Abstract
Flagellar assembly requires the expression of a large number of flagellum-specific genes. However, mutations in a number of other genes in Salmonella and Escherichia coli have been shown to have pleiotropic effects that affect flagellar assembly. FlgH (the L-ring subunit of the flagellar basal body) is a lipoprotein whose modification is important for L-ring assembly. We therefore tested whether the lack of motility of Salmonella mutants defective in lipoprotein biogenesis is a result of inability to modify FlgH. Our results show that temperature-sensitive apolipoprotein N-acyltransferase [lnt(Ts)] mutants are nonflagellate at 42°C. However, the flagellar assembly defect occurs at a much earlier step in the pathway than L-ring assembly. These mutants failed to assemble even an MS ring, presumably because of the observed decrease in transcription of fliF. In contrast, temperature-sensitive diacylglycerol transferase [lgt(Ts)] mutants were motile at 42°C, provided the strains carried an lpp (Braun lipoprotein) mutation to permit growth. We have isolated second-site mutants from an lgt(Ts) lpp+ strain that grow but are nonflagellate at 42°C. Thus, lipoprotein biogenesis is a factor that is important for flagellar assembly.
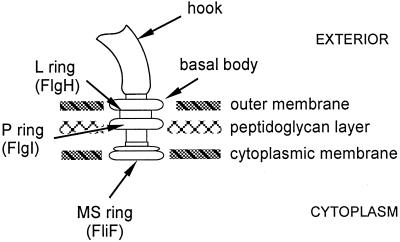
The morphological pathway of flagellar assembly in several gram-negative bacteria such as Salmonella and Escherichia coli is fairly well understood (1, 6, 28). Many of the genes encoding products for flagellar assembly have been identified, including more than 40 specific flagellar genes. A number of so-called housekeeping genes have been shown to be required for flagellar assembly also. These include genes which encode proteins such as H-NS (4, 15) and heat shock proteins (34), whose function seems to be required for transcription of the master operon genes flhD and flhC. Expression of the flhDC operon is also regulated by cyclic AMP (21, 35). The inability to express the positive regulators encoded by flhC and flhD (26) results in an inability to transcribe the other flagellum-specific genes such as fliF, which encodes the subunits of the MS ring. This ring, which is located in the cytoplasmic membrane (Fig. 1), is the first flagellar structure to be assembled in the morphogenetic pathway. However, some non-flagellum-specific gene products have been shown to be required for flagellar synthesis at a posttranscriptional level. For example, mutations in dsbA or dsbB affect flagellar assembly in E. coli at the level of the assembly of the periplasmic or P ring (Fig. 1). This apparently is the result of an inability to put a disulfide bond in the P-ring subunit, FlgI (9).
FIG. 1.
Cartoon of the flagellar basal body. The outer-membrane L-ring protein, FlgH, is lipoylated, while the periplasmic P-ring protein, FlgI, undergoes intramolecular disulfide bridge formation.
FlgH encodes the subunit of the outer membrane L ring (Fig. 1) and undergoes signal peptide cleavage (17). The FlgH sequence contains a signal peptidase II consensus cleavage motif or lipobox, LTG↓C, near its N terminus, suggesting that it is a lipoprotein (19), and this has been verified experimentally (32). Furthermore, mutations affecting the lipobox not only affect the ability of FlgH to be modified by the lipoprotein biogenic enzymes but also disrupt L-ring assembly. This led us to consider whether, conversely, mutations affecting lipoprotein biogenesis might cause an L-ring assembly defect.
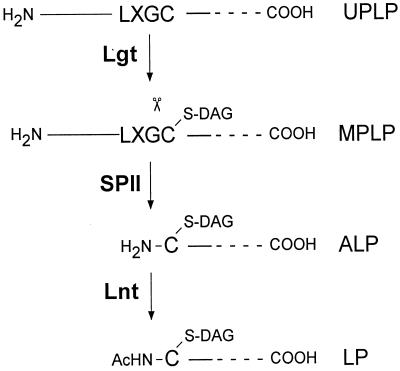
The pathway of lipoprotein biogenesis can be summarized as follows (Fig. 2) (for a review, see reference 14). Lipoproteins are synthesized with a signal sequence containing a lipobox motif (L-X-G/A-↓C) surrounding the signal peptidase II cleavage site. The unmodified prolipoprotein is first modified on the sulfhydryl group of the cysteine by the cytoplasmic membrane-associated enzyme prolipoprotein diacylglyceryl transferase (Lgt) to form modified prolipoprotein (MPLP). MPLP is cleaved by signal peptidase II (SPII, prolipoprotein signal peptidase) to generate apolipoprotein, which is then acylated on the amino group of the N-terminal cysteine by apolipoprotein N-acyltransferase (Lnt) to generate the mature lipoprotein. Mutations in genes coding for Lgt (12) and Lnt (13) have been described. Analysis of temperature-sensitive mutants of lgt and lnt (also referred to for E. coli as umpA and cutE, respectively [12, 31]) showed that both gene products are essential for growth of Salmonella. However, strains of Salmonella carrying lgt(Ts) or lnt(Ts) alleles can grow at 42°C if they also carry a mutation in the lpp gene, which encodes the major Braun lipoprotein (12, 13); this presumably is a consequence of removing the major enzymatic substrate load, namely, lipoylation of Braun lipoprotein. Thus, by using these double mutants, the effect of decreased activity of the lipoprotein biogenic enzymes on flagellar assembly could be studied. Our results show that although the lgt(Ts) allele studied in this work did not greatly affect flagellar assembly when analyzed in an lpp strain (and thus, the strain was motile, even at the restrictive temperature), the lnt(Ts) allele caused a flagellar assembly defect at the restrictive temperature (and thus, the strain was nonmotile). However, analysis of the assembly defect revealed that the lnt(Ts) strain was nonmotile because of an inability to assemble even an MS ring. This defect was likely the result of an inability to transcribe the gene for the subunit of the MS ring, fliF. Our results showed that the lnt mutation caused a decrease in expression of flhD, one of the positive activators for fliF (and other flagellar gene) expression.
FIG. 2.
Schematic representation of the pathway leading to formation of a lipoprotein (see the text). LXGC (or LXAC) is the sequence known as a lipobox. UPLP, unmodified prolipoprotein; MPLP, modified prolipoprotein; ALP, apolipoprotein; LP, mature lipoprotein; Lgt, prolipoprotein diacylglyceryltransferase; SPII, lipoprotein-specific signal peptidase; Lnt, apolipoprotein N-acyl transferase; scissors, potential site of cleavage by SPII; -S-DAG, diacylglycerol in thioether linkage to cysteine; AcHN-, N-acylation of terminal amino group.
Despite the fact that lgt(Ts) lpp strains were motile at 42°C, we were able to isolate second-site revertants of lgt(Ts) lpp+ strains that could grow at 42°C but were nonmotile. Complementation tests showed that the motility defect was due to the lgt(Ts) allele. Thus, both the synthesis of Lgt and that of Lnt affect flagellar assembly.
MATERIALS AND METHODS
Bacterial strains and strain construction.
All bacterial strains used in this study are derivatives of Salmonella enterica serovar Typhimurium LT2 (Table 1). P22 transductions were done as described earlier (10) by using P22HT105 int 1 phage. Tets strains were isolated from Tetr strains using Bochner plates as described previously (5). Plasmids were isolated using Wizard mini-prep kits (Promega), and transformation of recipients was achieved by the heat shock method described in reference 10. TOP1O (Invitrogen) and DH5α (BBL Microbiology Systems) were used as hosts for cloning. Second-site revertants of lgt(Ts) strains were isolated by plating approximately 108 cells on Luria-Bertani (LB) plates at 42°C and saving isolates that grew for further study.
TABLE 1.
Bacterial strains used in this studya
| Strain (original designation) | Relevant genotypeb | Source, reference, or construction |
|---|---|---|
| SJW1103 | Wild type | 39 |
| SE5221 | lgt(Ts) | 12 |
| SE5312 | lnt(Ts) | 13 |
| FDY3 (SE5221 lpp::Tn10) | lgt(Ts) lpp::Tn10 | 12 |
| FDY110 | lgt(Ts) lpp::Tn10 | P22(FDY3 × SE5221)c; this work |
| FDY7 (SE5312 zbf::Tn10) | lnt(Ts) zbf::Tn10 | 13 |
| FDY8 (SE5312 lpp::Tn10) | lnt(Ts) lpp::Tn10 | 13 |
| FDY40 | lnt(Ts) lpp | Tets of FDY8; this workd |
| FDY43 | lnt+zbf::Tn10 | P22(FDY7 × SJW1103); this work |
| FDY46 | lnt+lpp::Tn10 | P22(FDY43 × FDY40)c; this work |
| FDY50 | lnt+lpp | Tets of FDY46; this work |
| FDY113 | lgt(Ts) lpp | Tets of FDY110; this work |
| FDY115 | lgt(Ts) lnt(Ts) lpp zbf::Tn10 | P22(FDY7 × FDY113); this work |
| MH111 | fliC::Tn10 | M. Homma |
| FDY82 | lnt(Ts) lpp fliC::Tn10 | P22(MH111 × FDY40); this work |
| SJW1684 | fliF | S. Yamaguchi |
| KK1113 | fliF-lacZ | 25 |
| FDY105 | lnt(Ts) lpp fliF-lacZ | P22(KK1113 × FDY40); this work |
| FDY106 | lnt+lpp fliF-lacZ | P22(KK1113 × FDY50); this work |
| KK1107 | flhD-lacZ | 25 |
| FDY107 | lnt(Ts) lpp flhD-lacZ | P22(KK1107 × FDY40); this work |
| FDY108 | lnt+lpp flhD-lacZ | P22(KK1107 × FDY50); this work |
| SJW1469 | flgH | S. Yamaguchi |
| FDY126 | lgt(Ts) lpr-1 | Revertant of SE5221; this work |
| FDY127 | lgt(Ts) lpr-2 | Revertant of SE5221; this work |
| FDY131 | lpp::Tn10 of FDY126 | This work |
| FDY147 | lpp::Tn10 of FDY127 | This work |
| FDY9 | pLnt (lnt+)e | Tetr of SE5312 (13) |
| FDY4 | pSK004dp (lgt+)f | Ampr of SE5221 (12) |
| FDY135g | pFDY135 (lnt+)e | Ampr of DH5α; this work |
| FDY140g | pFDY140(lgt+)f | Ampr of DH5α; this work |
| FDY137 | pFDY135/lnt(Ts) | Ampr of SE5312; this work |
| FDY138 | pFDY135/lnt(Ts) lpp | Ampr of FDY8; this work |
| FDY144 | pFDY140/lgt(Ts) | Ampr of SE5221; this work |
| FDY145 | pFDY140/lgt(Ts) lpr-1 | Ampr of FDY126; this work |
| FDY146 | pFDY140/lgt(Ts) lpr-2 | Ampr of FDY127; this work |
| TOP1Og | Δ(mrr-hsdRMS-mcrBC) | Invitrogen |
| DH5αg | BBL |
All strains used in this study are derived from S. enterica serovar Typhimurium LT2, except where indicated otherwise.
Genotype is chromosomal, except when preceded by a plasmid name, in which case it is the plasmid-encoded genotype.
P22 transductions were done as described in reference 10 using P22HT105 int 1 phage.
Tets strains were selected on Bochner plates as described in reference 5.
pLnt is based on the medium-copy-number plasmid pACYC184 (Tetr). A 2.3-kb EcoRI fragment carrying the wild-type lnt allele from pLnt was subcloned into the low-copy-number plasmid pWSK29 (Ampr) (38) to give pFDY135.
pSK004dp is based on the high-copy-number plasmid pBluescript SK(+) (12). A 1.4-kb SalI-Xba I fragment containing the wild-type lgt allele was subcloned into the low-copy-number plasmid pWSK29 (Ampr) (38) to give pFDY140.
E. coli strains.
Hook-basal body preparations and electron microscopy.
Hook-basal body complexes were prepared for electron microscopy by the methods described in reference 11 on cells grown to an optical density at 600 nm (OD600) of 1.0 in LB at either 30 or 42°C. The samples were negatively stained with 2% (wt/vol) uranyl acetate, and the images were recorded on a Phillips model 301 or 420 electron microscope (80 kV; magnification, × 4,500).
Electrophoresis and immunoblotting.
Strains FDY40 and FDY50 were grown on LB at 30 or 42°C to an OD600 of 1.0 and then boiled for 10 min in protein sample buffer containing 1% sodium dodecyl sulfate before loading onto sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis gels. Proteins were transferred to polyvinylidene difluoride membranes (Millipore) with a Hoefer Transblot apparatus. Membranes were probed with a monoclonal antibody to FliF (16) diluted 1:10,000 and detected using an ECL Immunoblotting kit (Amersham International) (17).
β-Galactosidase assays.
Strains for β-galactosidase assays were grown to an OD600 of 0.5 in LB at either 30 or 42°C. Assays were performed by the method of Miller (29).
Characterization of motility and flagellation.
LB and tryptone broth (TB) swarm plates (0.35% agar) were used to characterize the chemotactic and motility phenotype of the strains. Strains were also grown in either LB or TB for visual examination of motility and visualization of flagella by high-intensity dark-field microscopy (27).
Cloning of lnt and lgt into low-copy-number plasmids.
lnt was subcloned into the low-copy-number plasmid pWSK29 (38) by cutting pLnt (a medium-copy-number pACYC184-based plasmid containing the wild-type lnt gene [13]) with EcoRI (New England Biolabs [NEB]), isolating the 2.9-kb fragment from a 0.8% agarose gel using a gel extraction kit (Qiagen) and ligating (T4 DNA ligase; NEB) into pWSK29. lgt was subcloned into pWSK29 by double digesting pSK004dp (a high-copy pBluescript-based plasmid containing the wild-type lgt gene (12) with SalI and XbaI (NEB), isolating the 1.4-kb fragment containing lgt, and ligating into pWSK29.
Chemicals.
All reagents used were of at least reagent grade.
RESULTS
Requirement for Lnt activity for flagellar assembly.
Strains of Salmonella carrying temperature-sensitive alleles for lgt or lnt were tested for their motility and chemotaxis phenotype at 30°C on swarm agar plates. Cultures were also grown in LB and examined by light microscopy. Strain SE5221 [lgt(Ts)] made smaller swarms than its lgt+ parent at 30°C; however, since liquid-grown cultures were very motile, the decrease in swarming is probably a consequence of the growth rate of the mutant being approximately 20% slower than that of the wild type at this temperature. Strain SE5312 [lnt(Ts)] was almost as motile and chemotactic as its lnt+ parent when tested at 30°C.
We next tested the effects of these mutations on motility at 42°C, at which temperature the enzymatic activities of Lgt and Lnt are severely reduced (12, 13). To do so, we used strains containing an additional mutation in the lpp (Braun lipoprotein) gene; this mutation has previously been shown to alleviate the growth defect of these strains at 42°C (12, 13). Strain FDY3 [lgt(Ts) lpp::Tn10] was motile and chemotactic at both 30 and 42°C. However, we discovered that this strain was also P22 resistant. Since it was possible that the motility phenotype was affected by the mutation conferring P22 resistance, we constructed strain FDY110 [lgt(Ts) lpp::Tn10], which is P22 sensitive. Strain FDY110, like FDY3, was motile at 42°C. In contrast to these strains, strain FDY8 [lnt(Ts) lpp::Tn10] showed near wild-type swarming behavior at 30°C but appeared nonmotile on swarm plates at 42°C. High-intensity dark-field-microscopic examination of strain FDY8 revealed that the strain was nonflagellate when grown at 42°C in LB.
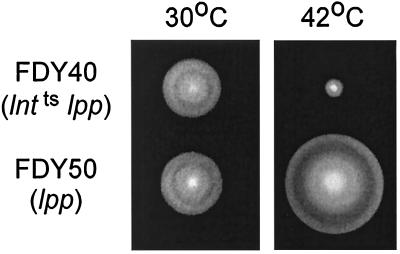
Since these temperature-sensitive mutants had been obtained by chemical mutagenesis (12, 13), it was possible that the parental strain SE5312 [lnt(Ts)] carried another mutation that affected flagellar assembly at 42°C. To establish whether the Lnt deficiency was the cause of the flagellar defect, we first constructed isogenic strains FDY40 [lnt(Ts) lpp] and FDY50 (lpp) by phage P22 transduction (Table 1). Results of tests on the motility phenotype of strains FDY40 and FDY50 on LB swarm agar at 30 and 42°C (Fig. 3) demonstrated that it was the Lnt deficiency, and not some unknown defect, that caused the motility defect at 42°C. As expected, dark-field light microscopy revealed an absence of flagella on FDY40 cells grown at this temperature. To further confirm that the motility defect of strain FDY40 was the result of the lnt(Ts) mutation, we attempted to complement the motility defect. We failed to do so with pLnt (a medium-copy-number pACYC184-based plasmid containing the wild-type lnt gene [13]) but succeeded with a lower-copy-number pWSK29-based (38) plasmid, pFDY135. This plasmid was also able to complement the growth defect of strain SE5312 [lnt(Ts)] at 42°C. Microscopic examination of strain FDY40 containing pLnt revealed a tendency for the cells to form chains. Possibly the higher-than-wild-type level of Lnt was the cause of the cell division defect and the inability to restore motility (13). We also constructed a triple mutant, strain FDY115 [lgt(Ts) lnt(Ts) lpp]. Comparison of this strain and FDY113 [lgt(Ts) lpp] showed that the introduction of the lnt(Ts) mutation to the lgt(Ts) strain caused a motility defect at 42°C, but not at 30°C. Although FDY115 cells grew well on TB agar at 42°C, they grew very poorly in liquid and tended to lyse. Cultures grown in liquid at 42°C were also nonmotile, as determined by light microscopy.
FIG. 3.
Swarming of FDY40 [lnt(Ts) lpp] and FDY50 (lpp) on soft agar plates after 11 h at 30 and 42°C.
Apolipoprotein N-acyltransferase-deficient strains fail to assemble an MS ring at 42°C.
Strains FDY40 [lnt(Ts) lpp], FDY50 (lnt+ lpp), and FDY82 (a fliC [flagellin−] derivative of FDY40) were grown in LB medium at 30 and 42°C, and hook-basal body complexes were prepared as described earlier (2, 11). Electron microscopy revealed wild-type structures present in the lnt+ strain FDY50 grown at either temperature and in the lnt(Ts) strain FDY40 or the fliC transductant FDY82 when grown at 30°C. Although samples prepared from the lnt(Ts) strains grown at 42°C revealed the presence of some wild-type structures and a few basal-body structures lacking the P and/or the L ring, the number of complete structures was at least 10-fold less than with the lnt+ strain. Thus, it appears that lnt(Ts) strains grown at 42°C are nonflagellate because of a very early flagellar assembly defect, possibly at the stage of MS-ring assembly.
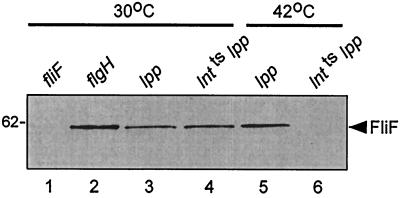
When we tested for the presence of the MS-ring protein FliF by immunoblotting using monoclonal anti-FliF antibody (16), we were unable to detect it in strain FDY40 grown at 42°C but detected it at wild-type levels when the strain was grown at 30°C (Fig. 4). Thus, an Lnt deficiency at the restrictive temperature causes a severe reduction in the amount of FliF present in the cells. Since an L-ring mutant strain SJW1469 (flgH) had wild-type levels of FliF, the decrease in FliF levels in strain FDY40 at 42°C cannot be simply the result of an inability to make the mature L ring.
FIG. 4.
Immunoblots, using monoclonal anti-FliF antibody, of strains SJW1684 (fliF), SJW1469 (flgH), FDY50 (lpp), and FDY40 [lnt(Ts) lpp], all grown at 30°C, and of strains FDY50 (lpp) and FDY40 [lnt(Ts) lpp] grown at 42°C.
fliF transcription is decreased at 42°C in an lnt(Ts) strain.
To see if fliF was transcribed at 42°C, we introduced a fliF-lacZ transcriptional fusion into strains FDY40 and FDY50 by phage P22-mediated transduction. β-Galactosidase assays (Table 2) of strains FDY105 and FDY106 showed that the lnt(Ts) mutation in an lpp strain causes about a six- to sevenfold decrease in lacZ expression at 42°C compared to that found in the isogenic lnt+ strain. This effect, though considerable, is not as extreme as the virtually total loss of FliF protein seen with the lnt(Ts) lpp strain at 42°C (Fig. 4); we are not sure of the reason for this quantitative discrepancy. We also found about a 10-fold decrease in expression of fliC (data not shown). Decreased expression of fliF in the lnt(Ts) strain at 42°C is therefore presumably the cause of the MS-ring assembly defect. However, it is possible that other factors contribute. Several groups have reported that mutations in pgsA (18, 20, 30, 37) and in psd and pss (33) affect flagellar assembly in E. coli. pgsA mutants are defective in phosphatidylglycerol synthesis, and psd or pss mutants are defective in phosphatidylethanolamine synthesis and cause a decrease in expression of flhD. These mutations affect membrane composition, and it has been proposed that this is the cause of the decrease in expression of flhDC. Since mutations in lipoprotein biogenesis also affect membrane composition, we looked at flhD expression by measuring β-galactosidase activity in strains carrying flhD-lacZ transcriptional fusions. Results (Table 2) showed that flhD expression at 42°C in the lnt(Ts) strain FDY107 was decreased compared to that seen with the isogenic lnt+ strain FDY108, but the difference was only about twofold, which may not be very significant since the growth rate of the lnt strains was about twofold slower at 42°C than that of the isogenic lnt+ strain. Hence, the reason for the greater decrease in expression of fliF is not clear.
TABLE 2.
Effects of the lnt(Ts) mutation on flagellar gene expression
| Strain | Relevant genotype | β-Galactosidase activity (Miller units)a
|
|
|---|---|---|---|
| 30°C | 42°C | ||
| FDY105 | lnt(Ts) lpp fliF-lacZ | 384 ± 3 | 131 ± 10 |
| FDY106 | lpp fliF-lacZ | 459 ± 7 | 983 ± 75 |
| FDY107 | lnt(Ts) lpp flhD-lacZ | 462 ± 1 | 288 ± 15 |
| FDY108 | lpp flhD-lacZ | 511 ± 41 | 615 ± 18 |
Assays were done as in reference 29. Means and standard deviations are for either duplicate or triplicate measurements. Cells were grown in LB until the OD600 reached 0.5 at the temperature indicated.
Isolation of revertants of lgt(Ts) strains which have a motility defect at 42°C.
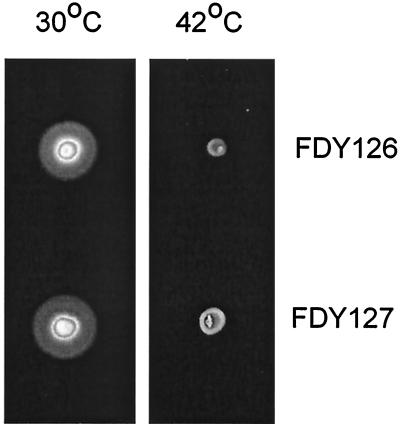
We wondered whether the reason why the lgt(Ts) lpp strains were motile might be that there was enough residual activity of Lgt at 42°C to permit flagellar assembly. We therefore sought to bypass the requirement for the lpp mutation for growth at 42°C by isolating pseudorevertants of SE5221 [lgt(Ts)] that were able to grow at 42°C. Twenty-two of these were tested for motility phenotype on TB motility plates at 30 and 42°C. Thirteen swarmed at both temperatures, but nine swarmed at 30°C but not at 42°C, even though they grew well at that temperature. We saved two of these nonmotile isolates, FDY126 and 127, for further study (Fig. 5). Light microscopy established that they were immotile when grown at 42°C, and high-intensity dark-field light microscopy showed that they were nonflagellate. We designated their genotype lgt(Ts) lpr (for lipoprotein requirement).
FIG. 5.
Swarming of FDY126 and FDY127 [both lgt(Ts) lpp+ lpr)] on soft agar plates after 8 h at 30 and 42°C.
Characterization of lgt(Ts) lpr strains.
The low-copy-number plasmid pFDY140 carrying lgt was able to complement the motility defects of the lgt(Ts) lpr strains FDY126 and FDY127 when grown in LB at 42°C. pFDY140 also complemented the growth defect of SE5221 [lgt(Ts)] at 42°C. (As was seen in the case of lnt, a high-copy-number plasmid [pSK004dp] carrying lgt was unable to complement the motility defect on LB swarm agar plates and the cells tended to form chains.) We transduced FDY126 and FDY127 to lpp to determine if the lpp mutation would restore motility at 42°C as it does to the lgt(Ts) parent. These lgt(Ts) lpr lpp transductants, such as FDY131 and FDY147, were poorly motile at 42°C. Thus, the two lgt(Ts) lpr strains studied are nonflagellate at 42°C because of the lgt(Ts) allele present and not because of the state of the lpp gene.
DISCUSSION
This study was designed to establish whether defects in lipoprotein biogenic enzymes would cause failure of the L-ring protein FlgH to function. Ironically, it was not possible to answer this question with respect to N-acyl lipoylation, because defects in this process resulted in a general failure of flagellar gene expression. A temperature-sensitive deficiency in Lnt activity results in a nonflagellate phenotype, as judged by both light and electron microscopy, and immunoblotting showed that the levels of the MS-ring subunit FliF were severely reduced (Fig. 4). The effect is predominantly at the transcriptional level: Transcription of fliF was reduced when the cells were grown at 42°C (Table 2), although the effect was not as pronounced as with the immunoblots. We found a slight decrease in expression of flhD, a positive activator of fliF expression (21). However, it is not clear that this is the sole or even the principal cause of the reduced transcription of fliF. Other than the gene products encoded by the flhDC operon, there are no other known specific regulators of fliF expression. Furthermore MS-ring assembly is not believed to require any other flagellar gene products (22, 24). Specifically, the phenotype of flgH mutants is that they produce a basal body structure consisting of the MS ring, rod, and P ring but, of course, lacking the L ring (36)
At the other extreme, introduction of lnt on medium-copy-number plasmids (but not low-copy-number ones) into strains FDY8 or FDY40 caused the cells to form chains, particularly at 42°C, and the cells remained nonmotile at this temperature. This suggests that too much Lnt activity in the cells interferes with cell division. Motile revertants of such strains often contained mutations in the lnt gene on the plasmid (our unpublished results). It is possible that this selection results in lnt mutations that reduce the activity of the enzyme and thus compensate for the higher-than-wild-type levels of expression.
We do not know why the Lgt deficiency in lpp strains FDY3 or FDY 110 does not cause a nonflagellate phenotype at the restrictive temperature. One possibility would be that enough Lgt activity is present at 42°C to allow expression and assembly of the flagellar apparatus. Gan et al. reported that their lgt(Ts) strain, which is the one we used in the present study, has a very low activity when grown at 42°C (12); nonetheless, as these authors suggest, there may be minor lipoproteins that are essential for viability, in which case some Lgt activity must remain at the restrictive temperature.
Analysis of second-site mutants of lgt(Ts) strains showed that, as the result of a defect in flagellar assembly in lpr strains, lgt activity is now required for motility. Strains FDY126 and FDY127 grow well at 42°C but are nonflagellate (Fig. 5). Introduction of wild-type lgt on a low-copy-number plasmid restored motility. It seems unlikely that the lpr mutation is an allele of lpp, since the motility of lgt(Ts) lpr lpp strains at 42°C was poor whereas the motility of lgt(Ts) lpp strains was good. We are now further analyzing the lpr mutations in order to understand why they allow lgt(Ts) strains to grow at 42°C yet do not allow motility.
The genetics of disulfide bond formation in eubacteria has been aided by the discovery that certain dsb mutations cause a nonmotile phenotype (3, 9). The discovery that certain mutations causing lipoprotein biogenic defects can also cause a nonmotile phenotype gives another powerful genetic tool that can aid in the analysis of these pathways.
Recent experiments (7, 8) have shown that InvH, a lipoprotein that may be homologous to FlgH, is required for assembly of type III secretion systems in Salmonella. Since type III secretion structures for virulence factors are homologous to flagellar basal structures (23), it is possible that the lipoprotein biogenesis mutants studied here may also show type III virulence factor secretion or assembly defects.
Acknowledgments
We dedicate this paper to the memory of Henry C. Wu, who made this work possible with his gift of strains. His contribution to the field of lipoprotein research is gratefully acknowledged. We also thank May Kihara and Gary Schoenhals for advice and help, Stanley Maloy for the gift of P22 phage, and Kazuhiro Kutsukake, Kelley Hughes, Sidney Kushner, and Shigeru Yamaguchi for gifts of strains. We thank Noreen Francis for her excellent help in performing the electron microscopy.
This work was supported by USPHS grant AI12202 to R.M.M., a Faculty Development Grant from the Dean of Faculty, College of Wooster, to F.E.D., and USPHS grant GM35433 to David J. DeRosier, whom we thank for access to the electron microscopy facility at Brandeis University.
REFERENCES
- 1.Aizawa, S.-I. 1996. Flagellar assembly in Salmonella typhimurium. Mol. Microbiol. 19:1–5. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, S.-I., G. E. Dean, C. J. Jones, R. M. Macnab, and S. Yamaguchi. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:836–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, J. C. A. 1994. Building bridges: disulphide bond formation in the cell. Mol. Microbiol. 14:199–205. [DOI] [PubMed] [Google Scholar]
- 4.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner, B. R., H.-C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crago, A. M., and V. Koronakis. 1998. InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30:47–56. [DOI] [PubMed] [Google Scholar]
- 8.Daefler, S., and M. Russel. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28:1367–1380. [DOI] [PubMed] [Google Scholar]
- 9.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270. [DOI] [PubMed] [Google Scholar]
- 12.Gan, K., S. D. Gupta, K. Sankaran, M. B. Schmid, and H. C. Wu. 1993. Isolation and characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in prolipoprotein modification. J. Biol. Chem. 268:16544–16550. [PubMed] [Google Scholar]
- 13.Gupta, S. D., K. Gan, M. B. Schmid, and H. C. Wu. 1993. Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J. Biol. Chem. 268:16551–16556. [PubMed] [Google Scholar]
- 14.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22:451–471. [DOI] [PubMed] [Google Scholar]
- 15.Hinton, J. C., D. S. Santos, A. Seirafi, C. S. Hulton, G. D. Pavitt, and C. F. Higgins. 1992. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol. Microbiol. 6:2327–2337. [DOI] [PubMed] [Google Scholar]
- 16.Homma, M., S.-I. Aizawa, G. E. Dean, and R. M. Macnab. 1987. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 84:7483–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homma, M., K. Ohnishi, T. Iino, and R. M. Macnab. 1987. Identification of flagellar hook and basal body gene products (FlaFV, FlaFVI, FlaFVII, and FlaFVIII) in Salmonella typhimurium. J. Bacteriol. 169:3617–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue, K., H. Matsuzaki, K. Matsumoto, and I. Shibuya. 1997. Unbalanced membrane phospholipid compositions affect transcriptional expression of certain regulatory genes in Escherichia coli. J. Bacteriol. 179:2872–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, C. J., M. Homma, and R. M. Macnab. 1989. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J. Bacteriol. 171:3890–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura, E., Y. Nakayama, H. Matsuzaki, K. Matsumoto, and I. Shibuya. 1994. Acidic-phospholipid deficiency represses the flagellar master operon through a novel regulatory region in Escherichia coli. Biosci. Biotechnol. Biochem. 58:2305–2307. [DOI] [PubMed] [Google Scholar]
- 21.Komeda, Y., H. Suzuki, J.-I. Ishidsu, and T. Iino. 1975. The role of cAMP in flagellation of Salmonella typhimurium. Mol. Gen. Genet. 142:289–298. [DOI] [PubMed] [Google Scholar]
- 22.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S.-I. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433–446. [DOI] [PubMed] [Google Scholar]
- 23.Kubori, T., A. Sukhan, S.-I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 97:10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubori, T., S. Yamaguchi, and S.-I. Aizawa. 1997. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J. Bacteriol. 179:813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutsukake, K., and T. Iino. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 176:3598–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macnab, R. M. 1976. Examination of bacterial flagellation by dark-field microscopy. J. Clin. Microbiol. 4:258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macnab, R. M. 1996. Flagella and motility, p.123–145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 29.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 30.Mizushima, T., R. Koyanagi, E. Suzuki, A. Tomura, K. Kutsukake, T. Miki, and K. Sekimizu. 1995. Control by phosphatidylglycerol of expression of the flhD gene in Escherichia coli. Biochim. Biophys. Acta 1245:397–401. [DOI] [PubMed] [Google Scholar]
- 31.Rogers, S. D., M. R. Bhave, J. F. Mercer, J. Camakaris, and B. T. Lee. 1992. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J. Bacteriol. 173:6742–6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoenhals, G. J., and R. M. Macnab. 1996. Physiological and biochemical analyses of FlgH, a lipoprotein forming the outer membrane L ring of the flagellar basal body of Salmonella typhimurium. J. Bacteriol. 178:4200–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, W., M. Bogdanov, W. Dowhan, and D. R. Zusman. 1993. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J. Bacteriol. 175:7711–7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi, W., Y. Zhou, J. Wild, J. Adler, and C. A. Gross. 1992. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J. Bacteriol. 174:6256–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman, M., and M. Simon. 1974. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J. Bacteriol. 120:1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, T., T. Iino, T. Horiguchi, and S. Yamaguchi. 1978. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J. Bacteriol. 133:904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomura, A., T. Ishikawa, Y. Sagara, T. Miki, and K. Sekimizu. 1993. Requirement of phosphatidylglycerol for flagellation of Escherichia coli. FEBS Lett. 329:287–290. [DOI] [PubMed] [Google Scholar]
- 38.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. [PubMed] [Google Scholar]
- 39.Yamaguchi, S., H. Fujita, K. Sugata, T. Taira, and T. Iino. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130:255–265. [DOI] [PubMed] [Google Scholar]