Abstract
Escherichia coli nucleoid-associated H-NS protein interacts with the Hha protein, a member of a new family of global modulators that also includes the YmoA protein from Yersinia enterocolitica. This interaction has been found to be involved in the regulation of the expression of the toxin α-hemolysin. In this study, we further characterize the interaction between H-NS and Hha. We show that the presence of DNA in preparations of copurified His-Hha and H-NS is not directly implicated in the interaction between the proteins. The precise molecular mass of the H-NS protein retained by Hha, obtained by mass spectrometry analysis, does not show any posttranslational modification other than removal of the N-terminal Met residue. We constructed an H-NS-His recombinant protein and found that, as expected, it interacts with Hha. We used a Ni2+-nitrilotriacetic acid agarose method for affinity chromatography copurification of proteins to identify the H-NS protein of Y. enterocolitica. We constructed a six-His-YmoA recombinant protein derived from YmoA, the homologue of Hha in Y. enterocolitica, and found that it interacts with Y. enterocolitica H-NS. We also cloned and sequenced the hns gene of this microorganism. In the course of these experiments we found that His-YmoA can also retain H-NS from E. coli. We also found that the hns gene of Y. enterocolitica can complement an hns mutation of E. coli. Finally, we describe for the first time systematic characterization of missense mutant alleles of hha and truncated Hha′ proteins, and we report a striking and previously unnoticed similarity of the Hha family of proteins to the oligomerization domain of the H-NS proteins.
The Hha protein from Escherichia coli (32) and the YmoA protein from Yersinia enterocolitica (9) belong to a new family of modulators of gene expression. Both are small (about 8.5 kDa), show extensive homology in their amino acid sequences (10), and are functionally interchangeable (3, 27). YmoA was characterized as a repressor of the yop virulence regulon in Y. enterocolitica (9, 22). Hha has been identified in E. coli as a thermo- and osmomodulator of the expression of the toxin α-hemolysin (7, 29). The expression of several other proteins is also altered in hha mutants, depending on the medium osmolarity (2). Homologues of Hha have also been found in conjugative plasmids, where they could be involved in the regulation of sexual transfer (34).
Protein-protein interaction studies performed by immobilizing His-tagged Hha on nickel-nitrilotriacetic acid (Ni2+-NTA) agarose led us to demonstrate that Hha interacts with the nucleoid-associated protein H-NS and that the Hha-H-NS complex is responsible for the thermo-osmotic modulation of the expression of the hemolysin operon in E. coli (33). H-NS (15.4 kDa) and homologues are widespread in enterobacteria and other genera of gram-negative bacteria (4). E. coli and Salmonella enterica serovar Typhimurium H-NS proteins are among the best-characterized examples of central modulators implicated in the response to changes in osmolarity and temperature (for reviews see references 1, 49, and 51). H-NS affects the expression of a large number of genes (1, 24), and it is considered a general negative regulator of transcription. This protein binds preferentially to curved DNA (52), and it is also able to generate bends in noncurved DNA (44). Three isoforms of the H-NS protein have been identified, although the biological significance of each isoform is unclear (43, 49).
We extend here our observations concerning the interaction between Hha and H-NS and present further evidence demonstrating a direct protein-protein interaction, even in the absence of DNA. We also show that this interaction is common to other members of both families of proteins: the use of His-tagged YmoA allowed us to identify the H-NS protein from Y. enterocolitica, thus extending this newly described mechanism of regulation of gene expression to other genera of gram-negative bacteria. Finally, the H-NS-binding properties of altered Hha proteins together with the amino acid sequence comparison presented here lead us to propose that Hha can be seen as an independent oligomerization domain of H-NS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used are listed in Table 1. Liquid Luria-Bertani (LB) medium (10 g of NaCl, 10 g of tryptone, and 5 g of yeast extract per liter) was used. Solid medium was LB medium plus 15 g of agar per liter. Antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 50 μg/ml; tetracycline, 12.5 μg/ml. β-Glucoside indicator plates with salicin were prepared as described previously (40).
TABLE 1.
Bacteria and plasmids used
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli 5K | F− rk− mk−rpsL thr thi leu ΔlacZ | 8 |
| E. coli BL21 (DE3) | hsdS gal (λcIts857 ind-1 Sam7 nin-5 lacUV5-T7 gene 1) | 45 |
| E. coli KL743 | Tcr | 14 |
| E. coli BSN26 | hns+ | 19 |
| E. coli BSN27 | Δhns | 19 |
| Y. enterocolitica Y754 | Spanish Type Cul-ture Collection | |
| E. coli H11 | 5K hha11 | This work |
| E. coli H18 | 5K hha18 | This work |
| E. coli H21 | 5K hha21 | This work |
| E. coli H29 | 5K hha29 | This work |
| E. coli H39 | 5K hha39 | This work |
| Plasmids | ||
| pLysE | Cmr | 45 |
| pET3b | Apr | 45 |
| pUC19 | Apr | 53 |
| pHly152 | hlyR+hlyC+hlyA+hlyB+hlyD+ | 35 |
| pANN202-312 | hlyC+hlyA+hlyB+hlyD+ | 14 |
| pETHHA-1 | Apr | 33 |
| pETHISHHA | Apr | 33 |
| pETHHAHIS | Apr | This work |
| pETHISHHA11 | Apr | This work |
| pETHISHHA18 | Apr | This work |
| pETHISHHA29 | Apr | This work |
| pETHISHHA39 | Apr | This work |
| pETHISHHAT1 | Apr | This work |
| pETHISHHAT2 | Apr | This work |
| pETHISHHAT3 | Apr | This work |
| pETHNSHIS | Apr | This work |
| pETHISYMOA | Apr | This work |
| pYNS-1 | Apr | This work |
Genetic and molecular procedures.
Isolation of plasmids, restriction digestion of DNA, ligation of DNA, and transformation were carried out by standard methods. Chromosomal DNA from Y. enterocolitica was isolated as described elsewhere (42). PCR amplification and sequencing of DNA were done according to standard methodology.
Plasmids for overexpression of proteins based on the T7 RNA polymerase system (45) were constructed as follows. For pETH-NSHIS, oligonucleotides used to amplify the hns gene of E. coli were H-NSNPRO (5′-GAGATTACTCATATGAGCG-3′) and HNSHISR (5′-CGGGATCCTATTAATGGTGATGGTGATGGTGTTGCTTGATCAGG-3′) which adds after the last hns codon six codons for His residues plus two stop codons and a BamHI site. The PCR fragment was cut with NdeI-BamHI and cloned into pET3b. For pETYMOA, oligonucleotides used to amplify the ymoA gene of Y. enterocolitica were YEYN (5′-GAAAAAACCATATGACAAAAACTGACT-3′), which creates an NdeI site overlapping the first codon of the ymoA gene, and YEYB (5′-CGATTATCGGATCCACGTTGTGT-3′), which creates a BamHI site 32 bp after the last codon of ymoA. The PCR fragment was cut with NdeI-BamHI and cloned into pET3b. For pETHISYMOA, plasmid pETYMOA was cut with NdeI and a His linker was inserted into it, as described previously (33).
Chemical mutagenesis of E. coli 5K (pANN202-312) was performed with diethyl sulfate by standard procedures. Colonies with the characteristic Hha phenotype (large hemolytic haloes) were purified as potential hha mutants. Cultures of the mutant strains previously obtained were infected with bacteriophage P1vir grown on E. coli KL743, which carries a Tcr marker close to the hha gene (14). Amplification of the hha gene and sequencing of the PCR band further analyzed the selected mutants. For both PCR amplification and DNA sequencing of the hha gene, we used oligonucleotides HHA2000 (5′-TCAGGTAATCGACTATTCCG-3′) and HHA2000R (5′-TGTGATAAAGATCACATAGGG-3′) of sequence accession X57977, which includes the hha gene. The open reading frames (ORFs) of hha′ genes in which base changes leading to amino acid substitutions could be identified were cloned in pET3b, and a histidine tag was added as previously described (33).
Shortened versions of the hha gene were obtained by PCR amplification and cloned in pET3b vectors. The oligonucleotides used correspond to sequence accession number X57977 as follows: for pETHHAT1, HHAT1 (5′-CTGCGGATCCTTAAAATACCGCC-3′) and HHANDE (33); for pETHHAT2, HHAT2 (5′-TTGTGGATCCGTTAATTCATGGT-3′) and HHANDE (33); for pETHHAT3, HHAT3 (5′-AGCGTCATATGGAGAAAAATAAATA-3′) and HHABAM (33). A histidine tag-encoding double-stranded oligonucleotide was added to the end of the truncated hha′ corresponding to the amino terminus of the protein, as previously described (33).
A derivative of Hha was constructed with a six-histidine tag at the carboxy-terminal end of the protein. This construct was obtained by PCR amplification with HHANDE (33) and HHACHIS (5′-CGGGATCCTATTAATGGTGATGGTGATGGTGGCGAATAAATTTCC-3′).
Overexpression of proteins by the T7 RNA polymerase system and purification of His-tagged proteins.
E. coli strain BL21(DE3)(pLysE) was used as a host for induction of expression of proteins. Plasmids containing the desired cloned genes (pET plasmids) were introduced into BL21(DE3)(pLysE) by transformation. One-liter cultures were grown at 30°C to an optical density at 600 nm of 1.0. At this point, IPTG (isopropyl-β-𝒟-galactopyranoside) was added to 0.5 mM, and incubation was carried out for 15 min. Cells were then pelleted by centrifugation and resuspended in 20 ml of buffer A (20 mM HEPES [pH 7.9], 10% glycerol, 100 mM KCl, 5 mM MgCl2, 50 mM imidazole). Clear cellular extracts were obtained as described previously (33). His-tagged proteins were purified by immobilized-metal affinity chromatography by using Ni2+-NTA technology (16, 17, 38) as described previously (33).
Electrophoresis and Western analysis of proteins.
Protein samples were analyzed in a Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system (16, 10, and 4%) (39) and stained with Coomassie blue. Bio-Rad protein standards and prestained protein standards were used for molecular weight estimation. Whole cells from overnight cultures of strains BSN27(pYHNS-1), BSN27(pUC19), and BSN26 were resuspended in standard denaturing sample buffer and analyzed by SDS-PAGE. Western blot analysis of proteins transferred to nitrocellulose membranes was performed with polyclonal antibodies raised against E. coli Hha and HNS. Hha-specific antibodies were prepared as previously described (7). H-NS specific antibodies were a kind gift from B. E. Uhlin (University of Umeå).
Dialysis and DNase treatment.
Spectra/Por 3.500 tubing was used to dialyze 1-ml fractions containing H-NS and Hha against HEPES (20 mM, pH 7.9)-10% glycerol-5 mM MgCl2-10 mM KCl overnight at 4°C. H-NS and His-Hha fractions were treated with fast protein liquid chromatography-purified DNase I (Amersham-Pharmacia) (specific activity, 135 U/mg). DNase was added to a final concentration of 20 U/ml, and samples were incubated at 37°C for 1 h. After DNase treatment, proteins and DNA in the samples were detected by SDS-PAGE and agarose electrophoresis, respectively.
DNA and protein sequences analysis.
DNA and protein sequences were obtained from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and European Bioinformatics Institute (www.ebi.ac.uk) sequence data banks via the Internet and analyzed locally with the Genetics Computer Group (Madison, Wis.) programs on a UNIX platform (Wisconsin package, version 10.1). The Yersinia pestis hns gene sequence was obtained from the University of Wisconsin genome project BLAST server (www.genome.wisc.edu) database yppv6 (4 Jan. 2001). The Boxshade program was used for display of alignments.
Mass spectrometry analysis.
Two separate experiments were performed with internal and external calibrations, with two proteins of known molecular mass. Matrix-assisted laser desorption ionization-time-of-flight (mass spectrometry) (MALDI-TOF) mass spectra were acquired on a Bruker Biflex spectrometer equipped with a pulsed nitrogen laser (337 nm) in linear, positive-ion mode, by using a 19-kV acceleration voltage. Sinapic acid (3,5-dimethoxy-4-hydroxycinnamic acid) was used as a matrix. Samples were prepared by mixing equal volumes of a saturated solution of the matrix in 0.1% trifluoroacetic acid in water-acetonitrile (2:1) and a protein solution with a concentration in the range of 1 to 10 μM.
Microsequencing of proteins.
N-terminal analysis of amino acid composition of proteins was performed by automatic Edman degradation. Prior to analysis, proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad) by semidry electroblotting. The membrane was then stained with Coomassie blue. The target protein was cut from the membrane and subjected to sequence analysis as previously described (36).
Measurement of hemolysin production.
Hemolysin in the culture supernatants was assayed by measuring hemolytic activity as previously described (30). Standard sheep blood agar plates were used to distinguish hemolytic phenotypes.
RESULTS
DNA is not required for the interaction between His-Hha and H-NS.
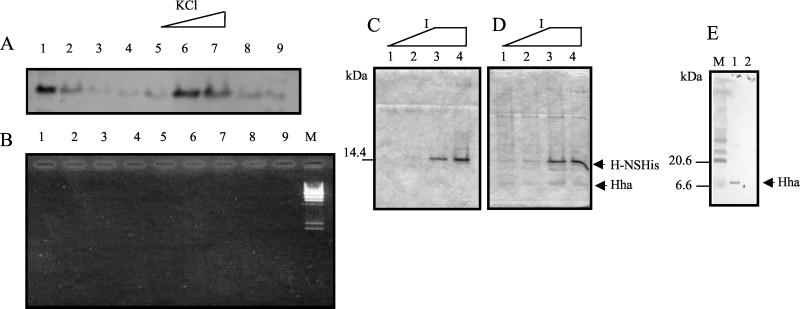
We previously showed in vitro interaction of Hha and H-NS by demonstrating that upon purification of His-tagged Hha by Ni2+-NTA technology, H-NS coeluted with His-Hha when the agarose matrix was washed with a buffer containing 200 mM imidazole (33). Prior to His-Hha elution with imidazole, H-NS can be separated from the Ni2+-NTA agarose matrix when the KCl concentration in the buffer increases (0.5 to 1 M) (33). This has been reported for other proteins retained by His-tagged partners bound to Ni2+-NTA agarose beads (37). When we further analyzed the fractions released upon KCl washing, we found that DNA was also present. This could be interpreted as His-Hha retaining DNA and as H-NS binding to DNA and not directly to His-Hha. To rule out this possibility, we tested the ability of His-Hha to bind H-NS in the absence of DNA. To do this, the fractions eluted at 0.5 and 1 M KCl (containing H-NS and DNA) were dialyzed and then treated with DNase. Samples that had been treated identically except for the DNase added were used as controls. Purified His-Hha bound to the agarose matrix was then mixed (2 h, 4°C) with H-NS in the presence and absence of DNA. Upon several washings of the agarose matrix with buffer A containing increasing KCl concentrations, H-NS could be visualized by immunodetection mainly in the fractions eluted at high salt concentration, thus demonstrating that the interaction between Hha and H-NS occurs in the absence of DNA (Fig. 1A and B).
FIG. 1.
In vitro interaction of Hha and H-NS. (A and B) Interaction in the absence of DNA; (C, D, and E) interaction of H-NS-His with Hha. (A) Immunodetection of H-NS in fractions obtained by washing with increasing KCl concentration in a Ni2+-NTA agarose matrix containing H-NS and His-Hha. H-NS and His-Hha preparations had been treated with DNase. Lanes: 1, unbound supernatant; 2 to 5, washes with buffer A; 6, buffer A and 0.5 M KCl; 7, buffer A and 1.0 M KCl; 8, buffer A; 9, buffer A and 200 mM imidazole. (B) Agarose electrophoresis plus ethidium bromide staining of the fractions in panel A. M, λ-HindIII molecular mass standard. (C and D) Coomassie blue-stained SDS-PAGE gel. (C) Progressive washes of a Ni2+-NTA agarose resin bound to a cleared extract of IPTG-induced cells expressing H-NS-His protein. (D) Fractions eluted from Ni2+-NTA agarose plus H-NS-His and a cleared lysate containing overexpressed Hha. Lanes: 1, 75 mM imidazole; 2, 100 mM imidazole; 3 and 4, 200 mM imidazole. (E) Immunodetection of Hha in fractions eluted with 200 mM imidazole. Lanes: 1, fraction 4 from panel D; 2, fraction 4 from panel C; M, prestained molecular mass standard.
Mass spectrometry analysis of the H-NS preparation copurifying with Hha.
Two or three H-NS isoforms, whose properties have not been described, can be observed by two-dimensional PAGE in cellular extracts (49). To analyze which of the H-NS isoforms copurifies with His-Hha, we determined its molecular mass by mass spectrometry analysis. We obtained masses of 15,407.8 and 15,410.1 Da in two separate experiments; these values closely resemble the mass calculated from the amino acid sequence (15,408.5 Da), without the first Met residue.
H-NS-His binds Hha.
Considering that the amino-terminal end of H-NS is responsible for protein-protein interaction and the carboxy-terminal end is responsible for protein-DNA interaction (13, 41, 48), we decided to add the six-His tag to the carboxy-terminal end of H-NS to test if, in vitro, immobilized H-NS is able to interact with Hha. The hns gene of E. coli was amplified with the oligonucleotides HNSNPRO and HNSHISR, which adds a six-His tag to the H-NS protein at the carboxy-terminal end, and then cloned in pET3b, yielding plasmid pETHNSHIS. The overexpressed fusion protein was purified by using Ni2+-NTA agarose. No other proteins copurified with H-NS-His (Fig. 1C). It is relevant that whereas H-NS is an abundant protein in the cell, this is not the case for Hha. When not overexpressed, this protein is barely detectable in SDS-PAGE gels (28). To overcome this problem, a cellular extract containing overexpressed Hha was prepared and mixed with Ni2+-NTA agarose already containing H-NS-His. A protein exhibiting a molecular mass similar to that of Hha copurified with H-NS (Fig. 1D). Western blot analysis demonstrated that this protein is Hha (Fig. 1E). It is thus apparent that, in vitro, H-NS-His is able to bind Hha too, as expected.
His-YmoA binds both Y. enterocolitica and E. coli H-NS.
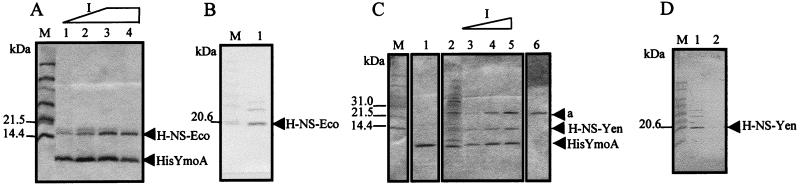
We next decided to test if the interaction between Hha and H-NS could be extended to other members of both families of proteins, particularly to YmoA and a hitherto-undescribed member of the H-NS family from Y. enterocolitica. The ymoA gene from Y. enterocolitica strain Y754 was first cloned in plasmid pET3b. Next, a histidine tag was added, yielding plasmid pETHISYMOA. His-YmoA was overexpressed in E. coli strain BL21(DE3)(pLysE) and purified by using Ni2+-NTA agarose. A second protein copurified with His-YmoA. As expected, this protein turned out to be E. coli H-NS (Fig. 2A and B). The next step was to release E. coli H-NS from the His-YmoA-H-NS complex by repeated washing of the Ni2+-NTA agarose matrix with buffer A containing 1 M KCl. The Ni2+-NTA agarose matrix containing His-YmoA was then split in two aliquots. One was used for a control experiment. The other was mixed with a Y. enterocolitica Y754 crude cell extract, and again, several washings with buffer A containing increasing concentrations of imidazole were performed. Two accompanying proteins coeluted with His-YmoA (Fig. 2C). One of them, with a larger molecular mass, was shown not to specifically interact with His-YmoA; it was also detected upon imidazole washing of a Ni2+-NTA agarose matrix with which a crude extract of the Y. enterocolitica culture was mixed (lacking bound His-YmoA) (Fig. 2C). The second protein, with a molecular mass of about 15 kDa, copurified specifically with His-YmoA and reacted with E. coli H-NS-specific antibodies (Fig. 2D). We suspected that this protein was Y. enterocolitica H-NS. This was confirmed by N-terminal microsequencing. The sequence obtained (SEALKILNNIRTLRAQAREXTLETLE) matched that of the E. coli H-NS, except for the Cys residue at position 20 (Cys residues are commonly not found in N-terminal Edman sequencing).
FIG. 2.
Interaction of His-YmoA with E. coli H-NS (A and B) and Y. enterocolitica H-NS (C and D). (A) Coomassie blue-stained SDS-PAGE gel of fractions eluted with increasing imidazole (75, 100, 200, and 200 mM) from Ni2+-NTA agarose resin bound to a cleared cellular extract from IPTG-induced E. coli BL21(DE3)(pLysE, pETHISYMOA). (B) Western blotting with H-NS-specific antibodies of fraction 4 from panel A. (C) Lane 1, fraction eluted with 200 mM imidazole from Ni2+-NTA-agarose resin containing bound His-YmoA protein. Interacting proteins (H-NS from E. coli) had been previously removed by repeated washing with buffer A-1.0 M KCl. Lanes 2 to 5, fractions eluted with buffer A with increasing concentrations of imidazole (50, 75, 100, and 200 mM) from Ni2+-NTA-agarose resin containing bound His-YmoA protein mixed with a cellular extract from Y. enterocolitica Y754. Lane 6 shows unspecific binding of an unknown protein (a) of Y. enterocolitica to the Ni2+-NTA-agarose resin. (D) Western blotting with E. coli H-NS-specific antibodies of fractions 5 and 1 from panel C (lanes 1 and 2, respectively). M, molecular mass standard (A and C) or prestained molecular mass standard (B and D).
Y. enterocolitica hns gene complements some hns-induced phenotypes in E. coli.
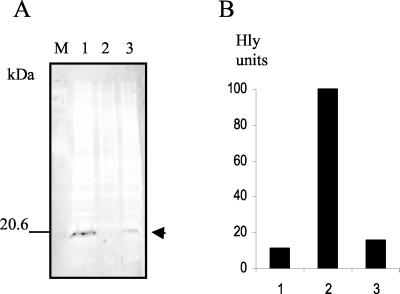
In order to confirm the identification in Y. enterocolitica crude cell extracts of a protein belonging to the H-NS family, we decided to clone the Y. enterocolitica hns gene and determine its nucleotide sequence. Chromosomal DNA was isolated from Y. enterocolitica Y754, subjected to partial Sau3A digestion, and then ligated to pUC19 plasmid previously digested with BamHI. This ligation was transformed into E. coli strain BSN27 (hns). About 4,000 colonies were lifted with toothpicks and streaked onto Bgl indicator plates, where hns+ colonies appear greenish and hns colonies appear yellow due to derepression of the bgl operon (15). One of the clones able to complement the Bgl phenotype was selected, and plasmid DNA was isolated from it. The resulting plasmid (pYNS-1) contained about 3 kbp of Y. enterocolitica DNA. Immunodetection of an ∼15-kDa protein in crude cell extracts from strain BSN27(pYNS-1) suggested that plasmid pYNS-1 carries the hns gene from Y. enterocolitica (Fig. 3A). This was confirmed by DNA sequencing (GenBank accession number AJ302081). Analysis of the DNA sequence showed an open reading frame whose translation closely matches the amino acid sequence of the H-NS proteins. Additionally, it perfectly matches the sequence of residues obtained in the N-terminal Edman degradation of the protein bound to His-YmoA. Other authors have identified the same Y. enterocolitica gene (5). We next tested whether the Y. enterocolitica hns gene is also able to complement other hns-related phenotypes of strain BSN27. Specifically, we tested the deregulation of hemolysin expression caused by the hns allele (33). E. coli BSN27(pHly152, pYNS-1) cells showed a significant decrease in hemolysin production compared to BSN27(pHly152, pUC19) cells (Fig. 3B), thus showing that the hns gene of Y. enterocolitica is also able to complement the deregulation of hemolysin expression caused by the hns allele of E. coli strain BSN27.
FIG. 3.
(A) Immunodetection of H-NS in cellular extracts corresponding to different E. coli strains. Lanes: M, prestained molecular mass standard; 1, BSN27(pYHNS-1); 2, BSN27(pUC19); 3, BSN26. Arrow, H-NS protein. (B) Evaluation of the hemolytic activity (Hly) in culture supernatants (expressed as percentages). Representative results are shown. 1, BSN27(pHly152)(pYHNS-1); 2, BSN27(pHly152)(pUC19); 3, BSN26(pHly152).
Isolation of single amino acid substitutions in Hha protein and truncated Hha proteins which affect binding to H-NS.
To gain information about the functional domain organization of the Hha protein we chose a genetic approach. We used mutagenesis to obtain single amino acid substitutions that showed the characteristic Hha− phenotype, i.e., derepression of the hly operon. E. coli 5K(pANN202-312) was used for chemical mutagenesis. Approximately 4 × 104 colonies on blood agar plates were screened, and a total of 42 clones were isolated which showed an increase in the hemolytic haloes (phenotype of hha mutants). In order to identify the mutations in the hha gene, these mutants were infected with bacteriophage P1 obtained on E. coli strain KL743 (which carries a Tcr marker close to the hha gene). Association of acquisition of Tcr and reversion to low hemolysin production (about 50% frequency) was observed in mutants H11, H18, H21, H29, and H39. Mutants showing no reversion at all, unstable phenotypes, or unclear results were not studied further.
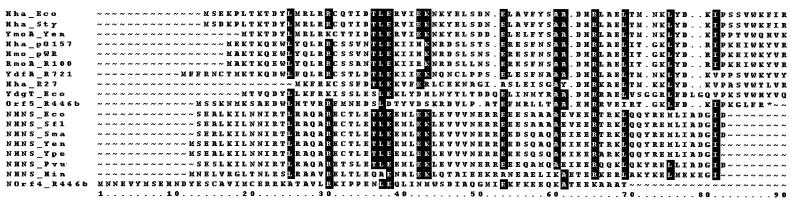
The hha genes of mutants H11, H18, H21, H29, and H39 were sequenced. All of them had single point missense mutations in the hha ORF which result in amino acid substitutions: hha11 and hha21, R50 to H (CGC to CAC); hha18, R16 to C (CGT to TGT); hha29, P64 to S (CCT to TCT); and hha39, P64 to L (CCT to CTT). In order to evaluate if binding to H-NS protein was affected by these mutations, we cloned the ORFs of hha11, hha18, hha29, and hha39 in pET3b and then added a six-histidine tag at the amino end. The recombinant mutant proteins were expressed in BL21(DE3)(pLysE) host cells and purified from the cleared cellular extracts by mixing with Ni2+-NTA agarose followed by elution with increasing concentrations of imidazole, Tricine-SDS-PAGE, and Western blotting with specific anti-H-NS antibodies. We found that the interaction with H-NS was impaired in all four His-Hha′ proteins (Fig. 4). The truncated His-HhaT1, His-HhaT2 and HisT3 proteins were analyzed in the same way. We found that none of the three truncated proteins retained H-NS in our assay. The addition of the six-histidine tag at the carboxy-terminal end of the Hha protein was evaluated and found to impair interaction with H-NS, too (Fig. 4). These results indicated that most of the Hha protein is involved in binding to H-NS and prompted us to take a closer look at the amino acid sequences of the Hha/YmoA family of proteins. We compared them to the oligomerization domain of the members of H-NS family (first 70 amino acids) and found a certain degree of similarity (Fig. 5) in the amino acid sequences. The similarity in length is also remarkable.
FIG. 4.
Amino acid substitutions obtained from missense mutations and sequences of the truncated Hha proteins. The histidine tags on Hha as well as the His-YmoA construct, at either the amino or carboxy terminal end, are shown. The effect on H-NS binding is indicated.
FIG. 5.
Comparison of known members of the Hha family of proteins and the oligomerization domain of members of the H-NS family of proteins. The accession numbers of the proteins are as follows. Hha_Eco (E. coli; P23870; Hha_Sty (S. enterica serovar Typhimurium), AF242359; YmoA_Yen (Y. enterocolitica; P27720), Hha_pO157 (plasmid pO157), Q9ZGV7; Hmo_pWR (plasmid pWR501), Q9AFJ8; RmoA_R100 (plasmid R100), O32581; YdfA_R721 (plasmid R721), 9971679; Hha_R27 (plasmid R27), Q9L5G3; YdgT_Eco (E. coli), P76179; Orf5_R446b (plasmid R446b), 312264. The oligomerization domains were extracted (first 70 amino acids in each case) from the sequences that follow: NHNS_Eco (E. coli; P08936); NHNS_Sfl (Shigella flexneri; P09120); NHNS_Sma (Serratia marcescens; P18955); NHNS_Yen (Y. enterocolitica; Q99QH3); NHNS_Ype (Y. pestis genome sequencing project BLAST server: www.genome.wisc.edu; NHNS_Pvu (Proteus vulgaris; P18818); NHNS_Hin (Haemophilus influenzae; P43841); NOrf4_R446b (plasmid R446b; 312263).
DISCUSSION
Interaction of H-NS with other proteins apart from itself or homologues (50) has been described in very few instances. H-NS has been proven to bind the bacteriophage T7 gene 5.5 protein product. The significance of this interaction seems clear, since it would favor bacteriophage T7 multiplication by inhibiting H-NS repressive function (25). H-NS also binds to the flagellar rotor protein FliG, but the significance of this interaction is unclear (11, 26). The significance of the interaction of H-NS with HF-I (21), an RNA-binding protein that participates in translation of rpoS RNA (31), is also unclear. Our previous observations about the interaction between Hha and H-NS (33) not only suggested the existence of an H-NS-Hha complex but also assigned a modulatory role to it: thermo- and osmomodulation of the expression of the toxin α-hemolysin. The data in this paper confirm a direct protein-protein interaction between Hha and H-NS and extend it to other members of both families of proteins. The protein complex can be evidenced in vitro in the absence of DNA, and it is also detected when HNS-His is the protein bound to the Ni2+-NTA agarose matrix. In this case, overexpression of Hha is required because of the relatively low abundance of Hha in the cell that could already be bound to native H-NS, being thus inaccessible for interaction with H-NS-His. This suggests that other low-abundance proteins interacting with tagged H-NS might not be apparent in similar copurification experiments.
The existence of H-NS isoforms has been previously documented (23, 43, 49). It has been suggested that the differences in isoelectric points detected between H-NS isoforms of similar molecular weights found in two-dimensional PAGE must correspond to posttranslational modifications located in the amino half of the protein (12), but the nature of such modifications remains unknown. The accuracy of MALDI-TOF permits the identification of changes in the molecular mass of the protein due to posttranslational modifications. Thus, the mass spectrometry data presented here allow us to assume that the H-NS isoform that interacts with Hha does not have any modification other than the removal of the initial Met residue.
E. coli H-NS and Y. enterocolitica H-NS have identical amino-terminal domains. This probably explains the ability of His-YmoA to bind E. coli H-NS and suggests that the H-NS proteins from both microorganisms have similar capacities for interacting with other protein partners. The strategy we used to clone the hns gene of Y. enterocolitica showed that it is able to complement in E. coli some of the defects caused by the hns mutation, such as the Bgl phenotype and the deregulation of hemolysin expression. The high degree of similarity of the H-NS proteins suggests that, as we show here for Y. enterocolitica H-NS, they are functionally interchangeable. The fact that H-NS of Y. enterocolitica interacts with YmoA, a known modulator of the expression of virulence genes (9), suggests that H-NS may play a significant role modulating expression of virulence in Y. enterocolitica.
Whereas the role of the N-terminal domain of H-NS in generating protein oligomers is well characterized (reviewed in reference 13), H-NS binding to DNA is the main factor considered in modeling H-NS-mediated regulation of gene expression (18, 20, 46, 47). Nevertheless, focusing on the H-NS modulation of the bgl operon, the need for H-NS to interact with other proteins (6) and the relevance of the N-terminal domain, perhaps to provide an anchoring point for other regulatory factors (48), have been pointed out. The results we present here further support the relevance of protein-protein interaction for H-NS-mediated modulation of gene expression.
The data obtained from the Hha′ proteins containing amino acid substitutions and truncations suggest that the entire protein is involved in the interaction with H-NS. Nevertheless, the carboxy-terminal end must be more closely implicated, since the addition of the six-histidine tag at the carboxy-terminal end severely impairs interaction with H-NS, which is not the case when the six-histidine tag is placed at the amino-terminal end. Additionally, the fact that YmoA, despite lacking the first five amino acid residues of Hha, still interacts with H-NS suggests that the very N-terminal end is not essential for Hha to interact with H-NS. These genetic and biochemical data are consistent with the results obtained from the alignment of the amino acid sequence of the different members of the Hha family and the oligomerization domain of the members of the H-NS family (Fig. 5). Apart from the similarity in length, it is apparent that there are identity boxes scattered along the sequence alignment.
Taking the results together, we conclude that Hha can be considered a specialized homologue of the amino-terminal oligomerization domain of H-NS and that this observation is very likely valid for the other members of the Hha/YmoA family of proteins. Furthermore, the interaction of Hha-type and H-NS-type proteins is probably present in many enteric bacteria as a mechanism of regulation of gene expression.
Acknowledgments
This work was supported by grants from the Ministerio de Ciencia y Tecnología (PB97-0950) and the CIRIT from the Generalitat de Catalunya (2000 SGR 00038). S. Rodríguez was the recipient of a F.I. fellowship from the Generalitat de Catalunya.
We thank F. Canals from the Universitat Autónoma de Barcelona for the mass spectrometry analysis and N-terminal protein sequencing.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7–17. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre, C., J. Johansson, B. E. Uhlin, A. Juárez, and F. Muñoa. 1999. Alterations in protein expression caused by the hha mutation in Escherichia coli: influence of the growth medium osmolarity. J. Bacteriol. 181:3018–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre, C., A. Juárez, C. Madrid, M. Mouriño, A. Prenafeta, and F. Muñoa. 1996. Complementation of the hha mutation in Escherichia coli by the ymoA gene from Yersinia enterocolitica: dependence on the gene dosage. Microbiology 142:1841–1846. [DOI] [PubMed] [Google Scholar]
- 4.Bertin, P., N. Benhabiles, E. Krin, C. Laurent-Winter, C. Tendeng, E. Turlin, A. Thomas, A. Danchin, and R. Brasseur. 1999. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in Gram-negative bacteria. Mol. Microbiol. 31:319–329. [DOI] [PubMed] [Google Scholar]
- 5.Bertin, P., F. Hommais, E. Krin, O. Soutourina, C. Tendeng, S. Derzelle, and A. Danchin. 2001. H-NS and H-NS-like proteins in Gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie 83:235–241. [DOI] [PubMed] [Google Scholar]
- 6.Caramel, A., and K. Schnetz. 1998. Lac and λ repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J. Mol. Biol. 284:875–883. [DOI] [PubMed] [Google Scholar]
- 7.Carmona, M., C. Balsalobre, F. J. Muñoa, M. Mouriño, Y. Jubete, F. De la Cruz, and A. Juárez. 1993. Escherichia coli hha mutants, DNA supercoiling and expression of the haemolysin genes from the recombinant plasmid pANN202–312. Mol. Microbiol. 9:1011–1018. [DOI] [PubMed] [Google Scholar]
- 8.Collins, J., and H. J. Brüning. 1978. Plasmids useable as gene-cloning vectors in an in vitro packaging by coliphage λ: “cosmids.” Gene 4:85–107. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., C. Sluiters, I. Delor, D. Gelb, K. Kaninga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michaelis. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023–1034. [DOI] [PubMed] [Google Scholar]
- 10.De la Cruz, F., M. Carmona, and A. Juárez. 1992. The Hha protein from Escherichia coli is highly homologous to the YmoA protein from Yersinia enterocolitica. Mol. Microbiol. 6:3451–3452. [DOI] [PubMed] [Google Scholar]
- 11.Donato, G. M., and T. H. Kawula. 1998. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J. Biol. Chem. 11:24030–24036. [DOI] [PubMed] [Google Scholar]
- 12.Donato, G. M., and T. H. Kawula. 1999. Phenotypic analysis of random hns mutations differentiate DNA-binding activity from properties of fimA promoter inversion modulation and bacterial motility. J. Bacteriol. 181:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman, C. J., J. C. D. Hinton, and A. Free. 1999. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 124:124–128. [DOI] [PubMed] [Google Scholar]
- 14.Godessart, N., F. J. Muñoa, M. Regué, and A. Juárez. 1988. Chromosomal mutations that increase the production of a plasmid-encoded haemolysin in Escherichia coli. J. Gen. Microbiol. 134:2779–2787. [DOI] [PubMed] [Google Scholar]
- 15.Higgins, C. F., C. J. Dorman, D. A. Stirling, L. Waddell, I. R. Booth, G. May, and E. Bremer. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569–584. [DOI] [PubMed] [Google Scholar]
- 16.Hochuli, E., W. Bannwarth, H. Dobeli, R. Gentz, and D. Stüber. 1988. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate absorbent. Bio/Technology 6:1321–1325. [Google Scholar]
- 17.Hoffman, A., and R. G. Roeder. 1991. Purification of His-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 19:6337–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulton, C. S. J., A. Seirafi, J. C. D. Hinton, J. A. M. Sidebotham, L. Waddell, G. D. Pavitt, T. Owen-Hughes, A. Spassky, H. Buc, and C. F. Higgins. 1990. Histone-like protein H1 (H-NS), DNA supercoiling and gene expression in bacteria. Cell 63:631–642. [DOI] [PubMed] [Google Scholar]
- 19.Johansson, J., B. Dagberg, E. Richet, and B. E. Uhlin. 1998. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 180:6117–6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordi, B., T. A. Owen-Hughes, C. Hulton, and C. F. Higgins. 1995. DNA twist, flexibility and transcription of the osmoregulated proU promoter of Salmonella typhimurium. EMBO J. 14:5690–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajitani, M., and A. Ishihama. 1991. Identification and sequence determination of the host factor gene for bacteriophage Qβ. Nucleic Acids Res. 19:1063–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert de Rouvroit, C., C. Sluiters, and G. R. Cornelis. 1992. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6:395–409. [PubMed] [Google Scholar]
- 23.Laurent Winter, C., P. Lejeune, and A. Danchin. 1995. The Escherichia coli DNA-binding protein H-NS is one of the first proteins to be synthesized after a nutritional upshift. Res. Microbiol. 146:5–16. [DOI] [PubMed] [Google Scholar]
- 24.Laurent-Winter, C., S. Ngo, A. Danchin, and P. Bertin. 1997. Role of Escherichia coli histone-like nucleotide-structuring protein in bacterial metabolism and stress response: identification of targets by two-dimensional electrophoresis. Eur. J. Biochem. 244:767–773. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Q., and C. Richardson. 1993. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1761–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marykwas, D. L., S. A. Schmidt, and H. C. Berg. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol. 256:564–576. [DOI] [PubMed] [Google Scholar]
- 27.Mikulskis, A. V., and G. R. Cornelis. 1994. A new class of proteins regulating gene expression in enterobacteria. Mol. Microbiol. 11:77–86. [DOI] [PubMed] [Google Scholar]
- 28.Mouriño, M., C. Balsalobre, C. Madrid, J. M. Nieto, A. Prenafeta, F. J. Muñoa, and A. Juárez. 1998. Osmolarity modulates the expression of the Hha protein from Escherichia coli. FEMS Microbiol. Lett. 160:225–229. [DOI] [PubMed] [Google Scholar]
- 29.Mouriño, M., C. Madrid, C. Balsalobre, A. Prenafeta, F. J. Muñoa, J. Blanco, M. Blanco, J. E. Blanco, and A. Juárez. 1996. The Hha protein as a modulator of expression of virulence factors in Escherichia coli. Infect. Immun. 64:2881–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouriño, M., F. J. Muñoa, C. Balsalobre, P. Díaz, C. Madrid, and A. Juárez. 1994. Environmental regulation of alpha-hemolysin expression in Escherichia coli. Microb. Pathog. 16:249–259. [DOI] [PubMed] [Google Scholar]
- 31.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10:1143–1151. [DOI] [PubMed] [Google Scholar]
- 32.Nieto, J. M., M. Carmona, S. Bolland, Y. Jubete, F. de la Cruz, and A. Juárez. 1991. The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285–1293. [DOI] [PubMed] [Google Scholar]
- 33.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juárez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes proteins Hha and H-NS. Mol. Gen. Genet. 263:349–358. [DOI] [PubMed] [Google Scholar]
- 34.Nieto, J. M., A. Prenafeta, E. Miquelay, S. Torrades, and A. Juárez. 1998. Sequence, identification and effect on conjugation of the rmoA gene of plasmid R100-1. FEMS Microbiol. Lett. 169:59–66. [DOI] [PubMed] [Google Scholar]
- 35.Noegel, A., U. Rdest, and W. Goebel. 1981. Determinations of the functions of hemolytic plasmid pHly152 of Escherichia coli. J. Bacteriol. 145:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Packman, L. C. 1993. Protein chemical methods for molecular biologists. Methods Mol. Cell. Biol. 4:189–198. [Google Scholar]
- 37.Pichoff, S., L. Alibaud, A. Guédant, M. P. Castanié, and J. P. Bouché. 1998. An Escherichia coli gene (yaeO) suppresses temperature-sensitive mutations in essential genes by modulating Rho-dependent transcription termination. Mol. Microbiol. 29:859–869. [DOI] [PubMed] [Google Scholar]
- 38.Porath, J., J. Carlsson, I. Olsson, and G. Belfrage. 1975. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 258:598–599. [DOI] [PubMed] [Google Scholar]
- 39.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379. [DOI] [PubMed] [Google Scholar]
- 40.Schnetz, K., C. Toloczyki, and B. Rak. 1987. Beta-glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J. Bacteriol. 169:2579–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shindo, H., T. Iwaki, R. Ieda, H. Kurumizaka, C. Ueguchi, T. Mizuno, S. Morikawa, H. Nakamura, and H. Kuboniwa. 1995. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS Lett. 360:125–131. [DOI] [PubMed] [Google Scholar]
- 42.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Spassky, A., S. Rimsky, H. Garreau, and H. Buc. 1984. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 12:5321–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for bending. EMBO J. 16:1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60–89. [DOI] [PubMed] [Google Scholar]
- 46.Tupper, A. E., T. A. Owen-Hughes, D. W. Ussery, D. S. Santos, D. J. P. Ferguson, J. M. Sidebotham, J. C. D. Hinton, and C. F. Higgins. 1994. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 13:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueguchi, C., and T. Mizuno. 1993. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 12:1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueguchi, C., T. Suzuki, T. Yoshida, K. Tanaka, and T. Mizuno. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263:149–162. [DOI] [PubMed] [Google Scholar]
- 49.Ussery, D. W., J. C. D. Hinton, B. J. A. M. Jordi, P. E. Granum, A. Seirafi, R. J. Stephen, A. E. Tupper, G. Berridge, J. M. Sidebotham, and C. F. Higgins. 1994. The chromatin-associated protein H-NS. Biochimie 76:968–980. [DOI] [PubMed] [Google Scholar]
- 50.Williams, R. M., S. Rimsky, and H. Buc. 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative mutants. J. Bacteriol. 178:4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the E. coli nucleoid protein H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175–185. [DOI] [PubMed] [Google Scholar]
- 52.Yamada, H., T. Yoshida, K. Tanaka, C. Sasakawa, and T. Mizuno. 1991. Molecular analysis of the Escherichia coli hns gene encoding a DNA binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 230:332–336. [DOI] [PubMed] [Google Scholar]
- 53.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequencing of the M13mp18 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]