright-field optics. Thin sections were cut with a diamond knife on an ultramicrotome, mounted on uncoated 200-mesh nickel grids, and stained sequentially with 0.5% aqueous uranyl acetate and 0.4% aqueous lead citrate. The sections were examined with a JEOL (Tokyo, Japan) 1200 EX transmission electron microscope at 80 kV.
Immunogold localization of NolX.
Thin sections of nodule primordia were collected on uncoated 200-mesh gold grids and blocked for 30 min with 1% BSA in TBS. The grids were incubated for 2 h in a 1:500 dilution of NolX antibodies in TBS with 1% BSA. The grids were washed five times at 5-min intervals and incubated with 1:10-diluted protein A-gold particles (10 nm in diameter) in 10 mM Tris-HCl (pH 7.5)-500 mM NaCl-0.1% Tween 20 (TBST) with 1% BSA. The grids were washed several times with TBST and water and stained with 0.5% aqueous uranyl acetate.
RESULTS
Specificity of NolX antibodies.
S. fredii USDA257 grown in liquid cultures produced several extracellular proteins (Fig. 1A, lane 1). The addition of genistein to the growth medium resulted in the production of additional proteins that were absent in noninduced cultures (Fig. 1A, lane 2). Prominent among them were the 64-, 39-, 36-, 21-, and 8-kDa proteins. In contrast to the case for USDA257, the induction of USDA257Mu78 (nolX mutant) with genistein did not alter the extracellular protein profile (Fig. 1A, lanes 3 and 4). It has been shown previously that NolX, which has a molecular size of 64 kDa, is an extracellular protein and is secreted through type III secretion machinery (34). Western blot analysis with affinity-purified NolX antibodies revealed a strong reaction against the 64-kDa extracellular protein of S. fredii USDA257 (Fig. 1B, lane 2). This protein was not detected in USDA257Mu78 (nolX mutant) or noninduced USDA257 cultures (Fig. 1B). The affinity-purified antibodies were highly specific for NolX and showed no cross-reaction against any other extracellular proteins (Fig. 1B).
FIG. 1.
Extracellular protein profile of S. fredii USDA257 and USDA257Mu78 (nolX mutant). Extracellular proteins were prepared from cells grown in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 1 μM genistein. The proteins were resolved on an SDS-15% polyacrylamide gel and stained with Coomassie brilliant blue (A) or electrophoretically transferred to nitrocellulose for immunological analysis with NolX-specific antibodies (B). Lanes 1 and 2, extracellular proteins from USDA257; lanes 3 and 4, extracellular proteins from USDA257Mu78 (nolX mutant); lanes 5, molecular size markers. The numbers between the panels indicate the sizes of the proteins in kilodaltons. The arrow in panel B points to NolX.
Immunocytochemical localization of NolX in soybean and cowpea nodules.
S. fredii USDA257 forms nitrogen-fixing nodules on both soybean and cowpea roots. Under the experimental conditions employed in this study, the inoculation of soybean roots with S. fredii USDA257 induced 3 to 4 nodule meristems within 4 to 8 days near the region that corresponded to the root tip mark at the time of inoculation. Under similar conditions, S. fredii USDA257 formed numerous (more than 12) nodule meristems on cowpea roots. During the early stages of nodule development (4 days after inoculation), cortical cells closest to the point of infection underwent rapid cell division. These cells had dense cytoplasm with prominent nuclei. Within a few days, these cells developed into a globular nodule meristem.
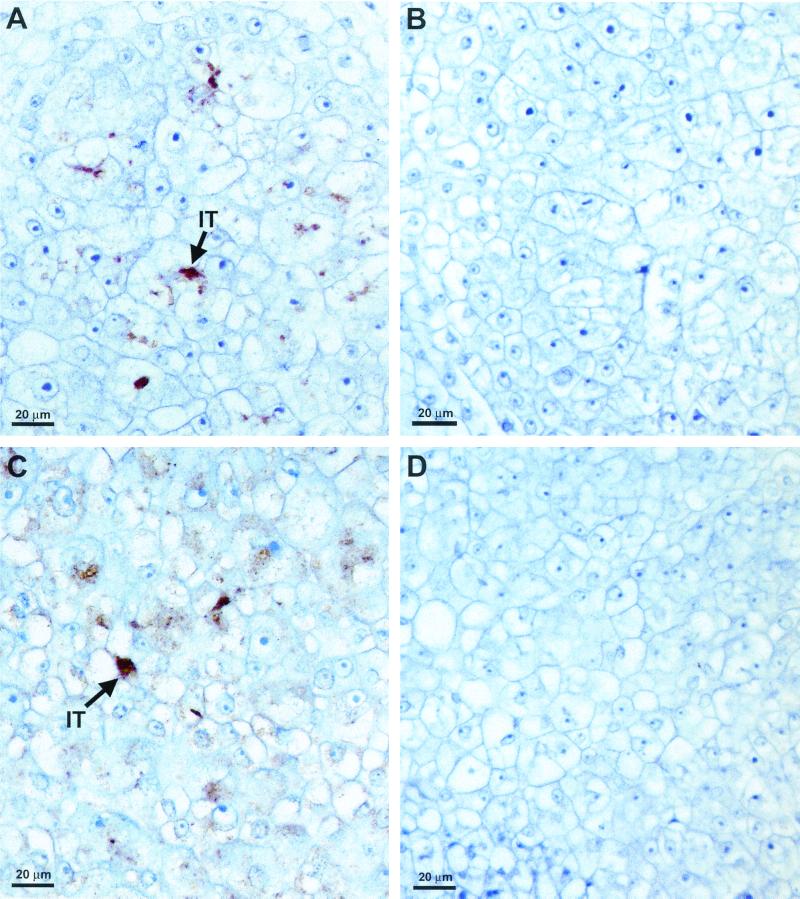
Cross sections of 6-day-old cowpea (Fig. 2A) and 8-day-old soybean (Fig. 2C) nodules revealed the spread of infection threads throughout the central meristematic region of the nodule. The infection threads were often seen in close proximity to the host cell walls (Fig. 2A and C). Paraffin-embedded sections of soybean and cowpea nodules, when incubated with NolX antibodies and streptavidin-conjugated horseradish peroxidase, clearly revealed specific staining in the infection threads (Fig. 2A and C). Brown staining, indicative of specific localization, was observed mostly on the infection threads, but a few other regions of the host cell also revealed brown staining (Fig. 2A and C). When preimmune serum was substituted for NolX antibodies, no staining was seen on the infection threads (Fig. 2B). Similarly, no specific localization was observed for the infection threads in cowpea nodules formed by USDA257Mu78 (nolX mutant) (Fig. 2D).
FIG. 2.
Immunohistochemical localization of NolX. Six-day-old cowpea nodules (A, B, and D) and 8-day-old soybean nodules (C) were embedded in paraffin, processed for immunohistochemical localization, and counterstained with hematoxylin. Paraffin-embedded sections were treated with NolX-specific antiserum (A, C, and D) or preimmune serum (B). Note the brown color (arrows) indicative of the specific labeling of NolX on the infection threads (IT) of both cowpea (A) and soybean (C) nodules. No specific labeling was detected in cowpea nodules formed by USDA257Mu78 (nolX mutant) (D).
Immunogold localization of NolX in cowpea and soybean nodules.
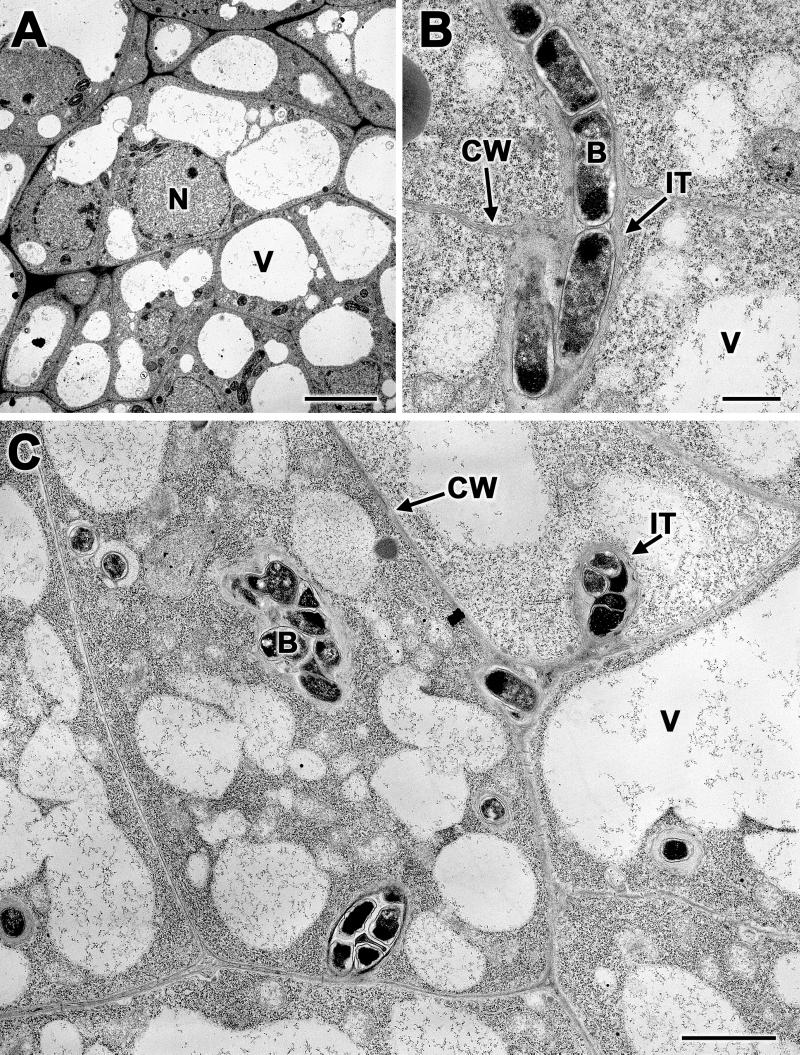
Transmission electron microscopic examination of 6-day-old soybean (Fig. 3A) and cowpea (Fig. 3C) nodules revealed cells with granular cytoplasm with several small vacuoles. The central regions of 6-day-old soybean nodules contained meristematic cells, but infection threads were not detected (Fig. 3A). Infection threads were found in 8-day-old soybean nodules (Fig. 3B). In contrast to soybean nodules, 6-day-old cowpea nodules contained several infection threads (Fig. 3C). In both soybean and cowpea, the infection threads were surrounded by fibrillar material (Fig. 3B and C). The bacteria within these infection threads stained prominently.
FIG. 3.
Low-magnification transmission electron micrographs of soybean and cowpea nodules. (A) Thin sections of 6-day-old soybean nodules revealed cells with prominent vacuoles, nuclei, and thickened cell walls, but infection threads were scarce. (B) A thin section of 8-day-old soybean nodule revealed infection threads near the host cell wall. (C) Numerous infection threads were found in 6-day-old cowpea nodules. The bacteria inside the infection threads were surrounded by fibrillar material. IT, infection thread; CW, cell wall; B, bacterium; V, vacuole; N, nucleus. Bars, 10 μm (A), 0.5 μm (B), and 2 μm (C).
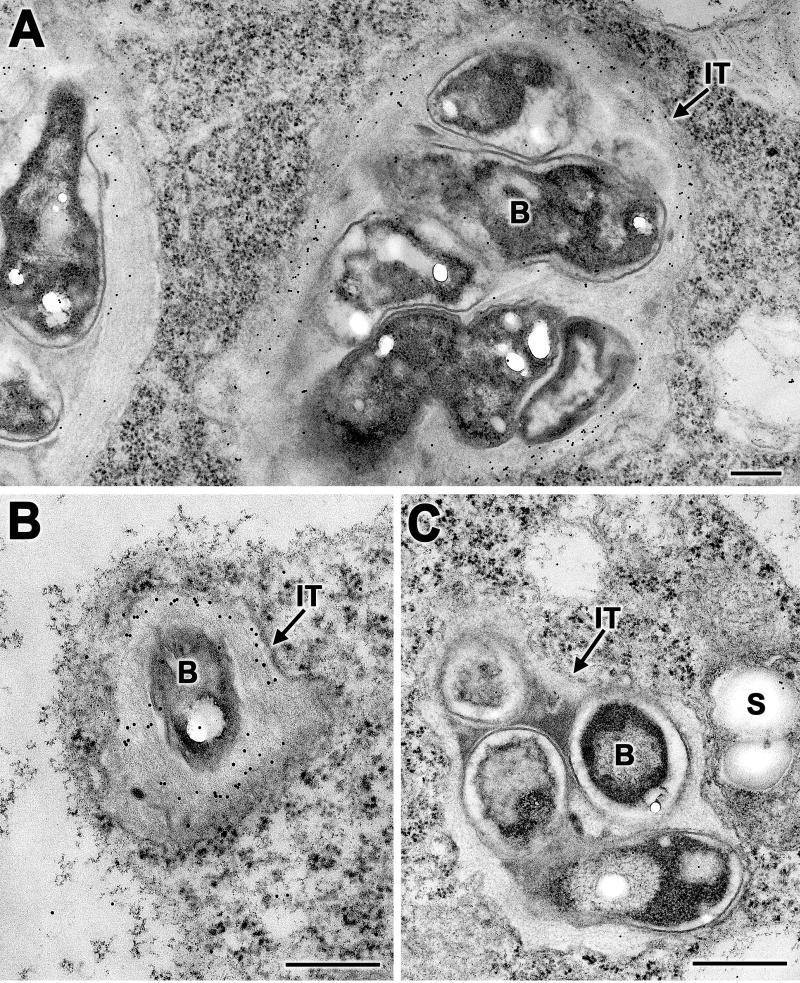
To investigate the subcellular location of NolX, we performed immunogold localization studies. Thin sections of 6-day-old cowpea nodules treated with NolX-specific antibodies and protein A-gold particles revealed specific deposition of gold particles on the fibrillar material present in the infection threads (Fig. 4A). In addition, a few gold particles were deposited on the bacteria (Fig. 4A). In a few instances, the host cytoplasm also contained gold particles, indicating that NolX may also be released from the infection threads (Fig. 4A). Since infection threads in 6-day-old soybean nodules were limited in number, 8-day-old nodules were used for the localization of NolX. As in the case of cowpea nodules, specific gold labeling of infection threads was observed in soybean nodules (Fig. 4B). The protein A-gold deposition on the infection threads, however, was not seen when the NolX antibodies were replaced with preimmune serum (Fig. 4C).
FIG. 4.
Immunogold localization of NolX in the infection threads. (A and B) Thin sections of 6-day-old cowpea nodules (A) and 8-day-old soybean nodules (B) incubated with NolX-specific antibodies and protein A-gold revealed specific labeling of infection threads. Note that the labeling was most heavily concentrated on the fibrillar material surrounding the bacteria. (C) Thin sections of cowpea nodules treated with preimmune serum failed to reveal specific labeling on the infection threads. IT, infection thread; B, bacterium; S, starch. Bars, 0.2 μm (A), 1 μm (B), and 0.25 μm (C).
NolX does not accumulate in mature soybean or cowpea nodules.
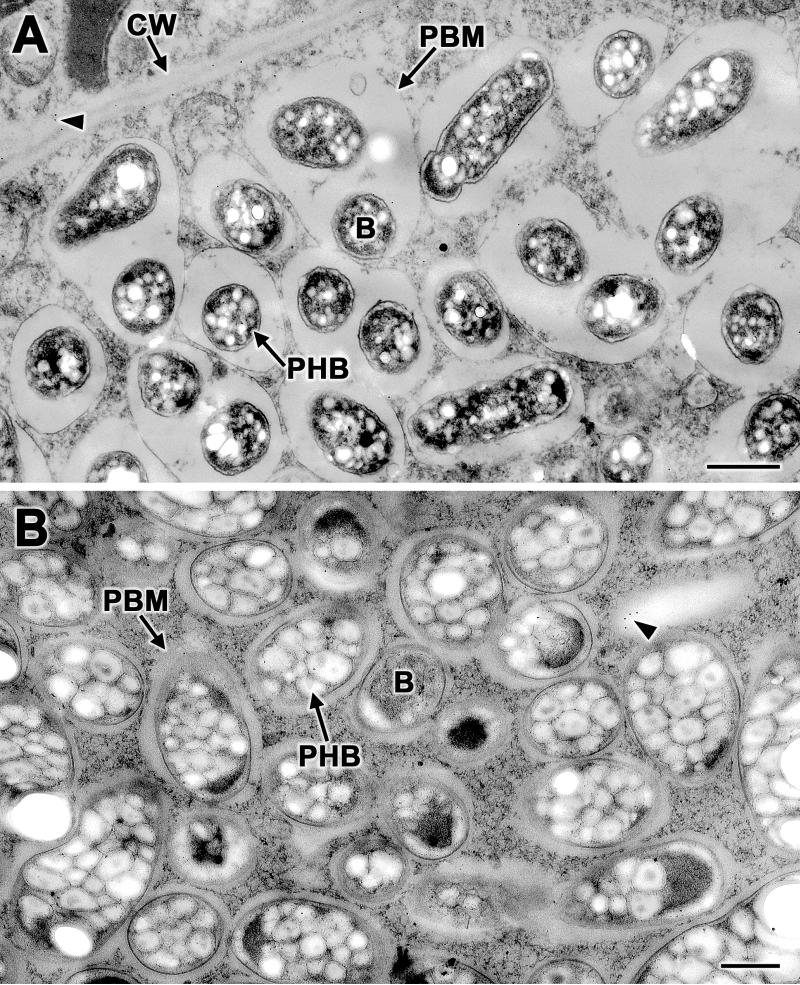
During nodule development, the bacteria are released from the infection threads into the host cell by endocytosis (7). The released bacteria are then surrounded by a host-derived membrane (peribacteroid membrane) and transformed into nitrogen-fixing bacteroids. To examine whether NolX is also present at the later stages of nodule development, we performed immunogold localization studies of 20-day-old soybean and cowpea nodules. Transmission electron microscopic examination of 20-day-old soybean (Fig. 5A) and cowpea (Fig. 5B) nodules revealed the presence of free-living bacteroids that were enclosed by peribacteroid membranes. Unlike the bacteria in the infection threads, the bacteroids in the mature nodules contained numerous poly-β-hydroxybutyrate granules (Fig. 5). When thin sections of the 20-day-old soybean and cowpea nodules were challenged with NolX antibodies and protein A-gold, no specific labeling was observed either on the bacteria or in the region enclosed by the peribacteroid membrane (Fig. 5).
FIG. 5.
Immunogold localization of NolX in mature nodules. Thin sections of 20-day-old soybean (A) and cowpea (B) nodules incubated with NolX-specific antibodies and protein A-gold revealed no specific labeling either on the bacteria or in the space enclosed by the peribacteroid membrane. A few randomly occurring protein A-gold particles can be seen (arrowheads). Note the presence of densely packed poly-β-hydroxybutyrate granules within the bacteroids. CW, cell wall; B, bacteroid; PBM, peribacteroid membrane; PHB, poly-β-hydroxybutyrate granule. Bars, 0.5 μm.
DISCUSSION
The infection of soybean roots by its microsymbionts has been studied in detail (3, 22, 27, 28). The initial interaction between soybean and S. fredii results in marked root hair curling and entrapment of the microsymbionts between appressed plant cell walls. Infection of the root hairs by the bacteria is initiated from such sites and is accompanied by changes in the plant cytoskeleton. The root hair cell walls invaginate to form an incipient tubule, the infection thread, which grows inward down the root hair via tip growth. Infection threads eventually ramify and grow toward actively dividing inner cortex cells. The rhizobia are released from the infection threads into the host cytoplasm by disintegration of the infection thread walls. This process ensures successful colonization of the nodule cortex by microsymbionts.
Bacterial genes involved in the synthesis of Nod factors (33) and exopolysaccharides (5, 23) play a crucial role in the initiation of infection threads. Recent studies have established that, in addition to bacterial genes, plant genes also play a key role in the initiation and elongation of infection threads (7, 29). Elegant ultrastructural and immunological studies have identified glycoproteins and repetitive proline-rich proteins as major components of infection threads (31, 34). The involvement of secreted proteins in infection thread formation comes from studies conducted on nodO of Rhizobium leguminosarum bv. vicae (32, 38). NodO, a secreted protein, forms pores in membranes and functions as a host range determinant (32, 37). A nodFEMNTLO deletion mutant of R. leguminosarum bv. vicae, which produces a Nod factor that is stripped of host-specific decorations, fails to initiate infection threads on its host vetch (38). Overexpression of nodO partially complements this defect. It is speculated that nodO rescues the phenotype by stimulating ion flow across the host membranes, resulting in the amplification of the defective Nod factor signal (7).
The localization of NolX within the infection threads indicates a role for this protein in infection thread growth. On soybean, NolX acts as the determinant of cultivar specificity, preventing S. fredii USDA257 from forming nitrogen-fixing nodules on cultivar McCall soybeans (2, 21). Transmission electron microscopy studies have shown that cultivar McCall soybean roots inoculated with S. fredii USDA257 form a few aberrant infection threads, but the bacteria are not released into the host cytosol (4). Coupled with the immunolocalization data, these observations suggest that NolX may have a role in the release of bacteria from the infection threads.
The present immunogold localization study demonstrates that NolX does not accumulate in bacteroids (Fig. 5). An earlier study also failed to detect the presence of SR3 and SR5, two flavonoid-inducible extracellular proteins of S. fredii USDA257, in mature soybean and cowpea nodules (18). Transcription analysis of the Rhizobium sp. strain NGR234 TTSS cluster indicated that most of the genes located in this region, including NolX, are expressed later than most nod genes involved in the synthesis and modification of Nod factors (6, 36). NolX transcription, however, was not detected in bacteroids of V. unguiculata or Cajanus cajan (36). This observation is consistent with our finding that NolX is not present in mature soybean or cowpea nodules.
Interestingly, mutations in the TTSS locus produce diverse symbiotic phenotypes. For example, Rhizobium sp. strain NGR234 rhcN and y4xI mutants showed increased nodulation, in comparison to that of the wild type, on Pachyrhizus tuberosus but diminished nodule formation on Tephrosia vogelii (36). This suggests that some of the proteins secreted via the TTSS can have either deleterious or beneficial effects on nodulation, depending on the host. Similarly, NolX appears to be beneficial to cultivar Peking nodulation but deleterious to cultivar McCall nodulation. Strong evidence supports the idea that the TTSS loci in animal and plant pathogens are involved in preventing host defense responses (8). NolX, which is encoded by the TTSS locus in S. fredii USDA257, may be perceived as pathogenic by McCall soybean roots, triggering a defense response that leads to the aborting of infection threads. In contrast, NolX may not be perceived as pathogenic by soybean cultivar Peking, and consequently, a successful interaction is established. The ability of legume hosts to recognize effector proteins such as NolX either as beneficial or detrimental to symbiosis may eventually be proven to determine whether an interaction will culminate in nodule formation or not.
Acknowledgments
I thank Laura Green and Larry Darrah for critical reading of this manuscript.
This work was supported by a Competitive Research Grant from the United States Department of Agriculture-National Research Initiative.
REFERENCES
- 1.Bec-Ferte, M.-P., H. B. Krishnan, D. Prome, A. Savagnac, S. G. Pueppke, and J.-C. Prome. 1994. Structures of nodulation factors from the nitrogen-fixing soybean symbiont Rhizobium fredii USDA257. Biochemistry 33:11782–11788. [DOI] [PubMed] [Google Scholar]
- 2.Bellato, C., H. B. Krishnan, T. Cubo, F. Temprano, and S. G. Pueppke. 1997. The soybean cultivar specificity gene nolX is present, expressed in a nodD-dependent manner, and of symbiotic significance in cultivar-nonspecific strains of Rhizobium (Sinorhizobium) fredii. Microbiology 143:1381–1388. [DOI] [PubMed] [Google Scholar]
- 3.Calvert, H. E., M. K. Pence, M. Pierce, N. S. A. Malik, and W. D. Bauer. 1984. Anatomical analysis of the development and distribution of Rhizobium infection in soybean roots. Can. J. Bot. 62:2375–2384. [Google Scholar]
- 4.Chatterjee, A., P. A. Balatti, W. Gibbons, and S. G. Pueppke. 1990. Interaction of Rhizobium fredii USDA257 and nodulation mutants derived from it with the agronomically improved soybean cultivar McCall. Planta 180:301–311. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, H.-P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature (London) 387:394–401. [DOI] [PubMed] [Google Scholar]
- 7.Gage, D. J., and W. Margolin. 2000. Hanging by a thread: invasion of legume plants by rhizobia. Curr. Opin. Microbiol. 3:613–617. [DOI] [PubMed] [Google Scholar]
- 8.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322–1328. [DOI] [PubMed] [Google Scholar]
- 9.Göttfert, M., S. Röthlisberger, C. Kündig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 11.Heron, D. S., and S. G. Pueppke. 1984. Mode of infection, nodulation specificity, and indigenous plasmids of 11 fast-growing Rhizobium japonicum strains. J. Bacteriol. 160:1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huguet, E., and U. Bonas. 1997. hrpF of Xanthomonas campestris pv. vesticatoria encodes an 87-kDa protein with homology to NolX of Rhizobium fredii. Mol. Plant-Microbe Interact. 10:488–498. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko, T., Y. Nakamura, S. Sato, et al. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331–338. [DOI] [PubMed] [Google Scholar]
- 14.Keyser, H. H., B. B. Bohlool, T. S. Hu, and D. F. Weber. 1982. Fast-growing rhizobia isolated from root nodules of soybean. Science 215:1631–1632. [DOI] [PubMed] [Google Scholar]
- 15.Kovacs, L. G., P. A. Balatti, H. B. Krishnan, and S. G. Pueppke. 1995. Transcriptional organization and expression of nolXWBTUV, a locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 17:923–933. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan, H. B., and S. G. Pueppke. 1993. Flavonoid inducers of nodulation genes stimulate Rhizobium fredii USDA257 to export proteins into the environment. Mol. Plant-Microbe Interact. 6:107–113. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan, H. B., V. R. Franceschi, and T. W. Okita. 1986. Immunocytochemical studies on the role of the golgi complex in protein-body formation in rice seeds. Planta 169:471–480. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan, H. B., C.-L. Kuo, and S. G. Pueppke. 1995. Elaboration of flavonoid-induced proteins by the nitrogen-fixing soybean symbiont Rhizobium fredii is regulated by both nodD1 and nodD2, and is dependent on the cultivar-specificity locus, nolXWBTUV. Microbiology 141:2245–2251. [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 20.Marie, C., W. J. Broughton, and W. J. Deakin. 2001. Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4:336–342. [DOI] [PubMed] [Google Scholar]
- 21.Meinhardt, L. W., H. B. Krishnan, P. A. Balatti, and S. G. Pueppke. 1993. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 9:17–29. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb, W., D. Sippel, and R. L. Peterson. 1979. The early morphogenesis of Glycine max and Pisum sativum root nodules. Can. J. Bot. 57:2603–2616. [Google Scholar]
- 23.Pellock, B. J., H.-P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips, D. A. 1992. Flavonoids: plant signals to soil microbes, p.201–231. In H. A. Stafford and R. K. Ibrahim (ed.), Phenolic metabolism in plants. Plenum Press, New York, N.Y.
- 25.Pueppke, S. G., and W. J. Broughton. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant-Microbe Interact. 12:293–318. [DOI] [PubMed] [Google Scholar]
- 26.Rossier, O., V. D. Ackerveken, and U. Bonas. 2000. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38:828–838. [DOI] [PubMed] [Google Scholar]
- 27.Roth, L. E., and G. Stacey. 1989. Bacterium release into host cells of nitrogen-fixing soybean nodules: the symbiosome membrane comes from three sources. Eur. J. Cell Biol. 49:13–23. [PubMed] [Google Scholar]
- 28.Roth, L. E., and G. Stacey. 1989. Cytoplasmic membrane systems involved in bacterium release into soybean nodule cells as studies with two Bradyrhizobium japonicum mutant strains. Eur. J. Cell Biol. 49:24–32. [PubMed] [Google Scholar]
- 29.Schauser, L., A. Roussis, J. Stiller, and J. Stougaard. 1999. A plant regulator controlling development of symbiotic root nodules. Nature (London) 402:191–195. [DOI] [PubMed] [Google Scholar]
- 30.Schlaman, H. R. M., R. J. H. Okker, and B. J. J. Lugtenberg. 1992. Regulation of nodulation gene expression by NodD in rhizobia. J. Bacteriol. 174:5177–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherrier, D. J., and K. A. VandenBosch. 1994. Localization of repetitive proline-rich proteins in the extracellular matrix of pea root nodules. Protoplasma 183:148–161. [Google Scholar]
- 32.Sutton, J. M., E. J. Lea, and J. A. Downie. 1994. The nodulation-signaling protein NodO from Rhizobium leguminosarum biovar viciae forms ion channels in membranes. Proc. Natl. Acad. Sci. USA 91:9990–9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timmers, A. C., M. C. Auriac, and G. Truchet. 1999. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126:3617–3628. [DOI] [PubMed] [Google Scholar]
- 34.VandenBosch, K. A., J. P. Knox, S. Perotto, G. W. Butcher, and N. J. Brewin. 1989. Common components of the infection thread matrix and the intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO J. 8:335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Oxford, England.
- 36.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381–1389. [DOI] [PubMed] [Google Scholar]
- 37.Vlassak, K. M., E. Luyten, C. Verreth, P. van Rhijn, T. Bisseling, and J. Vanderleyden. 1998. The Rhizobium sp. BR816 can function as a determinant for nodulation of Leucaena leucocephala, Phaseolus vulgaris and Trifolium repens by a diversity of Rhizobium spp. Mol. Plant-Microbe Interact. 11:383–392. [Google Scholar]
- 38.Walker, S. A., and J. A. Downie. 2000. Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal Nod factor specificity, but subsequent infection thread growth reguires nodO or nodE. Mol. Plant-Microbe Interact. 13:754–762. [DOI] [PubMed] [Google Scholar]