Abstract
The general stress resistance of Escherichia coli is controlled by the RpoS sigma factor (ϕS), but mutations in rpoS are surprisingly common in natural and laboratory populations. Evidence for the selective advantage of losing rpoS was obtained from experiments with nutrient-limited bacteria at different growth rates. Wild-type bacteria were rapidly displaced by rpoS mutants in both glucose- and nitrogen-limited chemostat populations. Nutrient limitation led to selection and sweeps of rpoS null mutations and loss of general stress resistance. The rate of takeover by rpoS mutants was most rapid (within 10 generations of culture) in slower-growing populations that initially express higher ϕS levels. Competition for core RNA polymerase is the likeliest explanation for reduced expression from distinct promoters dependent on ϕ70 and involved in the hunger response to nutrient limitation. Indeed, the mutation of rpoS led to significantly higher expression of genes contributing to the high-affinity glucose scavenging system required for the hunger response. Hence, rpoS polymorphism in E. coli populations may be viewed as the result of competition between the hunger response, which requires sigma factors other than ϕS for expression, and the maintenance of the ability to withstand external stresses. The extent of external stress significantly influences the spread of rpoS mutations. When acid stress was simultaneously applied to glucose-limited cultures, both the phenotype and frequency of rpoS mutations were attenuated in line with the level of stress. The conflict between the hunger response and maintenance of stress resistance is a potential weakness in bacterial regulation.
The heterogeneity of rpoS gene sequence and function is a puzzling feature of bacterial isolates from either natural populations or laboratory cultures of Escherichia coli (13–15, 18, 31–33) and Salmonella enterica (17). For example, 13 of 58 toxin-producing E. coli isolates from various sources contained mutated rpoS and exhibited increased acid sensitivity (32). Given that the RpoS sigma factor ϕS controls the general stress response of E. coli and related bacteria (11), there is a paradoxical handicap in bacteria losing resistance to environmental challenges. Yet even in batch culture with rich media, the “growth advantage in stationary phase,” or GASP, phenotype is associated with the acquisition of attenuated rpoS mutations (34). A possible explanation is that GASP rpoS mutations aid scavenging of recycled nutrients in stationary phase (9). In this study, we investigated the selective advantage of losing RpoS and whether RpoS contributes to fitness under nutrient limitation. Reduced growth rates controlled by the availability of glucose or a nitrogen source can be established in continuous cultures or chemostats. RpoS is expressed under these conditions, particularly at lower growth rates (μ ≈ 0.1 h−1) (27), so we compared wild-type and rpoS mutant bacteria under these suboptimal growth conditions. Interestingly, mutation of rpoS turned out to be a great advantage under steady-state nutrient limitation.
Glucose- or N-limited growth leads to a hunger response, with induction of high-affinity nutrient-scavenging mechanisms essential for competitive fitness under low-nutrient conditions (8, 35). The cellular components required for nutrient acquisition under glucose and N limitation are distinct, but both sets of proteins are expressed from genes transcribed by RNA polymerase primed with sigma factors other than ϕS. There is evidence that competition between the ϕS and ϕ70 sigma factors for core RNA polymerase affects the relative expression of several genes, as does competition between ϕ32 and ϕ70 (6, 22). Also, the anti-ϕ70 factor encoded by rsd (16) is under RpoS control and could contribute to competition for core RNA polymerase. Hence, a notion tested in this study is whether derepressed expression of nutrient-scavenging systems provides the selective advantage for the accumulation of mutations in rpoS under nutrient-limited conditions.
The hunger response in E. coli is induced by intermediate, micromolar levels of available glucose but is repressed under conditions of either nutrient excess or nutrient starvation (8). The induction of genes significant in transport under glucose limitation was therefore tested in the presence and absence of ϕS. The lamB and mglBAD genes contribute to the high-affinity pathway of glucose uptake (7), so their expression was tested in an rpoS mutant. Glucose limitation resulted in significantly greater induction of transporter genes in the rpoS mutant, providing a strong selective advantage to bacteria that lose RpoS at suboptimal nutrient levels.
MATERIALS AND METHODS
Bacterial strains.
All strains used were derivatives of the E. coli K-12 strain MC4100, which contains a wild-type rpoS gene (18). Strain BW2952 is an MC4100 derivative containing a malG-lacZ transcriptional fusion [genotype, F− araD139 Δ(argF-lac)U169 rpsL150 deoC1 relA1 thiA ptsF25 flbB5301 rbsR malG::λplacMu55 Φ(malG::lacZ)] (26). Derivatives of these two strains containing either rpoS::Tn5 or rpoS::Tn10 insertions were constructed by P1 transduction using P1 cml clr1000 grown on strain ZK1000 or ZK1171 (1), respectively, to create BW2938 (MC4100 rpoS::Tn5) and BW3511 (BW2952 rpoS::Tn10) by selection on plates containing kanamycin (30 μg/ml) or tetracycline (15 μg/ml). A strain (BW3245) containing an mgl(Con) mutation (mglD L126stop) in the MC4100 background (28) was also made RpoS− (BW3522) by introduction of the rpoS::Tn5 mutation from ZK1000.
Growth medium and culture conditions.
The medium used in chemostat cultures was minimal medium A (MMA) (25). The amount of ammonium sulfate in the medium was reduced from 1 to 0.04 g/liter in the nitrogen-limiting chemostats. The carbon source in all cases was glucose, which was present at 0.02% (wt/vol) in the feed medium in glucose-limiting experiments and at 0.2% (wt/vol) in nitrogen-limiting experiments. For batch cultures, glucose was included at a concentration of 0.2% (wt/vol). The medium pH was adjusted to 7 in standard cultures but to 6 or 5.5 in acid stress experiments. Eighty-milliliter chemostat cultures were set up as described previously (3, 21). Dilution rates were set to 0.1, 0.3, and 0.6 h−1 (doubling times of 1.15, 2.4, and 6.9 h, respectively) as specified. The culture densities were between 1.9 × 108 and 2.1 × 108 bacteria ml−1.
Detection of rpoS mutants.
rpoS partial and null mutants were distinguished from the wild type by staining colonies on Luria agar plates. Plates were incubated overnight at 37°C and then left at 4°C for 24 h before being flooded with concentrated iodine. Dark brown colonies were wild type, while pale brown or white colonies indicated partial or null mutants with different levels of glycogen (12, 33). Chemostat isolates were also tested qualitatively for catalase activity by applying 6% (wt/vol) H2O2 directly onto colonies on Luria agar plates. Vigorous bubbling indicated wild-type RpoS activity.
β-Galactosidase assay and transport studies.
Five-milliliter samples from chemostat cultures were removed, and β-galactosidase activity was measured as described by Miller (25) by using sodium dodecyl sulfate- and chloroform-treated cells. The initial rate of uptake of 1 μM [14C]galactose by the same chemostat samples was determined with bacteria resuspended in MMA to an optical density at 580 nm of 0.2 as described previously (3). The rate of transport was calculated in units of picomoles of sugar transported per minute per 108 bacteria.
rpoS amplification and DNA sequencing.
A 1,302-bp fragment containing the rpoS gene was amplified from chemostat isolates by PCR using two external primers, RpoSF1 (5′-CGGACCTTTTATTGTGCACA-3′) and RpoSR1 (5′-TGATTACCTGAGTGCCTACG-3′), and a gene-internal primer, RpoSI (5′-CTGTTAACGGCCGAAGAAGA-3′). The reaction profile consisted of 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). PCR products were purified directly with Wizard DNA Preps DNA purification system (Promega Corp., Sydney, Australia). The nucleotide sequence was determined using the primers described above and dye-terminator sequencing reactions on a Catalyst Robotic Workstation. Mutations in mutant sequences were located by alignment with the known rpoS sequence in the E. coli genome database, using software available in the Australian National Genomic Information Service, Sydney, Australia.
Thermotolerance assay.
The chemostat samples were diluted in 0.9% NaCl to a density of 5 × 103 cells/ml. One-milliliter samples were then transferred to prewarmed tubes held at 60°C. One hundred microliters was withdrawn at time intervals of 1.5, 3, 5, and 7.5 min and plated directly onto nutrient agar plates, and these were incubated overnight at 37°C. The initial 100% survival point was determined by plating a 100-μl suspension just before heat shock.
Hydrogen peroxide resistance.
The chemostat sample was harvested, washed twice in 0.9% (wt/vol) NaCl, and resuspended in 0.9% NaCl to a final optical density at 580 nm of 0.1. Ten microliters of freshly diluted H2O2 (final concentration, 75 mM) was added to 1 ml of culture. The suspension was held at room temperature, and 100-μl samples were taken at 5, 10, 15, and 25 min. Serial dilutions in 0.9% NaCl were plated on nutrient agar and incubated overnight at 37°C. The initial 100% survival point was determined by counting the culture before addition of the H2O2.
RESULTS
Loss of rpoS function during growth with a limiting glucose or nitrogen source.
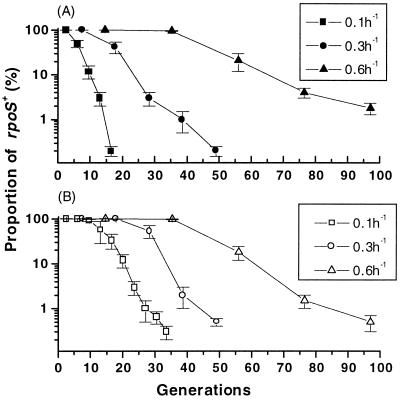
Wild-type bacteria were inoculated independently into glucose-limited chemostats growing with various steady-state growth rates. rpoS status was screened by plating organisms in the population and testing for glycogen in individual colonies (the synthesis of glycogen is under RpoS control [12]). As shown in Fig. 1A, the glycogen phenotype changed in every population established, and the proportion of wild-type bacteria decreased rapidly. The large majority of colonies in the populations after 4 days of culture also exhibited reduced bubbling with hydrogen peroxide, which is also indicative of a loss of katG expression in rpoS function (24). There was a 100% correlation between those colonies that lost iodine staining and those with reduced bubbling with peroxide. The rate of phenotypic change was remarkably reproducible in three independent populations, as indicated by the error bars in Fig. 1. The loss of glycogen production was most marked with slow-growing bacteria (doubling time of approximately 7 h) and less pronounced with faster-growing bacteria (doubling time of 69 min).
FIG. 1.
Appearance of rpoS mutations in chemostat cultures operating at various growth rates. Chemostat cultures of strain BW2952 growing at dilution rates of 0.1, 0.3, and 0.6 h−1 were monitored for the first 10 days after inoculation. Under each condition, three replicate populations were established. At the same time each day, samples were diluted in MMA, plated onto Luria agar plates, and incubated overnight at 37°C. Plates were stored at 4°C for another 24 h before being flooded with iodine to distinguish dark brown glycogen-containing cells (RpoS+), the proportion of which is shown in each graph. (A) Glucose-limiting cultures; (B) nitrogen-limiting cultures. Error bars indicate standard deviations.
The change in the glycogen phenotype shown in Fig. 1A was indeed due to a variety of rpoS mutations in the populations. Sequencing of the rpoS genes from 18 isolates with altered glycogen phenotypes showed that each isolate contained spontaneous rpoS changes (Table 1). Two of the mutations led to partial loss of function, as in the attenuated GASP mutants (34), but the majority led to more drastic changes to RpoS-regulated phenotypes. Hence, glucose-limited populations were rapidly taken over by mutants with rpoS mutations.
TABLE 1.
rpoS sequences in chemostat isolates
| Change in nucleotide sequencea | Change in amino acid sequence | Glycogen productionc | Catalase activityd | No. of isolates sequencede |
|---|---|---|---|---|
| Controls | ||||
| Wild type RpoS | ++++ | ++++ | ||
| rpoS::Tn10 | − | − | ||
| Isolatesb | ||||
| G to T at 297* | Leucine to phenylalanine at aaf 99 | ++ | ++ | 3 |
| T to A at 383* | Isoleucine to asparagine at aa 128 | ++ | ++ | 1 |
| T to G at 515 | Valine to glycine at aa 172 | − | − | 1 |
| G to T at 814* | Glutamic acid to stop codon at aa 272 | − | − | 1 |
| Δ484–487* | Truncation; retains first 161 native aa + 10 nonsense aa added | − | − | 1 |
| Δ151–155* | Truncation; retains first 50 native aa + 12 nonsense aa added | − | − | 1 |
| Δ393–397 | Truncation; retains first 131 native aa + 2 nonsense aa added | − | − | 6 |
| ΔA at 900 | Truncation; retains first 299 native aa + 23 nonsense aa added | − | − | 1 |
| A insertion at 571 | Truncation; retains first 190 native aa + 2 nonsense aa added | − | − | 2 |
| Δ249–345 | In-frame deletion of 32 aa (Δ44–75) | − | − | 1 |
Numbering of sequences is from the first nucleotide in the start codon
rpoS sequences are from unstained colonies screened with iodine for glycogen after 4 days of culture. The isolates were from two independent glucose-limited chemostats growing at a D = value of 0.3 h−1. The mutations from the first population are marked with asterisks.
Glycogen production was tested as described for Fig. 1.
Catalase activity was tested by visual comparison of the bubbling activity of colonies in the presence of H2O2.
Number of isolates with the same sequence analyzed.
aa, amino acid.
To check whether the loss of rpoS was peculiar to carbon limitation, similar experiments were initiated with cultures with glucose excess and N limitation. As shown in Fig. 1B, the rate of loss of RpoS function was comparable to that in glucose-limited populations. Here again, loss was more rapidly selected at low growth rates. The nature of the limitation was therefore not the determining factor in rpoS mutation selection, and every chemostat established contained >90% of rpoS mutants within 5 days of cultivation.
To test if selection was dependent on the strain background, mutation of rpoS was also monitored in ZK126, another rpoS+ strain (genotype, W3110 ΔlacU169 tna-2) (1). The kinetics of loss at dilution rates of 0.1 and 0.3 h−1 were slower than in the MC4100 strain (results not shown). There are numerous genotypic differences between BW3952 and ZK126, so it remains to be investigated how genetic background influences rpoS inactivation kinetics.
Loss of RpoS and expression of nutrient-scavenging pathways.
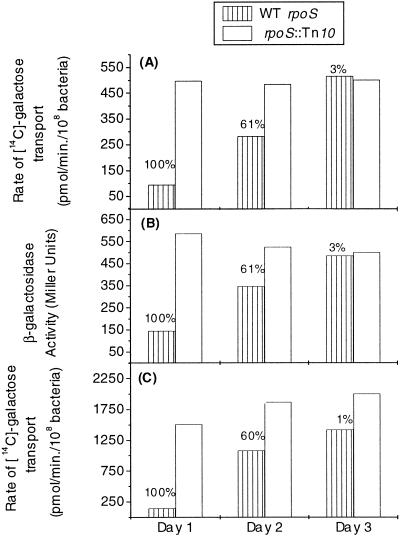
The results in Fig. 1 raised the question of why loss of RpoS should be strongly selected. The following experiments focused on glucose-limited cultures, in which the ability to scavenge glucose and the major determinants of fitness under chemostat conditions are well understood (7). The effect of rpoS status on glucose transporters was evaluated under glucose limitation. The high-affinity glucose uptake pathway involves expression of mglBAD genes as well as lamB in the separate mal regulon (7). In monitoring expression of these genes, wild-type and rpoS mutant strains were inoculated separately into chemostats, and expression of mgl and a transcriptional mal fusion were both assayed. As shown in Fig. 2, there was a strong difference in expression of both transport components (four- to fivefold) after 1 day of glucose limitation, with higher levels in the rpoS mutant. As the proportion of rpoS mutants in the wild-type population began to increase, as shown in Fig. 2, the expression of the mgl-mal systems converged to that found in the rpoS insertion mutant.
FIG. 2.
Effect of rpoS mutations on expression of the glucose hunger response. (A and B) Chemostat cultures operating at a dilution rate of 0.3 h−1 with 0.02% glucose in the feed medium were inoculated with either BW2952 (malG-lacZ) or BW3511 (malG-lacZ rpoS::Tn10) and monitored for the rate of uptake of [14C]galactose (A) and for β-galactosidase activity of the malG-lacZ fusion (B), as well as for the proportion of RpoS mutants (by iodine staining as described for Fig. 1). The culture of BW2952 showed decreasing proportions of wild-type rpoS colonies, with 100, 61, and 3% on days 1, 2, and 3, respectively. (C) Rate of uptake of [14C]galactose in cultures of a strain with an mgl(Con) mutation (mglD L126stop) (BW3245 rpoS+ and BW3522 rpoS::Tn5). Wild-type rpoS colonies dropped from 100% (day 1) to 60% (day 2) to <1% (day 3) in the BW3245 population. The pattern shown is representative of that for three or four replicates under each condition. WT, wild type.
To separate the possible effect of RpoS on transcriptional regulators (mglD and malT) from that on transcription of mal-mgl genes directly, the mgl assay was repeated with an mgl operator mutant fully constitutive for mgl expression. As shown in Fig. 2C, there was an even greater difference (sevenfold) in expression on day 1 as a result of RpoS, with the difference decreasing with increasing frequency of rpoS mutations in the population. A similar difference was seen with a malT(Con) mutant (result not shown). Hence, the rpoS effect was not on the inducibility of the hunger response genes but more likely on transcription directly.
A difference in mal expression between wild-type and rpoS mutant strains was not previously detected in an exponentially growing, glucose-excess batch culture (26). Likewise, we found differences of less than twofold in mgl expression between wild-type and rpoS mutant strains in glucose or glycerol batch culture (result not shown). Under these conditions of glucose excess, exponential-phase rpoS expression is quite low (11). Hence, there seems to be an inverse correlation between rpoS expression and the effect on mal-mgl expression. This trend extends to glucose-limited chemostats, where RpoS levels are much higher at lower growth rates (at D = 0.1 h−1) (27). These are precisely the conditions that provide the fastest loss of RpoS function (Fig. 1). As shown with the constitutive mgl mutant, the expression difference is elevated when transporter expression is high, as in Fig. 2C.
Loss of RpoS and effect on stress resistance.
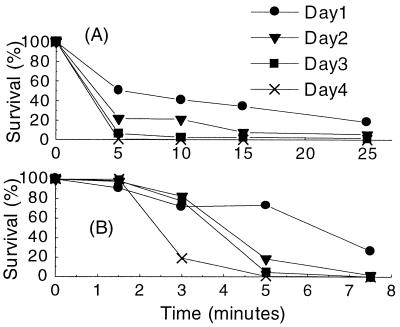
The mutations in rpoS affected the stress resistance of chemostat populations. In cultures assayed for susceptibility to elevated temperature and oxidative stress, as shown in Fig. 3, the general resistance properties of the cultures decreased day by day together with the proportion of bacteria carrying a wild-type rpoS gene. Hence, loss of RpoS caused by nutrient limitation makes these cultures more vulnerable to other environmental challenges.
FIG. 3.
Effect of rpoS mutations on stress resistance. Chemostat cultures of strain BW2952 growing at a dilution rate of 0.3 h−1 were sampled daily after inoculation and challenged for resistance to elevated temperature (A) and H2O2 (B). Viable bacteria were determined from plate counts. At the same time, on each day samples were tested for the proportion of glycogen-containing (rpoS+) cells as described for Fig. 1. Wild-type bacteria decreased from 100% (day 1) to 24% (day 2), 15% (day 3), and <1%(day 4). The pattern shown is representative of that for three replicate experiments under each condition.
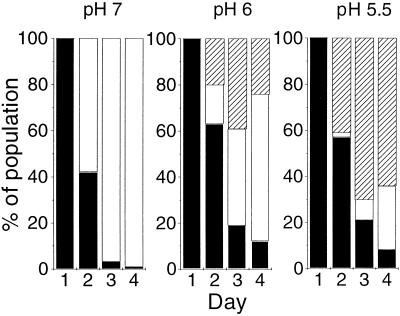
The selection conditions for Fig. 1 to 3 used a single stress, namely, nutrient limitation, and the environment was otherwise optimal for growth. To test whether an additional, secondary stress influenced the loss of RpoS, a series of experiments similar to those for Fig. 1Awere conducted in the presence of reduced pH. RpoS contributes to pH tolerance (2, 20), so the expectation was that mutations in rpoS may be less advantageous at low pH. With the chemostat medium adjusted to pH 6 or 5.5 instead of pH 7, but otherwise under the conditions used for Fig. 1A, the progression of changes in glycogen phenotype was determined. The altered environment affected both the rate of loss of rpoS and the proportion of partial mutants (Fig. 4). Partial mutants, a minority at pH 7, became dominant with acid stress. The partial mutants were confirmed to show intermediate resistance to general stresses (temperature or H2O2 stress) (results not shown). These findings suggest that E. coli can avoid extreme sensitivity to environmental challenge under nutrient limitation through these partial, attenuated mutants. Nevertheless, wild-type bacteria were displaced even at pH 5.5, so mutation of rpoS was still of selective advantage.
FIG. 4.
Effect of pH stress on rpoS mutations. Chemostat cultures of strain BW2952 growing at a dilution rate of 0.3 h−1 were grown in minimal medium whose pH was adjusted to pH 7, 6, and 5.5. At the same time each day, samples were tested for the proportion of glycogen-containing (rpoS+) cells as described for Fig. 1. Black bars represent wild-type bacteria, white bars represent null mutants, and hatched bars represent partial mutants. The pattern shown is representative of that for three replicate populations under each pH condition.
DISCUSSION
The rapid sweeps by rpoS mutants in nutrient-limited populations suggest a powerful selection pressure in chemostat culture. An implication of this finding is that most chemostat populations of E. coli used in fundamental studies or in industry are likely to be altered in RpoS status within days of inoculation. The sweep by rpoS mutants needs to be kept in mind when interpreting the results of earlier published chemostat experiments, including our own. Further studies are needed to test whether the same selection pressure exists for bacteria other than E. coli under nutrient limitation and why the kinetics of loss is affected by genetic background. As shown in Fig. 3, the loss of rpoS certainly has an impact on the general stress resistance of chemostat populations and may change other properties of continuously cultured organisms.
The simplest explanation for the lack of competitiveness of rpoS+ bacteria in chemostats is the reduced expression of transporter genes essential for nutrient scavenging. As shown in early studies (5), the selection in the chemostat environment is for a genotype that maintains the lowest equilibrium concentration of nutrient (10). Expression of mgl and mal genes is important in scavenging and is particularly sensitive to mutations increasing expression under glucose limitation (28, 29). The results in Fig. 2 suggest that an rpoS mutation will significantly enhance expression of these genes in chemostat cultures, and loss of rpoS during the culture of wild-type bacteria is associated with increased mal and mgl expression. Aside from LamB levels, outer membrane permeability is reduced in RpoS+ bacteria because the large-channel porin induced by the hunger response, OmpF (21), is also under negative control by RpoS (30). Hence, cell components involved in outer membrane permeability and nutrient scavenging are coordinately increased through rpoS mutation.
The effect of rpoS mutations on mal expression in glucose-excess batch cultures is not strong (less than twofold) (26), so mal genes were not previously considered to be rpoS regulated. However, we totally missed the difference in mal expression in chemostat culture in an earlier study (26). The error was that the “steady-state” samples for mal expression were taken after 3 days of continuous cultivation, by which time the original glucose-limited cultures must have been overrun by rpoS mutants (Fig. 1). This is a good illustration of the potential problems that RpoS status can cause in interpreting chemostat experiments.
Previous results (6, 22) strongly support the notion that competition between RpoS and other sigma factors offers a global explanation of why expression from non-RpoS promoters is reduced. The MalT activator and MglD repressor control the distinct and physically separate mal and mgl promoters, but both systems were influenced by loss of RpoS, with or without the induction process. This pleiotropic effect is consistent with the presence of competing RpoS having a global effect on ϕ70-dependent transcription. Similarly, under nitrogen limitation, additional competition with the ϕ54 regulating ammonia assimilation (23) may provide a comparable disadvantage, but further studies are needed to prove this point.
The results reported above throw new light on the central question of why rpoS mutations are present in many E. coli and Salmonella strains. An explanation can be based on the likelihood that many natural environments limit growth rates but that RpoS-mediated cross-resistance does not provide the means of overcoming the growth limitation. Indeed, in any situation where sigma factors other than RpoS are required for the regulatory response, there may well be a selective advantage in losing RpoS. Consistent with this notion, the GASP phenomenon is thought to be a reflection of the fact that higher growth rates in stationary phase require scavenging of micronutrients and are dependent on ϕ70 for expression. As noted by Finkel et al., increased scavenging of nutrients was a feature of GASP mutants (for amino acids in the case of GASP) (9), so the selection pressure for expression of scavenger systems is common to chemostats and stationary phase. Also, in some niches (19), cell components useful for survival, such as type 1 pili (4) may be useful but under negative control of RpoS, again leading to rpoS mutations.
The nature of the enriched rpoS mutations was not independent of the environment. Partial mutants were enriched in the presence of low-level acid stress (Fig. 4) and in stationary phase with the GASP phenotype (9). In the absence of stress, the chemostats were selecting drastic loss-of-function mutations. Likewise, in natural environments, the type of rpoS mutation selected may depend on whether there is a need for a partial stress response. The attenuation-type GASP mutation may already be an evolutionary adaptation to avoid complete loss of stress responses under conflicting selection conditions, as would occur in complex stationary-phase media.
Making the hunger response and the general stress response compete for core RNA polymerase appears to be a basic weakness in the regulatory circuits of E. coli and S. enterica. The strong selection pressure in bacteria dependent on ϕ70 or ϕ54-RNA polymerase for continued growth is bound to lead to rpoS loss or attenuation in many environments not requiring a stress response. Why did evolution commit bacteria to this competing form of regulation? A possible explanation is that ancient, free-living bacteria were rarely under purely hunger conditions as occur in chemostats or human-made environments and that RpoS was always essential in response to multiple stresses. Yet the frequency of rpoS mutations in natural populations argues that environmental situations where loss of RpoS is at least a short-term advantage do currently exist.
Acknowledgments
We thank Shona Seeto for help with some of the experiments.
We thank the Australian Research Council for grant support.
REFERENCES
- 1.Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel, M. M. Zambrano, and R. Kolter. 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of sigma 70. J. Bacteriol. 173:4482–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheville, A. M., K. W. Arnold, C. Buchrieser, C. M. Cheng, and C. W. Kaspar. 1996. RpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157–H7. Appl. Environ. Microbiol. 62:1822–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Death, A., L. Notley, and T. Ferenci. 1993. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J. Bacteriol. 175:1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dove, S. L., S. G. J. Smith, and C. J. Dorman. 1997. Control of Escherichia coli type 1 fimbrial gene expression in stationary phase—a negative role for RpoS. Mol. Gen. Genet. 254:13–20. [DOI] [PubMed] [Google Scholar]
- 5.Dykhuizen, D. E., and D. E. Hartl. 1983. Selection in chemostats. Microbiol. Rev. 47:150–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS—a case of sigma factor competition. Mol. Microbiol. 29:1039–1051. [DOI] [PubMed] [Google Scholar]
- 7.Ferenci, T. 1996. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol. Rev. 18:301–317. [DOI] [PubMed] [Google Scholar]
- 8.Ferenci, T. 1999. Regulation by nutrient limitation. Curr. Opin. Microbiol. 2:208–213. [DOI] [PubMed] [Google Scholar]
- 9.Finkel, S. E., E. R. Zinser, and R. Kolter. 2000. Long-term survival and evolution in the stationary phase, p.231–238. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 10.Hansen, S. R., and S. P. Hubbell. 1980. Single-nutrient microbial competition: qualitative agreement between experimental and theoretically forecast outcomes. Science 207:1491–1493. [DOI] [PubMed] [Google Scholar]
- 11.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161–178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 12.Hengge-Aronis, R., and D. Fischer. 1992. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol. Microbiol. 6:1877–1886. [DOI] [PubMed] [Google Scholar]
- 13.Herbelin, C. J., S. C. Chirillo, K. A. Melnick, and T. S. Whittam. 2000. Gene conservation and loss in the mutS-rpoS genomic region of pathogenic Escherichia coli. J. Bacteriol. 182:5381–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanova, A., M. Renshaw, R. V. Guntaka, and A. Eisenstark. 1992. DNA base sequence variability in katF (putative sigma factor) gene of Escherichia coli. Nucleic Acids Res. 20:5479–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jishage, M., and A. Ishihama. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jishage, M., and A. Ishihama. 1999. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 181:3768–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen, F., S. Leach, S. J. Wilde, A. Davies, G. Stewart, and T. Humphrey. 2000. Invasiveness in chickens, stress resistance and RpoS status of wild-type Salmonella enterica subsp enterica serovar Typhimurium definitive type 104 and serovar Enteritidis phage type 4 strains. Microbiology 146:3227–3235. [DOI] [PubMed] [Google Scholar]
- 18.Kaasen, I., P. Falkenberg, O. B. Styrvold, and A. R. Strom. 1992. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli: evidence that transcription is activated by katF (AppR). J. Bacteriol. 174:889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogfelt, K. A., M. Hjulgaard, K. Sorensen, P. S. Cohen, and M. Givskov. 2000. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect. Immun. 68:2518–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, I. S., J. S. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma factor sigma(s) (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155–167. [DOI] [PubMed] [Google Scholar]
- 21.Liu, X. Q., and T. Ferenci. 1998. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J. Bacteriol. 180:3917–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magasanik, B. 1996. Regulation of nitrogen utilization, p.1344–1356. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 24.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 173:4188–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Notley, L., and T. Ferenci. 1995. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient stressed Escherichia coli. Mol. Microbiol. 160:121–129. [DOI] [PubMed] [Google Scholar]
- 27.Notley, L., and T. Ferenci. 1996. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J. Bacteriol. 178:1465–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notley-McRobb, L., and T. Ferenci. 1999. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ. Microbiol. 1:33–43. [DOI] [PubMed] [Google Scholar]
- 29.Notley-McRobb, L., and T. Ferenci. 1999. The generation of multiple coexisting mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1:45–52. [DOI] [PubMed] [Google Scholar]
- 30.Pratt, L. A., W. H. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to OsmZ—multiple factors influence the synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911–917. [DOI] [PubMed] [Google Scholar]
- 31.Sutton, A., R. Buencamino, and A. Eisenstark. 2000. rpoS mutants in archival cultures of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:4375–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterman, S. R., and P. L. Small. 1996. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect. Immun. 64:2808–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei, B. D., S. Shin, D. LaPorte, A. J. Wolfe, and T. Romeo. 2000. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J. Bacteriol. 182:1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757–1760. [DOI] [PubMed] [Google Scholar]
- 35.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: Scavenging as a defense against nitrogen limitation. Proc. Nat. Acad. Sci. USA 97:14674–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]