Abstract
Cells of Escherichia coli take up vitamin B12 (cyano-cobalamin [CN-Cbl]) and iron chelates by use of sequential active transport processes. Transport of CN-Cbl across the outer membrane and its accumulation in the periplasm is mediated by the TonB-dependent transporter BtuB. Transport across the cytoplasmic membrane (CM) requires the BtuC and BtuD proteins, which are most related in sequence to the transmembrane and ATP-binding cassette proteins of periplasmic permeases for iron-siderophore transport. Unlike the genetic organization of most periplasmic permeases, a candidate gene for a periplasmic Cbl-binding protein is not linked to the btuCED operon. The open reading frame termed yadT in the E. coli genomic sequence is related in sequence to the periplasmic binding proteins for iron-siderophore complexes and was previously implicated in CN-Cbl uptake in Salmonella. The E. coli yadT product, renamed BtuF, is shown here to participate in CN-Cbl uptake. BtuF protein, expressed with a C-terminal His6 tag, was shown to be translocated to the periplasm concomitant with removal of a signal sequence. CN-Cbl-binding assays using radiolabeled substrate or isothermal titration calorimetry showed that purified BtuF binds CN-Cbl with a binding constant of around 15 nM. A null mutation in btuF, but not in the flanking genes pfs and yadS, strongly decreased CN-Cbl utilization and transport into the cytoplasm. The growth response to CN-Cbl of the btuF mutant was much stronger than the slight impairment previously described for btuC, btuD, or btuF mutants. Hence, null mutations in btuC and btuD were constructed and revealed that the btuC mutant had a strong impairment similar to that of the btuF mutant, whereas the btuD defect was less pronounced. All mutants with defective transport across the CM gave rise to frequent suppressor variants which were able to respond at lower levels of CN-Cbl but were still defective in transport across the CM. These results finally establish the identity of the periplasmic binding protein for Cbl uptake, which is one of few cases where the components of a periplasmic permease are genetically separated.
The outer membrane (OM) of gram-negative bacteria forms a permeability barrier which restricts passage of both nutrients and toxic environmental agents (19, 25). Most nutrients cross the OM into the periplasmic space by diffusion through general or specific porins, such as OmpF or LamB. Nutrients which are too large or scarce to enter efficiently through the porins are taken into the periplasm via specific, high-affinity active transport systems. The transport systems for passage across the OM of ferric iron complexed with siderophores, heme, or host iron-binding proteins, and of cobalamins (Cbls) such as vitamin B12 (CN-Cbl), consist of a substrate-specific TonB-dependent OM transporter, the transperiplasmic energy-coupling protein TonB, and its ancillary proteins ExbB and ExbD in the cytoplasmic membrane (CM).
Most nutrients are transported across the CM by active transport systems coupled to a transmembrane ion gradient or to pyrophosphate bond hydrolysis. The most common ATP-coupled transport systems in bacteria are the heteropentameric periplasmic permeases, which consist of a periplasmic substrate-binding protein, two transmembrane subunits, and two peripheral ATP-binding cassette (ABC) proteins. The latter two pairs of subunits can be the same or different proteins. TonB-dependent OM transport systems typically act in series with specific periplasmic permeases. For example, uptake of ferric enterobactin in Escherichia coli occurs by action of the TonB-dependent transporter FepA, the periplasmic binding protein FepB, and the periplasmic permease FepDGC2, in which FepD and FepG are the transmembrane components and FepC is the ABC protein (38). Ferric hydroxamate entry in E. coli uses multiple substrate-specific OM transporters: FhuA for ferrichrome, IutA for aerobactin, FhuE for rhodotorulic acid and coprogen, and FhuF for ferrioxamine B. Once in the periplasm, all of these ferric hydroxamates are transported by the periplasmic binding protein FhuD and the FhuBC2 permease complex (for a recent review, see reference 20). The component genes for each iron transport system are usually closely linked and are often coregulated in single or clustered operons in response to the level of internal iron bound to the Fur repressor.
The organization of the genes for transport of Cbls, such as CN-Cbl, differs from that of other TonB-dependent systems (32). The OM transporter BtuB is encoded by a Cbl-repressible gene at min 89.6 of the genetic map (2). Cbl transport across the CM requires the transmembrane protein BtuC and the ABC protein BtuD, encoded in the constitutively expressed btuCED operon at 38.6 min (14). No gene for a periplasmic Cbl-binding protein, which is expected to be a necessary component of any periplasmic permease, is linked to these btu genes. The btuE gene, which is related in sequence to glutathione peroxidase, plays no apparent role in Cbl transport or utilization (30). Assays of osmotic shock fluid revealed the presence of a periplasmic Cbl-binding activity, but its role in transport was not established (7). Two lines of evidence identified a candidate periplasmic Cbl-binding protein. The translated sequence of the yadT open reading frame (ORF) in the E. coli genome is related to the periplasmic iron-siderophore-binding proteins FhuD and FepB and thus could act in a similar manner (20). More directly, van Bibber et al. (42) found that mutations in the yadT orthologue in Salmonella enterica serovar Typhimurium conferred growth and Cbl transport phenotypes similar to those conferred by a btuC mutation. Suggesting that it has a role in CM transport, they termed this ORF btuF.
We demonstrate here that the E. coli BtuF protein is a periplasmic protein with a cleaved N-terminal signal sequence and a high affinity for CN-Cbl binding. The growth and transport phenotypes of null mutations in the btuF gene and its two flanking genes, pfs and yadS, showed that only BtuF participates in Cbl transport across the CM. However, the BtuF mutant phenotype was quite different from those previously described for btuC and btuD mutants of E. coli (15) and for the S. enterica serovar Typhimurium btuF mutants (42). Although Cbl transport into the cytoplasm was eliminated or greatly reduced in all of these mutants, the previously described btuC, btuD, and btuF mutants were only slightly impaired in their ability to use low levels of CN-Cbl in the usual growth response assay. This assay measures the ability of limiting amounts of CN-Cbl to replace the methionine requirement of metE mutants, which lack the Cbl-independent homocysteine transmethylase and thus require methionine or Cbl (12). The modest impairment in these mutants contrasts to the phenotype of btuB or tonB mutants, which are defective in Cbl transport across the OM and whose CN-Cbl utilization is reduced by at least 4 orders of magnitude. When we found here that the E. coli BtuF null mutant was highly impaired for CN-Cbl utilization and gave rise to suppressor variants at high frequency, we made a new set of null mutations in the genes of the btuCED operon. These new mutants showed that the absence of BtuC confers a strong growth phenotype like that of the btuF mutant. These results demonstrate the importance of CM transport for CN-Cbl utilization and reveal that frequent suppressor variants partially bypass this defect and allow more effective Cbl utilization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2, and their construction is described in the following sections. Unless otherwise specified, bacteria were grown in Luria-Bertani (LB) or minimal A salts medium (12, 24), supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), gentamicin (15 μg/ml), or chloramphenicol (20 μg/ml) when appropriate. Cultures were incubated at 37°C with vigorous aeration. Agar was added at 1.8% for solid media.
TABLE 1.
E. coli K-12 strains
| Strain | Genotypea | Reference or source |
|---|---|---|
| RK4353 | MC4100 [Δ(argF-lac)U169 araD139 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR22] non-9 gyrA219] | 37 |
| NC13 | RK4353 Δpfs(8-226)::Km | This study |
| NC14 | RK4353 ΔbtuF(11-239)::Km | This study |
| NC15 | RK4353 ΔyadS::Km | This study |
| RK4379 | RK4353 metE70 | 1 |
| NC16 | RK4379 Δpfs(8-226)::Km | This study |
| NC17 | RK4379 ΔbtuF(11-239)::Km | This study |
| NC18 | RK4379 ΔyadS(7-204)::Km | This study |
| RK4936 | RK4379 btuB::Tn10 | Lab strain |
| NC19 | RK4936 Δpfs(8-226)::Km | This study |
| NC20 | RK4936 ΔbtuF(11-239)::Km | This study |
| NC21 | RK4936 ΔyadS(7-204)::Km | This study |
| RK5015 | RK4379 ΔtonB | 30 |
| NC22 | RK5015 Δpfs(8-226)::Km | This study |
| NC23 | RK5015 ΔbtuF(11-239)::Km | This study |
| NC24 | RK5015 ΔyadS(7-204)::Km | This study |
| RK6049 | RK4379 zdh-1::Tn10 btuC456 | 15 |
| NC25 | RK6049 Δpfs(8-226)::Km | This study |
| NC26 | RK6049 ΔbtuF(11-239)::Km | This study |
| NC27 | RK6049 ΔyadS(7-204)::Km | This study |
| NC28 | RK4379 ΔbtuC::Gm | This study |
| NC29 | RK4379 ΔbtuE::Gm | This study |
| NC30 | RK4379 ΔbtuD::Gm | This study |
| JM109 | e14−(McrA−) recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Stratagene |
| BL21(DE3) | E. coli B F−dcm ompT hsdS(rB− mB−) gal λ(DE3) | Stratagene |
The numbers after the genes with deletions indicate the amino acid residues of the coding sequence which were removed in formation of each deletion.
TABLE 2.
Plasmids
| Plasmid | Characteristics | Reference or source |
|---|---|---|
| pET17b | Novagen | |
| pBR322 | rep(p15A); Ar, Tc | 4 |
| pT7-6 | ori(ColE1); Ap, T7p | 40 |
| pYadT2 | pfs-btuF-yadS in pT7-7; Ap | This study |
| pΔpfs | pYadT2 Δpfs(8-226)-btuF-yadS | This study |
| pΔbtuF | pYadT2 pfs-ΔbtuF(11-239)-yadS | This study |
| pΔyadS | pYadT2 pfs-btuF-ΔyadS(7-204) | This study |
| pKO3 | repA(pSC101-ts) sacB; Cm | 22 |
| pNC5 | pKO3 pfs-btuF-yadS; Cm | This study |
| pΔpfs::Km | pKO3 Δpfs(8-226)::Km-btuF-yadS; Cm | This study |
| pΔbtuF::Km | pKO3 pfs-ΔbtuF(11-239)::Km-yadS; Cm | This study |
| pΔyadS::Km | pKO3 pfs-btuF-ΔyadS(7-204)::Km; Cm | This study |
| pbtuCED | pBR322 btuC-btuE-btuD; Ap | This study |
| pΔbtuC | pBR322 ΔbtuC-btuE-btuD; Ap | This study |
| pΔbtuE | pBR322 btuC-ΔbtuE-btuD; Ap | This study |
| pΔbtuD | pBR322 btuC-btuE-ΔbtuD; Ap | This study |
| pNC6 | pKO3 btuC-btuE-btuD; Cm | This study |
| pΔbtuC::Gm | pKO3 ΔbtuC::Gm-btuE-btuD; Cm | This study |
| pΔbtuE::Gm | pKO3 btuC-ΔbtuE::Gm-btuD; Cm | This study |
| pΔbtuD::Gm | pKO3 btuC-btuE-ΔbtuD::Gm; Cm | This study |
| pBtuF-His | pET17b expressing BtuF-His6 under T7 promoter control; Ap | This study |
Plasmid construction.
Isolation and manipulations of DNA fragments were by standard protocols (33). Plasmid pYadT2 was constructed by PCR amplification and cloning of a 3,122-bp EcoRI-XbaI fragment from the E. coli chromosomal yad region into plasmid pT7-6 (40). The oligonucleotides used as primers for amplification introduced the indicated restriction sites and annealed to the regions ending 1,342 bp upstream and 980 bp downstream from the yadT (btuF) gene. Sequences of oligonucleotide primers are available upon request.
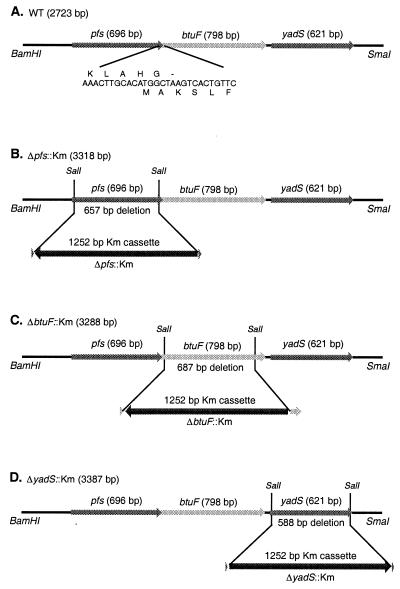
The insert in plasmid pYadT2 also carries the pfs and yadS genes on either side of btuF. For complementation and functional assays, in-frame deletion mutations which removed most of the coding regions for each of the three genes were prepared. The SalI site in the multiple cloning region of pYadT2 was first removed by digesting with SalI, filling with T4 DNA polymerase, and religating the blunt-ended plasmid, to yield plasmid pYadT3. Two in-frame SalI sites were introduced near each end of each of the three genes carried in pYadT3, using the QuickChange site-directed mutagenesis kit (Stratagene). The resulting plasmids were digested with SalI and religated to remove the region between the two SalI sites. As shown schematically in Fig. 1, plasmid pΔpfs carries an in-frame deletion which removed a 657-bp fragment corresponding to amino acids 8 to 226, out of the total of 232 residues. Plasmid pΔbtuF has an in-frame deletion of the btuF gene, which removed 687 nucleotides from the 798-bp gene and deleted amino acids 11 to 239, out of the total of 266 residues. Plasmid pΔyadS contains a 588-bp in-frame deletion in the 621-bp yadS gene, which removed amino acids 7 to 204, out of the total of 207 residues.
FIG. 1.
Representation of the pfs-btuF-yadS region of the E. coli chromosome map and description of deletion mutations. (A) The wild-type 2,723-bp region in plasmid pNC5. The expanded sequences show the overlap between the 3′ end of the pfs gene and the 5′ end of the btuF gene. Arrows indicate the direction of transcription. (B) The 3,318-bp insert in pΔpfs::Km, showing the 657-bp in-frame deletion of part of pfs gene and insertion of the Km cassette. (C and D) Structures of ΔbtuF::Km (C) and ΔyadS::Km (D).
For transfer of the pfs, btuF, and yadS null mutations to the chromosomes of strains carrying other btu lesions, another series of inserts were constructed in plasmid pKO3 (22). Plasmid pNC5 carries a 2,723-bp fragment with the pfs-btuF-yadS genes amplified from plasmid pYadT2 using primers which annealed to regions 1,076 bp upstream and 847 bp downstream from the btuF gene, respectively, and which introduced a BamHI site and a SmaI site at either end. This amplified fragment was digested with BamHI and SmaI and ligated into similarly digested pKO3. The in-frame deletions of the three genes described above were transferred into pNC5 by restriction fragment exchange. Each resulting plasmid was digested with SalI and ligated with the 1,252-bp aph (kanamycin resistance [Km]) cassette from pUC4kan digested with SalI, to yield plasmids pΔpfs::Km, pΔbtuF::Km, and pΔyadS::Km. The orientation of the Km cassette in each case was analyzed by digestion with NruI.
For preparation of a new set of btuCED deletion mutants, plasmid pLCD25 (15) containing the entire btuCED operon was digested with SmaI and BclI, releasing a 2,593-bp fragment. This insert was ligated into pBR322 digested with EcoRV and BamHI to create pbtuCED. The same fragment was ligated to pKO3 digested with SmaI and BamHI to form pNC6. Two series of plasmids were created using pNC6, one in which part of each of the btuCED genes was deleted and the other in which a gentamicin resistance (Gm) cassette from plasmid pUCGM (36) was inserted in place of the deleted regions. For the btuC deletion, pNC6 was digested with BssSI (bp 137) and BglII (bp 653) to remove a 516-bp fragment from the 978-bp coding region (17). Similarly, the btuE deletion was created by digesting pNC6 with PvuI (bp 165) and ApaLI (bp 404), which removed 239 bp of the 549-bp ORF. Finally, SexAI (bp 89) and MluI (bp 304 and 399) were used to remove 310 of the 747 nucleotides of btuD. These three deleted plasmids were blunt ended using T4 DNA polymerase and religated with or without the 855-bp SmaI-digested Gm cassette. The orientation of the cassette was determined by digestion with AvaII for btuC and btuE or with BglII for btuE and btuD. The plasmids with the Gm cassette were designated pΔbtuC::Gm, pΔbtuE::Gm, and pΔbtuD::Gm and were used for allelic replacement of the corresponding genes onto the chromosome of strain RK4379 (Table 1). The plasmids without the cassette were digested with AflII and StuI and cloned into pbtuCED by restriction fragment exchange, forming plasmids pΔbtuC, pΔbtuE, and pΔbtuD (Table 22), which were used in complementation assays.
A His-tagged version of BtuF under transcriptional control of the T7 promoter was constructed. The btuF gene was amplified using two oligonucleotide primers which introduced an NdeI site at the start of btuF and a His6-coding sequence followed by a termination codon and an EcoRI site at the 3′ end. The resulting fragment was digested with NdeI and EcoRI and ligated into similarly digested pET17b (Novagen) to yield plasmid pBtuF-His. The nucleotide sequence of the insert in plasmid pBtuF-His was verified by automated DNA sequence determination at the University of Virginia Biomolecular Resource Facility.
Strain construction.
The in-frame deletions of pfs, btuF, or yadS containing a Km cassette were transferred onto the chromosomes of various bacterial strains (wild type, metE, metE btuB, metE btuC, and metE tonB) using the pKO3 system (22). The same approach was used for allelic exchange of the deletions with an inserted Gm cassette in each gene of the btuCED operon. Plasmid pKO3 derivatives carrying the desired deletion-Km or Gm insertion mutations were introduced into the recipient strains by transformation, with selection on plates containing chloramphenicol and kanamycin or gentamicin. Transformants were transferred to chloramphenicol and kanamycin or gentamicin plates and grown at 42°C to select for Campbell-type integration of pKO3, taking advantage of the temperature-sensitive origin of replication of plasmid pKO3. Survivors were grown overnight in LB broth with kanamycin or gentamicin, diluted, and plated on LB medium with 5% sucrose and kanamycin or gentamicin to select for the second recombination event which removed plasmid sequences. Colonies which were resistant to kanamycin or gentamicin but sensitive to chloramphenicol were identified by replica plating. Each recombinant was tested by PCR using the primer pairs that were originally used to amplify the 2,723-bp insert in pNC5 or the btuCED operon. The amplified products gave the expected restriction fragments when digested with NruI or SmaI, respectively.
Purification of BtuF-His.
Cells of E. coli strain BL21(DE3) carrying plasmid pBtuF-His were grown at 37°C in 500 ml of LB broth or minimal medium with ampicillin to an optical density at 595 nm of between 0.5 and 0.7, induced with 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubated for an additional 3 h. Cells were harvested by centrifugation (6,500 × g) for 5 min, suspended in 20 ml of ice-cold wash buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl) containing 10 mM imidazole, and lysed in a French pressure cell at 18,000 lb/in2 in the presence of the protease inhibitor phenylmethylsulfonyl fluoride (17 μg/ml). DNase and RNase were added to the lysate (10 μg/ml), and unlysed cells and debris were removed by centrifugation at 14,500 × g for 10 min. One milliliter of Ni-agarose (Qiagen) was added to half of the cleared lysate and mixed gently for 2 h at 4°C for binding of His-tagged BtuF. The slurry was transferred to a small column, and the column was washed six times with 5 ml of ice-cold wash buffer containing 20 mM imidazole. Proteins were eluted by washing with 1-ml fractions of ice-cold wash buffer containing increasing amounts of imidazole from 100 mM to 1 M.
Preparation of periplasmic proteins.
For osmotic shock the method of Nossal and Heppel (26) was used with the modifications recommended in the Qiagen protocol for Ni affinity purification. Briefly, cells grown as described above were harvested by centrifugation (6,500 × g) for 5 min and suspended in 20 ml of ice-cold buffer (30 mM Tris-HCl, 20% sucrose [pH 8.0]). EDTA was slowly added to a concentration of 1 mM, and the suspension was swirled gently on ice for 10 min. After centrifugation at 9,000 × g for 5 min, the cells were suspended in 20 ml of 5 mM MgSO4 and gently agitated on ice for 10 min. The osmotic shock fluid was collected as the supernatant after centrifugation at 9,000 × g for 5 min.
SDS-PAGE and Western immunoblot analysis.
Whole cells and protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 13% polyacrylamide (wt/vol) gels using the discontinuous buffer system of Laemmli (21). Resolved proteins were transferred to a nitrocellulose membrane (Bio-Rad) by electrophoresis for 1 h at 500 mA in buffer consisting of 25 mM Tris-HCl (pH 8.3), 192 mM glycine, and 20% (vol/vol) methanol (41). The membrane was then blocked for 1 h to overnight in phosphate-buffered saline (PBS)−3% bovine serum albumin and incubated for 1 h with Tetra-His antibody (Qiagen) diluted 1:25,000 in the same buffer. The membrane was washed extensively in PBS−0.02% Tween 20, blocked again for 1 h in PBS−5% dried nonfat milk, and incubated for 1 h with affinity-purified horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G secondary antibodies (Jackson Immunoresearch Laboratories) diluted 1:5,000 in the same buffer. After extensive washing in PBS−0.02% Tween 20, the immunoblot was developed using the chemiluminescent substrate LumiGlo (Kirkegaard & Perry Laboratories) and exposed to X-ray film (Kodak XAR).
Protein sequencing.
The N-terminal protein sequences of the precursor and mature forms of BtuF-His protein were obtained. The precursor form was transferred by electrophoresis from an SDS-PAGE electropherogram onto a polyvinylidene difluoride Polyscreen membrane (NEN Research Products), and the mature form of the protein was supplied as the purified protein. These proteins were sequenced at the University of Virginia Biomolecular Research Facility by Edman degradation on an Applied Biosystems Procise protein sequencer.
Binding of CN-Cbl to BtuF-His.
Two methods were used to measure the binding of CN-Cbl to BtuF-His. The binding of radiolabeled CN-Cbl used a modification of the charcoal-binding assay of Gottlieb et al. (18). A charcoal suspension was prepared by mixing equal volumes of 1% bovine serum albumin (Sigma catalog no. A3902), which is deficient in Cbl and Cbl-binding proteins, and 5% neutralized charcoal (Sigma catalog no. C5385). An 800-μl volume of this suspension was filtered in Spin-X centrifuge filter tubes (Costar) to leave 20-mg layers of charcoal on the filters. The binding mixture contained 0.8 μg of BtuF-His and variable amounts of CN-[57Co]Cbl in 100 mM potassium phosphate at pH 6.6. After incubation at room temperature for 5 min, 800-μl samples were transferred to the charcoal-containing Spin-X tubes and centrifuged immediately at 8,000 rpm for 15 s in a Sorvall Biofuge. Free Cbl was bound by the charcoal layer, and the filtrate contained protein-bound Cbl. The Cbl in the filtrates was measured by counting the radioactivity in a Beckman LS6500 liquid scintillation counter. Blank values, obtained from binding mixtures which lacked the BtuF protein, were subtracted from the experimental values.
Isothermal titration calorimetry assay of the binding of CN-Cbl to BtuF-His was performed using a MicroCal System MCS ITC (MicroCal Inc.). Purified BtuF-His was dialyzed extensively against 20 mM Tris-HCl (pH 8.0) buffer, and the final dialysate was used to prepare CN-Cbl solutions and adjust the protein concentration. The protein concentration was based on the extinction coefficient calculated from the amino acid composition. Protein sample and CN-Cbl solutions were clarified by passage through a sterile 0.22-μm-pore-size filter and then degassed. Each experiment consisted of 30 injections of 8 μl each of a CN-Cbl (410 to 450 μM) solution into a sample cell (volume = 1.334 ml) containing BtuF-His (41 to 44 μM). Each 8-μl injection was made for a period of 20 s, with a 210-s interval between injections. The sample cell was stirred continuously at 400 rpm. Three separate experiments were performed using two different batches of the purified BtuF-His protein, and the temperatures for the runs were 21.87, 22.94, and 21.62°C. Control experiments were carried out by diluting the CN-Cbl into buffer. For data analysis, the CN-Cbl dilution enthalpies were subtracted from the titration with BtuF-His, and the data were fitted to a one-site model using Origin (MicroCal Inc.).
CN-Cbl growth phenotype.
All strains constructed in this study were tested for their ability to grow on two types of media. What we term the methionine assay uses minimal A salts agar supplemented with 0.02% glucose, 0.01% arginine, and various concentrations of CN-Cbl (0.1 to 5,000 nM) to test the ability of metE mutants to use CN-Cbl for methionine synthesis (1). What we term the ethanolamine assay uses the medium described by Scarlett and Turner (34) supplemented with glycerol (0.5%), ethanolamine-HCl (1 mg/ml), arginine and methionine (both at 5 μg/ml), and the indicated concentrations of CN-Cbl to test for the ability of cells to acquire CN-Cbl for conversion to the cofactor needed for use of ethanolamine as a nitrogen source (23). Growth phenotypes are determined from the colony sizes after 48 h of incubation at 37°C. Plasmids carrying the intact pfs-btuF-yadS region or the btuCED region, and derivative plasmids carrying deletions in each of the genes, were tested for their ability to complement any growth defect in the same assays following their transformation into each host strain.
CN-Cbl uptake assay.
Strains were tested for their ability to transport radiolabeled CN-Cbl. Preparation of CN-[57Co]Cbl was previously described (9). Cells were grown at 37°C in minimal medium A (12) containing glucose and methionine, harvested in mid-exponential phase, washed, and suspended in 100 mM potassium phosphate (pH 6.6)−1% glucose (5). Cells were incubated with CN-[57Co]Cbl (10 nM; ca. 1,000 cpm/pmol), and 1-ml samples were removed at intervals, collected on Millipore filters (0.45-μm pore size), washed twice with 10 ml of 100 mM LiCl, and dried. Radioactivity retained on the filters was determined by liquid scintillation counting, and uptake is expressed as picomoles of CN-Cbl taken up per 109 cells.
RESULTS
Cloning and expression of E. coli btuF.
The three to five proteins which comprise periplasmic permeases are typically encoded in operons or closely linked gene clusters. Hence, for characterization of the role of the BtuF protein in Cbl transport, the btuF gene and its flanking genes, pfs and yadS, were cloned from the E. coli chromosome by PCR amplification (3). The pfs product is a nucleosidase which releases adenine from S-adenosylhomocysteine (SAH) and methylthioadenosine, the two metabolic products formed from S-adenosylmethionine (SAM) during methyl transfer reactions and polyamine synthesis, respectively (11). No function has been assigned to the yadS product. All three genes in this cluster are transcribed in the same direction and are in close proximity, with the 3′ end of pfs overlapping the 5′ end of btuF and the start codon of yadS lying 38 bp from the stop codon of btuF (Fig. 1A). Because these genes may be cotranscribed, a 3,122-bp fragment containing all three genes was amplified from the chromosome and cloned into pT7-6 to form pYadT2. The nucleotide sequence of the insert in pYadT2 was shown to match exactly the genomic sequence (3). A shorter insert of 2,723 bp containing all three genes was subcloned into pKO3.
Properties of His-tagged BtuF protein.
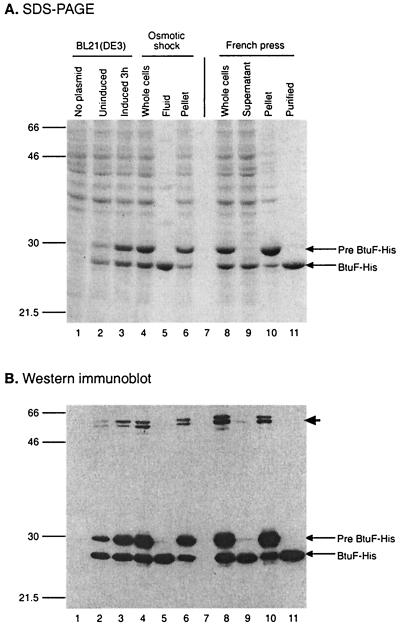
To facilitate biochemical analysis of the BtuF protein, we inserted into the pET17b expression plasmid a version of the btuF gene encoding a BtuF protein with a C-terminal six-histidine extension, termed BtuF-His. This His-tagged version of BtuF was able to complement the growth defect of a ΔbtuF::Km strain (described below). Induction by IPTG of BtuF-His expression in strain BL21(DE3) grown in LB medium resulted in strong amplification of two protein bands on SDS-PAGE which were absent in the strain with the empty vector (Fig. 2A, lane 1). Substantial amounts of both plasmid-specified proteins were produced in the absence of IPTG induction, but their levels increased further after induction (lanes 2 and 3). Since BtuF was expected to be a periplasmic protein, these two bands probably represent the precursor and mature forms of BtuF-His. The mobility of the larger protein band on SDS-PAGE matched closely the molecular mass of 30.19 kDa predicted from the btuF nucleotide sequence. Western immunoblot analysis of a duplicate gel, detected with a tetra-His-directed monoclonal antibody (Qiagen), showed that both bands contained the C-terminal His-tag (Fig. 2B). Two additional immunoreactive bands of roughly double the molecular mass were seen, but they disappeared when the samples were boiled in sample buffer for >5 min (data not shown).
FIG. 2.
Expression, purification, and cellular localization of BtuF-His protein. (A) SDS-PAGE analysis with Coomassie blue staining; (B) Western immunoblot with primary tetra-His monoclonal antibody (Qiagen). Whole-cell samples suspended and boiled in sample buffer were obtained from strain BL21(DE3) carrying no plasmid (lanes 1), the pBtuF-His plasmid without induction (lanes 2), or the pBtuF-His plasmid 3 h after induction with 0.25 mM IPTG (lanes 3). IPTG-induced cells were analyzed before osmotic shock treatment (lanes 4), and following osmotic shock, samples from the osmotic shock fluid (lanes 5) and pellet (lanes 6) were run. Induced cells were taken before (lanes 8) and after disruption in French pressure cell. The lysate was subjected to centrifugation, and the supernatant (lanes 9) and pellet (lanes 10) were resolved. Lanes 11 show the affinity-purified BtuF-His protein following elution from an Ni-nitrilotriacetic acid affinity matrix. On the left are shown the mobilities of molecular weight standards (in thousands), and on the right are indicated the positions of the precursor and mature forms of BtuF-His. The arrow points to oligomeric forms which are lost upon prolonged heating in sample buffer.
The cellular locations of both BtuF-His proteins were investigated by using osmotic shock for release of periplasmic proteins. Most of the putative mature form of BtuF-His was released in the osmotic shock fluid (Fig. 2, lanes 5 and 6), whereas the putative precursor form remained in the cell pellet after osmotic shock. When the cells were lysed by passage through a French pressure cell, the mature form of the protein was found mainly in the soluble fraction, whereas the putative precursor form was found in the low-speed pellet, probably in inclusion bodies (Fig. 2, lanes 9 and 10). The soluble form of the BtuF-His protein was purified from the French press supernatant by elution from an Ni-agarose matrix (Qiagen) (Fig. 2, lanes 11).
To confirm the identities of the two His-tagged protein bands and determine the site of signal sequence cleavage, the N-terminal sequences of both polypeptides were determined. For the putative BtuF-His precursor, the insoluble fraction was resolved by SDS-PAGE and the appropriate protein band was transferred to a polyvinylidene difluoride membrane (Fig. 2, lanes 10). Purified soluble protein was the source of the mature form of BtuF-His (Fig. 2, lanes 11). In agreement with the translated sequence, the N-terminal sequence of the BtuF-His precursor was (M)AKSLF. About 80% of the polypeptides lacked the N-terminal Met residue. The N-terminal sequence of the mature BtuF-His protein was APRVI, indicating that the signal sequence was cleaved after Ala-22 in the leader peptidase-1 recognition sequence Leu-Asn-Ala. The mature protein is predicted to have a molecular mass of 27.78 kDa, in agreement with that observed on SDS-PAGE (Fig. 2). Thus, mature BtuF was shown to be a periplasmic protein with a cleaved N-terminal signal sequence.
Cbl binding by BtuF.
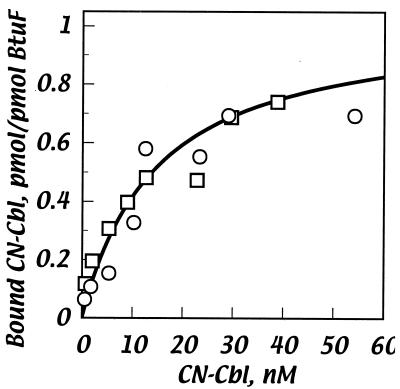
The binding of CN-Cbl to purified BtuF-His was measured by two methods. In the first, BtuF-His was incubated with varied concentrations of CN-[57Co]Cbl, and samples were filtered through a charcoal pad to rapidly remove unbound Cbl. The amount of protein-bound label in the filtrate was determined. The amount of bound Cbl was plotted against the unbound concentration, and the data points were fit to a hyperbolic binding curve (Fig. 3). The parameters determined from the curve fitting indicated a maximal stoichiometry of roughly 1 mol of CN-Cbl bound per mol of BtuF-His and a high affinity, with a Kd of around 15 nM.
FIG. 3.
Binding of CN-[57Co]Cbl to purified BtuF-His, measured in the charcoal filtration assay. Binding is plotted as the number of picomoles of CN-Cbl bound to BtuF as a function of the concentration of CN-Cbl added. The two symbols represent results from separate experiments. The curve was fit to a one-site hyperbolic model by the DeltaGraph curve-fitting feature.
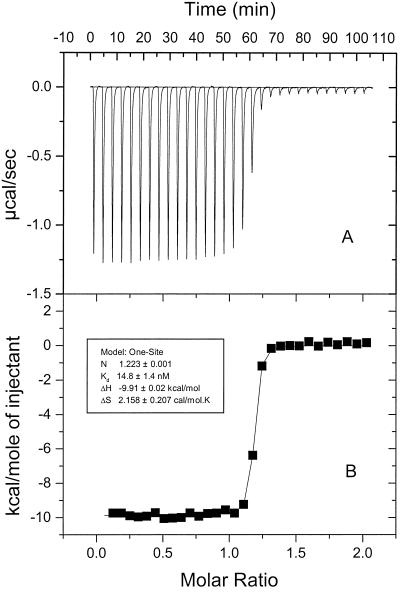
Binding was also measured by isothermal titration calorimetry, in which the heat evolved following addition of incremental portions of CN-Cbl was measured and plotted against the CN-Cbl concentration (Fig. 4). Correction was made for the heat of mixing when CN-Cbl was added to buffer alone. In three experiments, the binding constant, Kd, averaged 14.8 ± 1.4 nM, which is in excellent agreement with the radiolabel-binding assay. The thermodynamic parameters were ΔH = −9,910 cal/mol and ΔS = 2.2 cal/(mol × K), indicating that the binding reaction is almost entirely enthalpy driven. These results confirm the prediction that BtuF is the periplasmic Cbl-binding protein.
FIG. 4.
Isothermal titration calorimetry of BtuF-His with CN-Cbl. (A) Specific heat versus time of titration of CN-Cbl into purified BtuF-His. The heat of dilution of CN-Cbl into the buffer has been subtracted. (B) Enthalpies per mole of CN-Cbl injected versus molar ratio (CN-Cbl/BtuF-His).
Growth phenotypes of btuF mutants.
There are three assays for Btu function, all of which require intact cells. Transport of radiolabeled CN-Cbl measures total uptake into the cell. The amount of label bound to BtuB or accumulated in the periplasm is estimated from its ability to be released during chase with excess unlabeled CN-Cbl and can represent a substantial proportion of the total. CN-Cbl taken into the cytoplasm is retained during chase (29). This transport assay distinguishes wild-type transport from defects in OM transport in btuB or tonB mutants and from defects in CM transport, but it is unable to detect low-level transport across the CM. The growth response in the methionine assay measures the ability of CN-Cbl to support growth of a metE mutant in place of methionine. The response measures entry of Cbl into the cytoplasm and is very sensitive, since entry of around 25 molecules of Cbl suffices for a cell doubling (16). Thus, a positive response in this assay can be seen even with very low CM transport activity. The ethanolamine growth assay measures the ability of cells to grow with ethanolamine as a nitrogen source. Growth requires activity of adenosyl-Cbl-dependent ethanolamine ammonia-lyase, and a positive response in this assay requires uptake of much larger amounts of Cbl than does the methionine assay, estimated at 500 molecules per cell (6).
The mutant strains described in this study were tested in all three phenotypic assays of Btu function. Null mutations in pfs, btuF, and yadS carrying an in-frame deletion of the bulk of each coding sequence were prepared in plasmid pYadT2, as described in Fig. 1. Deletions which also carried a kanamycin resistance aph cassette for selection were prepared in plasmid pKO3. The chromosomal alleles of pfs, btuF, and yadS in wild-type, metE, btuB, tonB, and btuC strains were replaced with the aph-marked deletion allele by homologous recombination using the pKO3 system (22). Growth phenotypes were determined to test whether these mutations affected CN-Cbl utilization, and complementation behavior was tested by introduction of plasmids carrying the wild-type or deletion versions of each gene.
The ΔbtuF::Km mutation strongly interfered with the growth response of metE strains to CN-Cbl (Table 3), but growth with methionine was not affected. In the metE strain RK4379, which grew well with 0.5 nM CN-Cbl, the presence of the ΔbtuF::Km mutation prevented utilization of CN-Cbl at concentrations of 500 nM or less, and the growth response was impaired even at 5 μM or higher. On plates with CN-Cbl concentrations of above 5 nM, large colonies frequently appeared against the background of weak or minimal growth, presumably as the result of secondary suppressor mutations. Combination of the ΔbtuF::Km mutation with null mutations in btuB (NC20) or tonB (NC23) resulted in complete loss of the ability to utilize CN-Cbl at all tested concentrations up to 500 μM. This is the same response displayed by btuB btuC or tonB btuC double mutants (1). The very poor utilization of CN-Cbl by the ΔbtuF::Km mutant contrasted with the behavior of the E. coli btuC strain RK6049 (Table 3) (15) or of S. enterica serovar Typhimurium btuC or btuF mutants (42), which were only slightly impaired in CN-Cbl utilization.
TABLE 3.
Growth on CN-Cbl of the btuF and other btu mutants
| Strain | Growtha on:
|
||||||
|---|---|---|---|---|---|---|---|
| Met | CN-Cbl at (nM):
|
||||||
| 0 | 0.5 | 5 | 50 | 500 | 5,000 | ||
| RK4353 (wild type) | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| NC14 (RK4353 ΔbtuF) | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| RK4379 (metE) | ++ | − | + | ++ | ++ | ++ | ++ |
| NC17 (RK4379 ΔbtuF) | ++ | − | − | −b | −b | −b | ±b |
| RK4936 (btuB) | ++ | − | − | − | − | ± | + |
| NC20 (RK4936 ΔbtuF) | ++ | − | − | − | − | − | − |
| RK5015 (tonB) | + | − | − | − | − | − | + |
| NC23 (RK5015 ΔbtuF) | + | − | − | − | − | − | − |
| RK6049 (btuC) | ++ | − | − | + | + | + | + |
| NC26 (RK6049 ΔbtuF) | ++ | − | − | + | + | + | + |
| NC28 (RK4379 ΔbtuC) | ++ | − | − | −b | −b | −b | ±b |
| NC29 (RK4379 ΔbtuE) | ++ | − | ++ | ++ | ++ | ++ | ++ |
| NC30 (RK4379 ΔbtuD) | ++ | − | −b | + | + | + | + |
Growth is indicated as colony size following 48 h at 37°C on minimal A agar plates containing the indicated supplements, relative to the size of colonies formed by strain RK4379 on methionine, designated ++.
Large colonies appear throughout the streaks (suppressors).
The ΔbtuF::Km strain NC17 was unable to use ethanolamine as a nitrogen source even in the presence of 5 μM CN-Cbl (Table 4), whereas the parental strain RK4379 grew optimally at 50 nM CN-Cbl and partially at 5 nM. This defect of the mutant for ethanolamine utilization confirms the strong deficiency for Cbl transport across the CM. The growth defects in both assays of the ΔbtuF::Km mutant were fully complemented by plasmid pYadT2 containing the entire insert, as well as by the pΔpfs and pΔyadS plasmids, but not by the pΔbtuF plasmid (Table 4). Thus, the observed phenotypes are specific for the deleted btuF gene and are independent of the presence of its flanking genes.
TABLE 4.
Complementation of growth defects on CN-Cbl and ethanolamine
| Strain and plasmid | Growtha in:
|
||
|---|---|---|---|
| Methionine assay with:
|
Ethanolamine assay with 5 μM CN-Cbl | ||
| 0.5 nM CN-Cbl | 5 μM CN-Cbl | ||
| RK4379 (metE) | + | ++ | + |
| NC17 (RK4379 ΔbtuF) | − | ±b | − |
| pYadT2 | + | ++ | + |
| pΔpfs | + | ++ | + |
| pΔbtuF | − | ±b | − |
| NC28 (RK4379 ΔbtuC) | − | ±b | − |
| pbtuCED | + | ++ | + |
| pΔbtuC | − | ±b | − |
| pΔbtuE | + | ++ | + |
| pΔbtuD | + | ++ | + |
| NC29 (RK4379 ΔbtuE) | + | ++ | − |
| pbtuCED | + | ++ | + |
| pΔbtuC | + | ++ | + |
| pΔbtuE | + | ++ | + |
| pΔbtuD | + | ++ | − |
| NC30 (RK4379 ΔbtuD) | −b | + | − |
| pbtuCED | + | ++ | + |
| pΔbtuC | + | ++ | + |
| pΔbtuE | + | ++ | + |
| pΔbtuD | −b | + | − |
Growth is indicated as colony size following 48 h at 37°C on minimal A agar plates containing the indicated suppplements, relative to the size of colonies formed by strain RK4379 on methionine or on medium A for reference to the ethanolamine assay, designated ++.
Large colonies appear throughout the streaks (suppressors).
Effect of the btuF mutation on CN-Cbl transport.
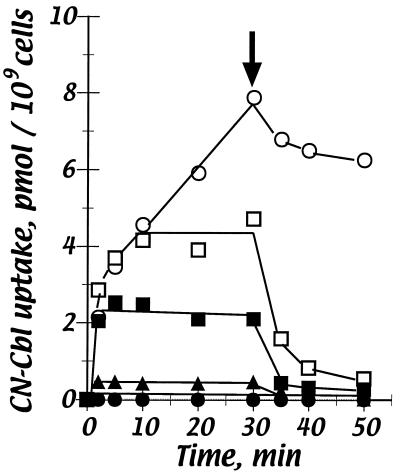
The CN-Cbl transport activities of all strains constructed in this study were determined (examples are shown in Fig. 5 and 6). The wild-type strain, as expected, exhibited rapid binding to the OM transporter BtuB, followed by relatively slow accumulation of label. Chase with nonradioactive CN-Cbl resulted in release of only a small fraction of the label, which had not entered the cytoplasm and was bound to BtuB or in the periplasm. As control strains, the btuB mutant showed no CN-Cbl binding or transport and the tonB mutant showed only binding to BtuB and no detectable accumulation (Fig. 5). In contrast, the btuC mutant RK6049 showed substantial accumulation of labeled CN-Cbl (ca. 1,500 molecules per cell), but this label was retained in the periplasm as indicated by its extensive release upon chase. The ΔbtuF::Km strain NC17 showed behavior similar to that of the btuC mutant but achieved about half of the steady-state level of periplasmic accumulation of the former strain. These results indicate that Cbl is accumulated in the periplasm in the absence of BtuF and thus that substantial uptake into the cytoplasm requires BtuC and BtuF.
FIG. 5.
Uptake of CN-[57Co]Cbl. Strains assayed were RK4379 (metE) (○), RK4936 (btuB) (•), RK5015 (tonB) (▴), RK6049 (btuC) (□), and NC17 (ΔbtuF:: Km) (▪). Results are expressed as picomoles of Cbl taken up per 109 cells. At the time indicated by the arrow (30 min), a 100-fold molar excess of unlabeled CN-Cbl was added. Transport by the newly constructed btuC and btuD mutants was similar to that shown here for the btuC mutant.
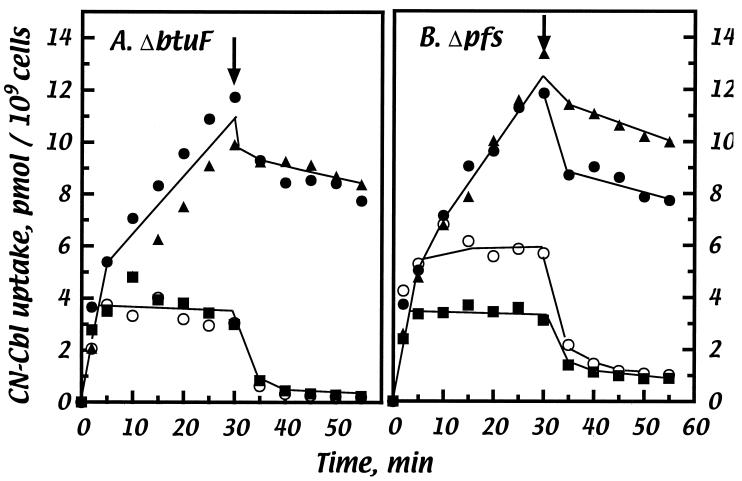
FIG. 6.
Complementation of the defect in CN-Cbl transport in btuF and pfs mutants by deletion plasmids. Host strains were NC17 (metE ΔbtuF::Km) (A) and NC16 (metE Δpfs::Km) (B). Host strains carried the following plasmids: no plasmid (○), pYadT2 (pfs-btuF-yadS) (•), pΔpfs (▴), or pΔbtuF (▪). Experimental conditions were as described for Fig. 5.
Reevaluation of the phenotypes of btuCED mutants.
The weak growth phenotype previously described for the mutants defective in the Cbl periplasmic permease was surprising in comparison to the greatly impaired transport across the CM and the strong phenotype of mutants affected in transport across the OM. It was also puzzling that the btuC ΔbtuF::Km double mutant displayed the same weak growth defect as the btuC mutant rather than the marked growth defect of the ΔbtuF::Km single mutant (Table 3). This double mutant strain was constructed by allelic exchange of the ΔbtuF::Km allele into btuC strain RK6049, and it was possible that the growth phenotype was complicated by the presence of compensatory suppressor mutations.
Hence, we constructed a set of new null mutations in each gene of the btuCED operon by an approach similar to that described above. A portion of each gene sequence was deleted using existing restriction sites within each gene and a Gm cassette was inserted in place of the deleted sequences to allow genetic selection. The mutations were transferred onto the chromosome of metE strain RK4379 by allelic replacement using the pKO3 system and were verified by PCR tests. These new null mutants defective in btuC (NC28), btuE (NC29), or btuD (NC30) were tested in both growth assays with CN-Cbl (Tables 3 and 4). In contrast to the previous findings, the ΔbtuC::Gm strain NC28 had the same phenotype as the ΔbtuF::Km mutant, was strongly impaired for utilization of 5 μM CN-Cbl in the methionine assay, and was completely impaired in the ethanolamine assay (Table 4). As was seen with the ΔbtuF mutant, suppressor colonies arose throughout the streaks on CN-Cbl plates. The ΔbtuE::Gm mutation had no effect on the response to CN-Cbl in the methionine assay, as expected (30), but this mutant was defective in the ethanolamine assay. The ΔbtuD::Gm strain NC30 had a phenotype similar to that previously reported (15); i.e., it was impaired in utilization of CN-Cbl at concentrations of below 50 nM in the methionine assay (Table 3). This mutant was defective in the ethanolamine assay. No substantial difference in CN-Cbl uptake between the newly constructed btuC and btuD null mutants and those isolated previously was seen (data not shown). The steady-state accumulation of periplasmic CN-Cbl was variable in different strains and experiments, but all of these mutants were defective in transport across the CM.
In tests of complementation of the growth defects in these mutants, the ΔbtuC::Gm mutant phenotype was fully restored by complementation with the pbtuCED plasmid carrying the intact locus, as well as by the pΔbtuE and pΔbtuD plasmids, but not at all by the pΔbtuC plasmid (Table 4). In contrast, the defective growth on ethanolamine of the ΔbtuE::Gm strain NC29 was complemented by the plasmids carrying the complete region or those with deletions in btuC or btuE but not by the plasmid with deletion of btuD. This complementation pattern indicates that the growth defect in the ΔbtuE::Gm mutant is the result of the polar effect on the mutation on btuD expression rather than of the involvement of the btuE product in Cbl transport. Finally, the defect in ethanolamine utilization and the slight impairment in the methionine assay of the ΔbtuD::Gm mutation were corrected by all plasmids except the one with deletion of btuD. We can thus conclude that the periplasmic permease components BtuF and BtuC are crucial for CN-Cbl utilization and transport into the cytoplasm, that BtuD is needed for a wild-type level of transport, and that BtuE is not involved.
As a test for the presence of suppressor mutations in the previously described strains, the btuC allele was transduced from RK6049 into RK4379 by linkage with the adjacent zdh-1::Tn10 marker. The tetracycline-resistant transductants exhibited two growth phenotypes. Some showed the same wild-type behavior, and others showed the same phenotype as the newly constructed ΔbtuC mutants, namely, an impaired response in the methionine assay with 5 μM CN-Cbl with generation of frequent suppressor variants. We thus conclude that the previously reported phenotype was complicated owing to the presence of suppressor mutations which improved transport across the CM of low levels of Cbl sufficient to allow a response in the methionine assay.
Properties of suppressor variants.
To initiate study of the suppressor variants, 20 large colonies which arose from the ΔbtuC::Gm and ΔbtuF::Km strains on CN-Cbl plates were tested for their growth phenotypes. All suppressor isolates remained auxotrophic for methionine or Cbl. None was able to grow on 0.5 nM CN-Cbl, which the btu+ strain could use efficiently. The minimal concentration of CN-Cbl which allowed good growth varied among the suppressors and did not correlate with the concentration on which they were selected. After passage on nonselective LB agar, many of the suppressor isolates were unstable and reverted to the original phenotype, i.e., poor growth on CN-Cbl with frequent appearance of large suppressor variants. This instability limits the ability to determine the basis for the suppression.
Growth phenotypes of pfs and yadS mutants.
To test the involvement of the genes flanking btuF in CN-Cbl utilization, the phenotypes of the null mutations in the respective genes were examined. The ΔyadS::Km mutation had no detectable effect on growth with CN-Cbl of any of the host strains on any medium tested (Table 5) and had no effect on CN-Cbl transport activity (data not shown).
TABLE 5.
Growth on CN-Cbl of the Δpfs::Km and ΔyadS::Km mutants
| Strain | Growtha on:
|
||||||
|---|---|---|---|---|---|---|---|
| Met | Cn-Cbl at (nM):
|
||||||
| 0 | 0.5 | 5 | 50 | 500 | 5,000 | ||
| RK4353 (wild type) | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| NC13 (RK4353 Δpfs) | ± | − | − | − | − | − | − |
| NC15 (RK4353 ΔyadS) | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| RK4379 (metE) | ++ | − | + | ++ | ++ | ++ | ++ |
| NC16 (RK4379 Δpfs) | ± | − | − | − | − | − | − |
| NC18 (RK4379 ΔyadS) | ++ | − | + | ++ | ++ | ++ | ++ |
| RK4936 (btuB) | ++ | − | − | − | − | ± | + |
| NC19 (RK4936 Δpfs) | ± | − | − | − | − | − | − |
| NC21 (RK4936 ΔyadS) | ++ | − | − | − | − | ± | + |
| RK5015 (tonB) | + | − | − | − | − | − | + |
| NC22 (RK5015 Δpfs) | ± | − | − | − | − | − | − |
| NC24 (RK5015 ΔyadS) | + | − | − | − | − | − | + |
| RK6049 (btuC) | ++ | − | − | + | + | + | + |
| NC25 (RK6049 Δpfs) | + | − | − | − | − | − | − |
| NC27 (RK6049 ΔyadS) | ++ | − | − | + | + | + | + |
Growth is indicated relative to that of the parental strain on minimal medium with ammonium as a nitrogen source (++).
In contrast, the Δpfs::Km mutation strongly affected bacterial growth. Colonies of all strains carrying the Δpfs::Km mutation on LB agar were much smaller than those of isogenic pfs+ strains. Growth was even more strongly reduced on minimal medium supplemented with methionine and was undetectable on minimal medium lacking methionine or supplemented with 5 μM CN-Cbl (Table 5). This growth defect occurred even in the metE+ strain NC13, which is not a methionine auxotroph. This strong growth impairment of the Δpfs::Km strains was completely reversed by the presence of the pfs+ plasmids pYadT2, pΔbtuF, and pΔyadS but not by the plasmid carrying the Δpfs allele (data not shown). Thus, the absence of pfs strongly reduces cell growth, but this deficiency is not related to CN-Cbl transport or metabolism, and it is not the result of polar effects on expression of the distal genes.
The physiological basis for the growth impairment in Δpfs strains is not obvious. The Pfs protein carries out a step in the recycling of the SAM derivatives which are produced during reactions of methyl transfer and spermidine synthesis. Pfs was recently found to carry out an essential step in the biosynthesis of a putative interspecies signaling molecule known as autoinducer-2 (AI-2) (35). Lack of AI-2 synthesis owing to a defect in the luxS product has substantial effects on cell physiology in E. coli but does not result in the growth inhibition seen with the Δpfs mutant (13). In tests of whether supplementation with products of SAM metabolism might correct the defect in the Δpfs strain, we found that addition of spermidine, the common nucleosides or bases, or a mixture of all amino acids did not improve growth on minimal medium of strain NC13 at all. In contrast, supplementation with a mixture of vitamins stimulated growth almost to the level of the wild-type pfs+ strain. Supplementation with the individual vitamins showed that the component able to restore near-normal growth on plates was biotin at concentrations of as low as 1 ng/ml (Table 6). Although we cannot yet explain why supplemental biotin circumvents the requirement for Pfs function, this observation could facilitate studies of AI-2 synthesis and the properties of the pfs-defective strain.
TABLE 6.
Complementation behavior of Δpfs::Km
| Strain and plasmid | Growtha in:
|
||||||
|---|---|---|---|---|---|---|---|
| Methionine assay with:
|
Ethanolamine assay with biotin and 5 μM CNCbl | ||||||
| No biotin
|
Biotin at 10 ng/ml
|
||||||
| Met | 0.5 nM CN-Cbl | 5 μM CN-Cbl | Met | 0.5 nM CN-Cbl | 5 μM CN-Cbl | ||
| RK4379 | ++ | + | ++ | ++ | ++ | ++ | + |
| NC16 (RK4379 Δpfs) | ± | − | − | ++ | + | + | − |
| pYadT2 (pfs btuF) | ++ | + | ++ | ++ | ++ | ++ | + |
| pΔpfs | ± | − | − | ++ | + | + | ± |
| pΔbtuF | ++ | + | ++ | ++ | ++ | ++ | − |
Growth is indicated relative to that of the parental strain on minimal medium with ammonium as a nitrogen source (++).
By using biotin supplementation to improve cell growth, we found that the Δpfs::Km defect had little effect on CN-Cbl utilization in the methionine assay over the full range of concentrations but that the strain was defective in the ethanolamine assay (Table 6 and data not shown). CN-Cbl transport assays (Fig. 6) also showed that the Δpfs::Km strain had a transport deficiency similar to that of the btuC or btuF strain. These growth and transport defects were complemented by pYadT2 or by pΔpfs but not by pΔbtuF. The Km cassette in the chromosomal Δpfs::Km mutation is oriented oppositely to the direction of pfs transcription. Thus, it is likely that this cassette inserted in pfs confers a polar effect that decreases btuF expression enough to impair transport and ethanolamine utilization but not so completely as to block CN-Cbl utilization in the very sensitive methionine assay. This polar effect is not seen with the plasmid-borne Δpfs allele which lacks the Km cassette. Taken together, these results show that the pfs and yadS gene products are not directly involved in CN-Cbl transport or utilization and suggest that pfs and btuF are cotranscribed.
DISCUSSION
Our results demonstrate that btuF, the E. coli ORF designated yadT in the genomic sequence (3), encodes the periplasmic Cbl-binding and Cbl transport protein, as was suggested from the growth and transport properties of mutants affected in the orthologous gene in S. enterica serovar Typhimurium (42). It is shown here that BtuF is a periplasmic protein, which is released from the cell by osmotic shock and possesses a typical cleaved N-terminal signal sequence. The purified protein with a C-terminal His6 tag binds CN-Cbl with high affinity in the range of 15 nM, which agrees closely with the Kd determined previously for binding to crude osmotic shock fluid (7). Similar binding constants were obtained by two independent methods which measured the binding of radiolabeled substrate and heat evolution in isothermal titration calorimetry. The substrate-binding affinity of BtuF is weaker than the 0.3 nM Kd of BtuB (7). However, the affinity is high enough for its role in transport because its substrate is accumulated in the periplasm by the action of BtuB.
The substrate specificity and affinity of several of the periplasmic binding proteins for iron or siderophore uptake have been determined. Hydroxamate binding to FhuD was measured by quenching of protein tryptophan fluorescence and ranged from 300 to 400 nM for coprogen and aerobactin to 1 μM for ferrichrome and to around 40 μM for ferrioxamines (31). The affinity constants for binding of ferric enterobactin to FepB ranged from 135 nM when assayed by gel filtration to 30 nM when assayed by quenching of tryptophan fluorescence (39). The structures of the periplasmic iron-binding protein Hit from Haemophilus influenzae (8) and the ferrichrome-binding protein FhuD from E. coli (10) have been determined. FhuD is fairly closely related in sequence to BtuF (20). The structure of FhuD exhibits several notable differences from that of periplasmic binding proteins for sugars and amino acids (28). FhuD has the typical bilobed structure present in other binding proteins, but its substrate-binding cleft is shallower and the hinge structure linking the two lobes is formed by a single bent α-helix rather than the typical three flexible β-strands (10). It was proposed that FhuD might undergo less extensive conformational movement upon substrate binding than do the classical proteins (10). It will be interesting to determine whether the structure of BtuF resembles that of FhuD.
CN-Cbl transport assays showed that the E. coli btuF mutant was extremely defective for uptake into the cytoplasm, similar to the previously described defect in the btuC and btuD mutants (15). The transport assays revealed that the btuC, btuD, and btuF mutants display substantial substrate accumulation into the periplasm above the level of binding to the receptor shown in the tonB mutant. The lack of Cbl transport into the cytoplasm is judged from the constant steady-state level of this material, the rapid and extensive release of label upon chase, and the defective growth responses to CN-Cbl. Previous studies showed that cells of a btuC mutant were unable to convert exogenous CN-Cbl to the intracellular coenzyme species (29). These results confirm that the BtuB/TonB-dependent system mediates active accumulation of CN-Cbl in the periplasmic space. The BtuF-defective strain typically accumulates a smaller amount of label than did the mutants defective in the other components in the permease. This difference was as much as a factor of 2, but it varied in different assays. The observed periplasmic accumulation of CN-Cbl could reflect its binding to BtuF, but this explanation would require the presence of at least 500 molecules of BtuF, which seems unlikely since there are only 200 to 300 molecules of BtuB in the OM and roughly 3 molecules of the binding activity per cell in osmotic shock fluid (7). A possibility to be explored is that BtuF helps in the release of CN-Cbl from BtuB.
The unexpected but satisfying growth phenotypes of the btuF null mutant emphasized the importance of testing all three assays of Btu function. Previous studies indicated that mutations affecting the Cbl-specific periplasmic permease in E. coli and S. enterica serovar Typhimurium caused only a slight impairment in utilization of CN-Cbl for the methionine replacement assay of growth of metE mutants (1, 15, 42). This phenotype contrasted with the strong defect in transport of radiolabeled CN-Cbl across the CM and with the response in the ethanolamine growth assay. Explanations for this behavior invoked the higher sensitivity of the methionine assay than the other assays and the possible existence of a minor route for entry of Cbl from the OM directly into the cytoplasm (19).
Unlike these previously described permease mutations, the ΔbtuF::Km mutation conferred highly defective CN-Cbl utilization except when the mutation was transferred into the previously studied btuC mutant strain. The discrepancy can be explained by the frequent appearance of suppressor variants better able to utilize CN-Cbl for growth. Evidence supporting this hypothesis came from analysis of a new set of null mutations in the btuCED genes. The newly constructed ΔbtuC mutant showed a marked deficiency in CN-Cbl utilization similar to that of the btuF mutant and also gave rise to large suppressor variant colonies. The ΔbtuE::Gm mutation did not affect CN-Cbl utilization in the methionine assay, as expected (30), but interfered with the ethanolamine and transport assays. This behavior was shown to be due to the polar effect of the resistance cassette on the expression of btuD. Surprisingly, the new ΔbtuD variant was not as severely affected as the ΔbtuC mutant, although it lacked substantial transport into the cytoplasm. Suppressor-containing strains able to utilize CN-Cbl arose at an appreciable frequency on methionine assay plates. It is important to test growth responses by streaking colonies on selection plates so that the occurrence of suppressors can be detected. They provide a bypass or alternative route across the CM. Because they do not arise in btuB or tonB mutants alone or when combined with the btuCDF mutants, the suppressors do not alter the requirement for Cbl or provide an alternative route across the OM. The suppressor mutations have not been mapped yet owing in part to their instability. Preliminary tests of CN-Cbl binding and transport suggest that some suppressor variants exhibit increased binding and periplasmic accumulation of CN-Cbl relative to the unsuppressed mutants. We also found that plasmid-encoded overexpression of BtuB improves CN-Cbl utilization in ΔbtuF::Km and ΔbtuC::Gm strains. We suggest that some suppressors result in increased expression of BtuB or increased transport of CN-Cbl into the periplasm. The elevated concentration of periplasmic CN-Cbl might allow some transport across the CM by a permease for a structurally related substrate.
An unusual feature of the Btu transport system is the absence of genetic linkage of the transport genes, which is found for most periplasmic permeases, especially those for OM-dependent transport systems. Since each permease component is required for effective Cbl transport, this genetic dispersal must reduce the ability for horizontal gene transfer of Cbl transport genes. The btu orthologues in the genome sequences of related bacteria are dispersed in all cases examined. This circumstance is interpreted to indicate that the btu genes were already scattered on the chromosome of the ancestor common to all strains which possess them. Köster (20) analyzed the phylogenetic relationships of the periplasmic permease components acting on iron and related substrates. There are multiple subfamilies of these transport systems, but the phylogenetic relationship of each component gene shows a tree similar to those of the other components. This result indicates that each periplasmic permease system has evolved jointly, without evidence of independent acquisition and adaptation of any individual component. This joint evolution of all components also appears to apply to the btu genes, whose scattered genetic location reduces the likelihood that any one component could be acquired and function independently. Further analysis of the phylogenetic relationships of btu sequences and map locations is ongoing.
The expression of btuB is controlled by the cellular level of adenosyl-Cbl through a complex process involving ribosome binding and RNA stability (27). Characterization of lac fusions indicated that the btuCED operon is not affected by Cbl supplementation (15). No information is available yet regarding the location of the transcriptional start site or the regulation of btuF expression. However, it is noteworthy that the btuF coding sequence overlaps the upstream pfs gene. The apparent polar effect of a Km cassette in pfs on btuF function suggests that btuF might be transcribed from a promoter upstream of pfs. Finally, the function of the pfs gene product is of considerable interest. The Pfs protein is involved in the recycling of the two products of SAM metabolism, SAH and methylthioadenosine. SAH interferes with SAM-dependent reactions. The importance of the recycling process is indicated by the marked growth impairment of the pfs mutant. Recently, it was shown that the product of the action of Pfs on SAH is the substrate for the synthesis by the widely distributed LuxS protein of the interspecies signaling molecule known as AI-2 (35). We tested whether any products of SAM metabolism might reverse the growth impairment in the pfs mutant and found unexpectedly that biotin was able to restore near-normal growth. The explanation for this finding is not apparent yet, but this observation should facilitate further study of the role of Pfs in bacterial metabolism.
Acknowledgments
This work was supported by National Institutes of Health research grants GM19078 (to R.J.K.) and DK59999 (to M.C.W.) and by Deutsche Forschungsgemeinschaft grant Ko967/7 (to W.K.).
REFERENCES
- 1.Bassford, P. J., Jr., and R. J. Kadner. 1977. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J. Bacteriol. 132:796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlyn, M. K. B., K. B. Low, and K. E. Rudd. 1996. Linkage map of Escherichia coli K-12, edition 9, p.1715–1902. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. E. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mao, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474. [DOI] [PubMed] [Google Scholar]
- 4.Bolivar, F., R. L. Rodriquez, P. J. Greene, M. C. Betlach, H. L. Heyneker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113. [PubMed] [Google Scholar]
- 5.Bradbeer, C. 1993. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175:3146–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradbeer, C. 1979. Transport of vitamin B12 in Escherichia coli, p.711–723. In B. Zagalak and W. Friedrich (ed.), Vitamin B12. Walter de Gruyter, Berlin, Germany.
- 7.Bradbeer, C., J. S. Kenley, D. R. DiMasi, and M. Leighton. 1978. Transport of vitamin B12 in Escherichia coli. Corrinoid specificities of the periplasmic B12-binding protein and of energy-dependent B12 transport. J. Biol. Chem. 253:1347–1352. [PubMed] [Google Scholar]
- 8.Bruns, C. M., A. J. Norwalk, A. S. Arvai, M. A. McTigue, K. G. Vaughan, T. A. Mietzner, and D. E. McRee. 1997. Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily. Nat. Struct. Biol. 4:919–924. [DOI] [PubMed] [Google Scholar]
- 9.Cadieux, N., C. Bradbeer, and R. J. Kadner. 2000. Sequence changes in the Ton box region of BtuB affect its transport activities and interaction with TonB protein. J. Bacteriol. 182:5954–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, T. E., S.-Y. Ku, D. R. Dougan, H. J. Vogel, and L. W. Tari. 2000. The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat. Struct. Biol. 7:287–291. [DOI] [PubMed] [Google Scholar]
- 11.Cornell, K. A., and M. K. Riscoe. 1998. Cloning and expression of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: identification of the pfs gene product. Biochim. Biophys. Acta 1396:8–14. [DOI] [PubMed] [Google Scholar]
- 12.Davis, B. D., and E. Mingioli. 1950. Mutants of Escherichia coli requiring methionine or B12. J. Bacteriol. 60:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLisa, M. P., C.-F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeVeaux, L. C., D. S. Clevenson, C. Bradbeer, and R. J. Kadner. 1986. Identification of the BtuCED polypeptides and evidence for their role in vitamin B12 transport in Escherichia coli. J. Bacteriol. 167:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeVeaux, L. C., and R. J. Kadner. 1985. Transport of vitamin B12 in Escherichia coli: cloning of the btuCD region. J. Bacteriol. 162:888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiGirolamo, P. M., R. J. Kadner, and C. Bradbeer. 1971. Isolation of vitamin B12 transport mutants of Escherichia coli. J. Bacteriol. 106:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich, M. J., L. C. DeVeaux, and R. J. Kadner. 1986. Nucleotide sequence of the btuCED genes involved in vitamin B12 transport in Escherichia coli and homology with components of periplasmic-binding-protein-dependent transport systems. J. Bacteriol. 167:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb, C., K.-S. Lau, L. R. Wasserman, and V. Herbert. 1965. Rapid charcoal assay for intrinsic factor (IF), gastric juice unsaturated B12-binding capacity, antibody to IF, and serum unsaturated B12-binding capacity. Blood 25:875–884. [PubMed] [Google Scholar]
- 19.Kadner, R. J. 1990. Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol. Microbiol. 4:2027–2033. [DOI] [PubMed] [Google Scholar]
- 20.Köster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152:291–301. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 22.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundrigan, M. D., and R. J. Kadner. 1989. Altered cobalamin metabolism in Escherichia coli btuR mutants affects btuB regulation. J. Bacteriol. 171:154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675–681. [DOI] [PubMed] [Google Scholar]
- 26.Nossal, N. G., and L. A. Heppel. 1966. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J. Biol. Chem. 241:3055–3062. [PubMed] [Google Scholar]
- 27.Nou, X., and R. J. Kadner. 1998. Coupled changes in translation and transcription during cobalamin-dependent regulatioh of btuB expression in Escherichia coli. J. Bacteriol. 180:6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiocho, F. A., and P. S. Ledvina. 1996. Atomic structure and specificity of bacteria periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 20:17–25. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds, P. R., G. P. Mottur, and C. Bradbeer. 1980. Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of btuC and tonB. J. Biol. Chem. 255:4313–4319. [PubMed] [Google Scholar]
- 30.Rioux, C. R., and R. J. Kadner. 1989. Vitamin B12 transport in Escherichia coli K12 does not require the btuE gene of the btuCED operon. Mol. Gen. Genet. 217:301–308. [DOI] [PubMed] [Google Scholar]
- 31.Rohrbach, M. R., V. Braun, and W. Köster. 1995. Ferrichrome transport in Escherichia coli K-12: altered substrate specificity of mutated periplasmic FhuD and interaction of FhuD with the integral membrane protein FhuB. J. Bacteriol. 177:7186–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137–181. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Scarlett, F. A., and J. M. Turner. 1976. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J. Gen. Microbiol. 95:173–176. [DOI] [PubMed] [Google Scholar]
- 35.Schauder, S., K. Shokat, M. G. Surette, and B. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum sensing signal molecule. Mol. Microbiol. 41:463–476. [DOI] [PubMed] [Google Scholar]
- 36.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831–834. [PubMed] [Google Scholar]
- 37.Shattuck-Eidens, D. M., and R. J. Kadner. 1981. Exogenous induction of the Escherichia coli hexose phosphate transport system defined by uhp-lac operon fusions. J. Bacteriol. 148:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shea, C. M., and M. A. McIntosh. 1991. Nucleotide sequences and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol. Microbiol. 5:1415–1428. [DOI] [PubMed] [Google Scholar]
- 39.Sprencel, C., Z. Cao, Z. Qi, D. C. Scott, M. A. Montague, N. Ivanoff, J. Xu, K. M. Raymond, S. M. C. Newton, and P. E. Klebba. 2000. Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J. Bacteriol. 182:5359–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehlin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Bibber, M., C. Bradbeer, N. Clark, and J. R. Roth. 1999. A new class of cobalamin transport mutants (btuF) provides genetic evidence for a periplasmic binding protein in Salmonella typhimurium. J. Bacteriol. 181:5539–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]