Abstract
We describe here the identification and characterization of two Listeria monocytogenes (Tn917-LTV3) relA and hpt transposon insertion mutants that were impaired in growth after attachment to a model surface. Both mutants were unable to accumulate (p)ppGpp in response to amino acid starvation, whereas the wild-type strain accumulated (p)ppGpp within 30 min of stress induction. The induction of transcription of the relA gene after adhesion was demonstrated, suggesting that the ability to mount a stringent response and undergo physiological adaptation to nutrient deprivation is essential for the subsequent growth of the adhered bacteria. The absence of (p)ppGpp in the hpt mutant, which is blocked in the purine salvage pathway, is curious and suggests that a functional purine salvage pathway is required for the biosynthesis of (p)ppGpp. Both mutants were avirulent in a murine model of listeriosis, indicating an essential role for the stringent response in the survival and growth of L. monocytogenes in the host. Taken as a whole, this study provides new information on the role of the stringent response and the physiological adaptation of L. monocytogenes for biofilm growth and pathogenesis.
The ubiquitous gram-positive pathogen L. monocytogenes is responsible for the clinical syndromes of listeriosis in humans and animals (27, 28). The principle mode of transmission of the organism to humans is believed to be the consumption of contaminated food (44). Normally, the consumption of food-borne L. monocytogenes does not result in overt gastrointestinal disease. Rather, in serious cases, listeriosis presents as bacteremia, meningitis, or miscarriage (30).
The ability of L. monocytogenes to adhere to and colonize surfaces during food preparation and storage is important in the contamination of food products prior to consumption. While much is known about the virulence of L. monocytogenes (reviewed in references 7 and 24), detailed knowledge about the mechanisms of attachment and proliferation of L. monocytogenes on surfaces is scant. Previously, the adhesion of L. monocytogenes to plant surfaces has been shown to be a consequence of hydrophobic bonds between the plant surface and outer surface components of the bacterium rather than a specific ligand-receptor binding process (3).
It is known that biofilm formation is a complex process, and a number of cell surface structures have been implicated in the initial stages of biofilm formation. In Pseudomonas aeruginosa these include flagella (motility) (35) and the expression of type IV pili for microcolony formation (36). In streptococci a number of cell surface adhesins, together with autolysins, have been identified as being important in biofilm development (12, 17, 19, 21, 26). The subsequent development and differentiation of an ordered three-dimensional biofilm requires the expression of extracellular polysaccharides (9) and may involve cell-to-cell signaling molecules (11, 26). During the initial establishment of a sessile community, it is likely that bacteria have to adapt their cellular physiology in response to environmental changes and the availability of nutrients. It is the ability to respond to such changes that is pivotal in the successful establishment of a biofilm.
In this study we describe the identification and characterization of relA and hpt mutants of L. monocytogenes defective in growth after attachment to an inert surface. Both the relA and the hpt mutants were unable to synthesize (p)ppGpp in response to nutritional starvation. In addition, we show transcription of the relA gene in adherent bacteria, indicating the induction of a stringent response after adhesion. Finally, both mutants were severely attenuated in a mouse model of infection, although the levels of listeriolysin were unaffected. These data indicate that the ability to synthesize (p)ppGpp and mount a stringent response is an essential physiological adaptation that is required for the initial growth of adhered bacteria and virulence.
MATERIALS AND METHODS
Bacterial strains and plasmids.
L. monocytogenes C52 has been described previously (22). Cells were cultured in either brain heart infusion (BHI) broth (Merck) or in tryptone soy broth (TSB; Oxoid) at 37°C with shaking (at 200 rpm) unless otherwise stated. Escherichia coli MC1061 was the host for rescue of DNA proximal to the site of transposon insertion. Either DH5α or XL1-Blue (Stratagene, Amsterdam, The Netherlands) cells were used for the transformations. E. coli strains were cultured in Luria-Bertani (LB) broth at 37°C. Where appropriate, agar was added to broth at a concentration of 1.5% (wt/vol) to make solid medium. Plasmid pLTV3 (4) harboring Tn917-LTV3 was maintained in E. coli HB101. Plasmids pDG148relA and pDG148 were gifts from Thomas Wendrich at the University of Marburg, Marburg, Germany. E. coli strains CF1943, CF1944 (ΔrelA::kan), and CF1946 (ΔrelA::kan ΔspoT::cm) (45) were a gift from Mike Cashel (National Institutes of Health, Bethesda, Md.).
DNA manipulations, transformations, and Southern hybridization.
DNA manipulations, plasmid DNA isolation, and transformation of E. coli were carried out by standard procedures (39). Nucleotide sequencing was done by using the BigDye Terminator sequencing kit for cycle sequencing (Perkin-Elmer Biosystems). Custom primers were obtained from Sigma-Genosys. Chromosomal DNA was prepared from L. monocytogenes according to the method of Flamm et al. (13). Electrotransformation of L. monocytogenes was achieved according to the method of Park and Stewart (37). For Southern hybridization, chromosomal DNA was digested with HindIII and probed with a 5-kb XbaI Tn917-LTV3 fragment. Probes were labeled and hybridized according to the Gene Images Random Prime Labeling Module (Amersham Life Sciences), and hybrids were detected by using the Gene Images CDP-star Detection Module (Amersham Life Sciences).
Insertional mutagenesis.
L. monocytogenes C52 harboring pLTV3 (4) was grown overnight at 30°C in BHI medium containing tetracycline (12.5 μg ml−1), erythromycin (1 μg ml−1), and lincomycin (25 μg ml−1). The culture was diluted 1:800 into fresh broth and grown at 40°C until stationary phase before the subculture was repeated. Cells were plated on BHI agar containing erythromycin and lincomycin only and grown at 40°C for 48 h.
Adhesion and growth on microtiter plates.
The method of Genevaux et al. (15) was used with the following modifications. A single colony was inoculated into 200 μl of TSB in the wells of a 96-well plate (tissue culture treated) (Corning Costar) and left to grow statically at 37°C. This plate served as a master plate and was subsequently stored at −80°C after the addition of sterile glycerol to a final concentration of 20% (vol/vol). After overnight culture in TSB, 5 μl was inoculated into 200 μl of fresh TSB in the wells of a hydrophobic polystyrene 96-well plate (Greiner) and left to grow for 24 h at 37°C. Planktonic bacteria were then removed, and the wells were washed twice with an equal volume of sterile phosphate-buffered saline (PBS). Wells were air dried, and adhered cells were heat fixed at 80°C before being stained in situ with 0.1% (wt/vol) crystal violet. A decolorizing solution (ethanol-acetone [80:20]) was added, and the absorption of the eluted stain was measured with a Micro-ELISA Autoreader MR7000 (Dynatech) at 590 nm. Putative biofilm mutants were selected from the master plate and rescreened in triplicate. The crystal violet staining was confirmed by disrupting the adhered bacteria by violent agitation, followed by colony counting.
To demonstrate the growth of adhered bacteria, bacteria were inoculated in TSB as described above and left to grow and adhere for 1 h. After this time, the planktonic bacteria were removed, and the wells were washed twice before the addition of fresh TSB. The growth of the adhered bacteria was quantified at hourly intervals by crystal violet staining as described above.
Rescue of flanking DNA.
Chromosomal DNA from insertion mutants was rescued as described previously (4). Briefly, this involved digesting chromosomal DNA with either Asp718 (in the case of Sag-1) or XbaI (in the case of Sag-2), followed by self-ligation before transformation into E. coli MC1061. Transformants were recovered on LB agar containing kanamycin (50 μg ml−1).
Murine infections.
Female MF1 outbred mice, ca. 30 to 35 g, were obtained from Harlan Olac, Ltd. (Bicester, United Kingdom). To prepare a standardized inoculum of in vivo-passaged bacteria, 200 μl of an overnight culture (2 × 107 viable bacteria) was inoculated intravenously (i.v.) into mice, and bacteria were recovered 48 h later on tryptone soy agar from the spleens (40). The passaged bacteria were grown in TSB in stationary culture at 37°C for 18 h, harvested by centrifugation, and resuspended in TSB containing 10% (vol/vol) glycerol. Portions of this suspension were then stored at −70°C. When required, the suspension was thawed, and the bacteria were harvested by centrifugation before resuspension in sterile PBS to the required concentration.
For virulence testing, groups of five mice were inoculated by the i.v. route. For i.v. injection, a dose of ca. 5 × 105 viable bacteria contained in a total volume of 100 μl of distilled water was injected into the tail vein. The number of bacteria inoculated was confirmed by plating serial dilutions on tryptone soy agar. After infection, mice were observed for 96 h before being killed by cervical dislocation. The spleen and liver were removed and homogenized, and bacteria were enumerated as described by Stephens et al. (40).
Accumulation of (p)ppGpp.
Bacteria were grown overnight on LB agar supplemented with the appropriate antibiotics. Subsequently, individual colonies were inoculated to give an optical density at 600 nm (OD600) of 0.5 in 100 μl of unlabeled morpholinepropanesulfonic acid (MOPS) starvation medium (5) before being serially diluted in labeled MOPS medium containing 0.2% (wt/vol) dextrose, serine hydroxamate (1 mg ml−1), l-valine (0.5 mg ml−1), and [32P]H3PO4 (5 μCi ml−1). Cells were then incubated for 30 min at 37°C before the addition of 100 μl of 13 M formic acid. Samples were frozen on dry ice and thawed at room temperature for two cycles. Cell extracts were then centrifuged for 2 min at 10,000 × g, and 5 μl of the supernatant was spotted onto 20-by-20-cm polyethylenemine cellulose plates (Sigma) for separation by thin-layer chromatography of the phosphorylated guanosine nucleotides in 1.5 M K2HPO4. Nucleotides were visualized by autoradiography.
β-Galactosidase assays.
After the careful removal of the planktonic culture, the microtiter well was washed twice with PBS prior to removal of the adhered bacteria in 100 μl of PBS by violent agitation. Subsequently, the level of β-galactosidase in the planktonic and adhered bacteria was measured by using a chemiluminescent assay according to the manufacturer’s instructions (Roche Molecular Biochemicals) and expressed as relative light units (RLU) per milligram of protein in the cell lysate. The mean value from five separate experiments was determined, and the standard error of the mean (SEM) was established.
RESULTS
Isolation of mutants defective in growth after adhesion.
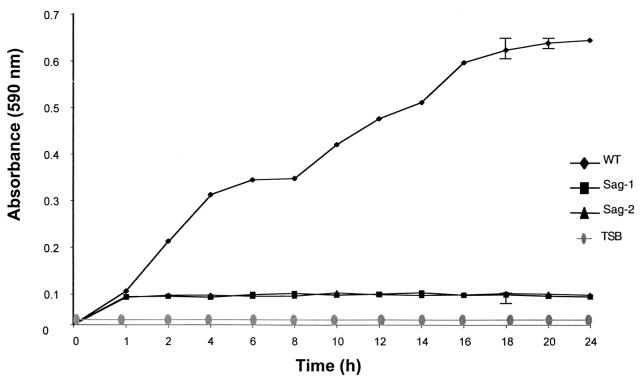
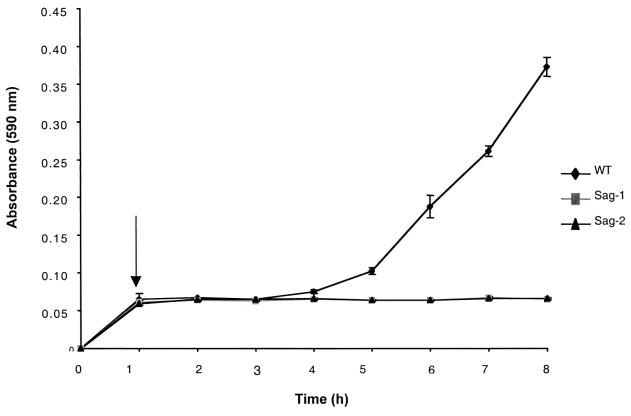
Tn917-LTV3 insertional mutagenesis was carried out after the introduction of plasmid pLTV3 harboring the transposon (4) into L. monocytogenes C52. Insertion mutants were generated in liquid culture at 40°C in the presence of erythromycin and lincomycin. A total of 98% of erythromycin- and lincomycin-resistant colonies were sensitive to tetracycline, indicating transposition of Tn917-LTV3 into the genome, with loss of the plasmid. The frequency of transposition was 1.5 × 10−3 per recipient and random insertion of the transposon into the genome was confirmed by Southern analysis (data not shown). Five thousand Tn917-LTV3 insertion mutants were screened in the microtiter plate assay. This analysis identified two mutants, designated Sag-1 (for surface-attached growth) and Sag-2, that adhered to the microtiter plates to a degree comparable to that of the wild type during the first hour of incubation, but subsequently the attached bacteria apparently were unable to grow (Fig. 1). The first hour is the time taken for the irreversible adhesion to the microtiter plates since washing after 1 h fails to remove the adhered bacteria (Fig. 1). Viable counts of the adhered bacteria after 1 and 24 h were performed. After 1 h the number of adhered bacteria was 5 × 105 (mean, n = 3) for the wild type, 3.7 × 105 (mean, n = 3) for Sag-1, and 3.5 × 105 (mean, n = 3) for Sag-2, with no statistical difference (P > 0.05) between the wild-type, Sag-1, and Sag-2 strains. After 24 h the number of viable adhered wild-type bacteria had risen to 1.6 × 109 (mean, n = 3), whereas the numbers of viable adhered Sag-1 and Sag-2 cells were unaltered at 3.7 × 105 (mean, n = 3) and 3.6 × 105 (mean, n = 3), respectively. This confirms the crystal violet staining (Fig. 1), demonstrating that there was no discernible difference in the initial attachment to the microtiter plate and that differences at 24 h reflect an inability of the adhered Sag-1 and Sag-2 mutants to grow. To confirm that the adhered Sag-1 and Sag-2 were impaired in surface-attached growth, the planktonic bacteria were removed after 1 h from the microtiter well and replaced by fresh medium (Fig. 2). Immediately after replacement with fresh medium, the number of viable adhered bacteria was 3.6 × 105 (mean, n = 3) for both Sag-1 and Sag-2, indicating that replacing the planktonic culture with fresh medium was not disrupting the adhered bacteria. After a lag, the adhered wild-type cells began to grow, reaching 6.4 × 108 (mean, n = 3) after 8 h, whereas both Sag-1 and Sag-2 showed no detectable growth with the total viable counts remaining at 3.6 × 106 (mean, n = 3) for both mutants (Fig. 2).
FIG. 1.
Adhesion and growth of L. monocytogenes in microtiter plates. Wild-type strain C52 (⧫), Sag-1 (▪), and Sag-2 (▴) were grown statically at 37°C in TSB in the wells of a microtiter plate. At each time point irreversibly attached bacteria were quantified by crystal violet staining as described in Materials and Methods. No change in crystal violet staining was observed in wells containing only TSB (•). The error bars represent the SEM (n = 6).
FIG. 2.
Growth of the adhered bacteria in a microtiter plate. Wild-type strain C52 (⧫), Sag-1 (▪), and Sag-2 (▴) were grown statically at 37°C in TSB in the wells of a microtiter plate for 1 h. At this point (depicted by the arrow) the planktonic bacteria were gently removed from the well and replaced by an equal volume of fresh TSB. Subsequently, at each time point attached bacteria were quantified by crystal violet staining. The error bars represent the SEM (n = 6).
The planktonic cultures of Sag-1 and Sag-2 in microtiter plates had growth rates that were indistinguishable from that of the wild-type strain (data not shown), and after 24 h the final OD630 was the same for all three cultures. Therefore, differences in the ability of the adhered bacteria to grow are not a consequence of a generalized reduction in growth rate or viability of the mutants in planktonic culture.
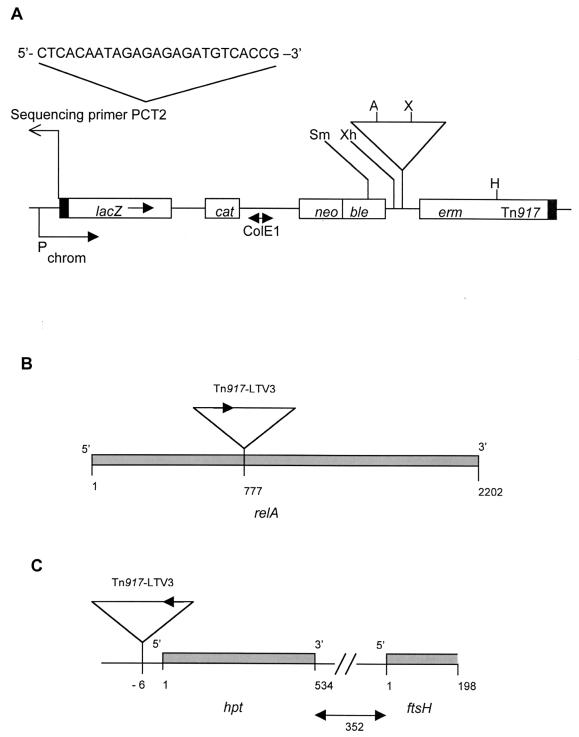
Southern blot analysis with a Tn917-LTV3-specific probe showed that each insertion mutant was different and carried a single copy of Tn917-LTV3 (data not shown). Genomic DNA flanking the lacZ-proximal end of Tn917-LTV3 of each mutant was recovered in E. coli after self-ligation of Asp718-digested Sag-1 chromosomal DNA and XbaI-digested Sag-2 chromosomal DNA (Fig. 3).
FIG. 3.
(A) Hypothetical chromosomal insertion of Tn917-LTV3. In this case a transcriptional fusion between the lacZ gene and a chromosomal promoter is shown. Only unique sites which can be used to clone sequence flanking the lacZ end are shown. Restriction enzyme abbreviations: X, XbaI; A, Asp7I8; Sm, SmaI; Xh, XhoI; H, HindIII. Antibiotic resistance genes: cat, chloramphenicol; neo, kanamycin-neomycin; ble, bleomycin; erm, erythromycin-lincomycin. (B) Site of Tn917-LTV3 insertion in mutant Sag-1. Nucleotide positions given are based on similarity with the relA gene of B. subtilis. (C) Site of Tn917-LTV3 insertion in mutant Sag-2. In panels B and C, the black arrows indicate the orientation of the lacZ gene of Tn917-LTV3.
Identification of the site of insertion in Sag-1 and Sag-2.
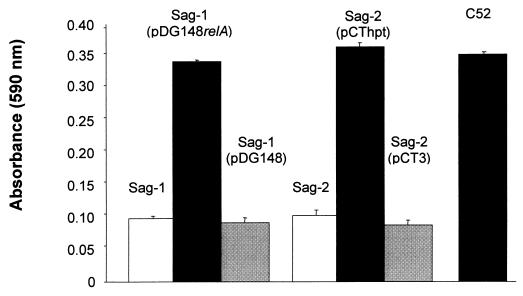
The nucleotide sequence of genomic DNA at the transposon junction in Sag-1 and Sag-2 was determined with primer PCT2 (Fig. 3A). In the case of Sag-1 more than 800 bp of sequence was obtained, and a BLASTX analysis (2) revealed that the predicted amino acid sequence had 79% similarity over 211 amino acids to the N terminus of the stringent response protein (p)ppGpp synthetase (RelA) of Bacillus subtilis. Based on this similarity, we can predict that, in strain Sag-1, Tn917-LTV3 has inserted 777 bp from the 5′ end of the L. monocytogenes relA gene (Fig. 3B). To confirm this conclusion, plasmid pDG148relA containing only the B. subtilis relA gene and its promoter (43) was introduced into strain Sag-1. The plasmid was able to complement the mutation and restored the biofilm formation of Sag-1 to that of the wild type (Fig. 4). The plasmid vector pDG148 (43) was unable to restore the ability of Sag-1 to form a biofilm (Fig. 4) and had no effect on the planktonic growth (data not shown). This result confirms that the inability of Sag-1 to replicate following adhesion is a consequence of the disruption of the L. monocytogenes relA gene and not due to polarity of the insertion on genes 3′ to the relA gene.
FIG. 4.
Complementation of the mutations in Sag-1 and Sag-2. Sag-1 was complemented with the relA gene of B. subtilis on plasmid pDG148relA. Sag-2 was complemented with plasmid pCThpt containing the wild-type hpt gene with a constitutive XylA promoter. The graph shows the growth of adhered bacteria quantified after 6 h as described in Materials and Methods. The error bars represent the SEM (n = 6).
In the case of Sag-2, more than 1,000 bp of sequence was obtained, and a BLASTX analysis (2) identified an open reading frame of 178 amino acids immediately 3′ of the transposon, the predicted amino acid sequence of which had 75% similarity to the Hgprt enzyme of B. subtilis. The sequence analysis indicates that in strain Sag-2 the Tn917-LTV3 has inserted in the promoter region, 6 bp from the 5′ end of the L. monocytogenes hpt gene (Fig. 3C). A second partial open reading frame of 66 amino acids was identified 352 bp 3′ to the hpt gene (Fig. 3C) which had 50% similarity to the FtsH protein of B. subtilis (34). The L. monocytogenes hpt gene was amplified and cloned into the shuttle plasmid pCT3 (41) to generate pCThpt. Plasmid pCThpt was introduced into strain Sag-2 and restored the ability to form a biofilm to that of wild type (Fig. 4). The plasmid vector pCT3 (41) was unable to restore the ability of Sag-2 to form a biofilm (Fig. 4) and had no effect on the planktonic growth of Sag-2 (data not shown). This result confirms that the inability of Sag-2 to grow after adhesion is a consequence of the disruption of the L. monocytogenes hpt gene and not due to polarity of the insertion on genes 3′ to the hpt gene.
Mutations in relA and hpt abolish the production of (p)ppGpp in response to amino acid starvation, and relA is transcribed during surface-attached growth.
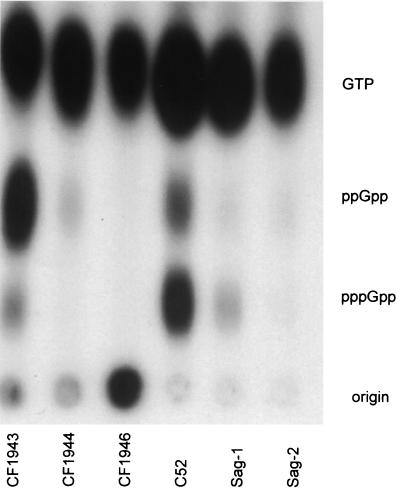
To confirm that the relA mutation abolished (p)ppGpp synthesis in response to nutritional stress, (p)ppGpp accumulation was measured in cells labeled with [32P]H3PO4 during amino acid starvation. The wild-type strain C52 accumulated both pppGpp and ppGpp in response to amino acid starvation, thus confirming the induction of a stringent response. In contrast, the relA mutation in Sag-1 dramatically reduced the synthesis of (p)ppGpp to barely detectable levels (Fig. 5). Unexpectedly, the hpt mutation in Sag-2 also abolished the synthesis of any detectable (p)ppGpp. As a control, the pattern of (p)ppGpp accumulation in an E. coli wild-type strain (CF1943), together with an isogenic relA mutant (CF1944) and a relA spoT double mutant (CF1946), was analyzed (Fig. 5). As predicted, strain CF1943 accumulated ppGpp and, to a lesser extent, pppGpp, whereas CF1944 had reduced levels of ppGpp and undetectable pppGpp, with the double mutant lacking any detectable pppGpp or ppGpp (Fig. 5).
FIG. 5.
Accumulation of (p)ppGpp after amino acid starvation. Bacteria were labeled with [32P]H3PO4 in MOPS starvation medium lacking phosphate and amino acids, but containing serine hydroxamate (1 mg ml−1) and ℒ-valine (0.5 mg ml−1), for 30 min at 37°C. An equal volume of 13 M formic acid was added and the samples were subjected to freeze (dry ice)-thawing twice. Acid extracts were centrifuged briefly, and the supernatants were spotted onto polyethyleneimine cellulose for thin-layer chromatography in 1.5 M K2HPO4 (pH 3.4). Strains: CF1943, E. coli W3110 parental strain; CF1944, E. coli CF1943 (ΔrelA::kan); CF1946, E. coli CF1943 (ΔrelA::kan ΔspoT::cm).
To establish that a stringent response is induced after attachment, the level of transcription of the relA gene in adhered cells was measured. To achieve this, the level of β-galactosidase was assayed in Sag-1, in which the lacZ gene in Tn917-LTV3 is in the same orientation as the relA gene (Fig. 3B). After 1 h, the level of β-galactosidase in both the planktonic and adhered Sag-1 cells in the microtiter plate was measured. Barely detectable β-galactosidase activity was found in the planktonic cells in the microtiter well at 4 ± 0.06 RLU mg−1 (mean ± SEM), whereas the adhered cells expressed much higher levels of β-galactosidase activity (931 ± 45 RLU mg−1). This demonstrates that during the period immediately after adhesion the adhered cells undergo a stringent response and transcription of the relA gene takes place.
Mutations in relA and hpt cause avirulence in a murine model of infection.
After i.v. inoculation with C52, high viable counts were detected in both the livers and spleens of infected animals after 96 h (Table 1). All five animals showed visible symptoms of infection, namely, a hunched appearance with ruffled fur. In contrast, no viable Sag-1 was detected in the livers of animals 96 h after infection, and low numbers were detectable in the spleens of two of the five infected animals (Table 1). Likewise, low numbers of Sag-2 were detectable in the livers and spleens of two of the five infected animals (Table 1). No overt sign of infection was detected in any animal during the 96 h after inoculation with either Sag-1 or Sag-2.
TABLE 1.
Mouse infection data for wild-type (C52) and Sag-1 and Sag-2 strains
| Mouse type and no. | Log10 CFU/mg of organ tissue (SEM)a
|
|
|---|---|---|
| Spleen | Liver | |
| Wild-type | ||
| 1 | 4.38 (2.84) | 3.30 (2.37) |
| 2 | 4.23 (3.52) | 3.34 (2.17) |
| 3 | 3.53 (2.35) | 2.92 (2.24) |
| 4 | 4.25 (3.52) | 3.25 (1.57) |
| 5 | 4.38 (3.61) | 3.40 (2.33) |
| Sag-1 | ||
| 1 | ND | ND |
| 2 | ND | ND |
| 3 | ND | ND |
| 4 | 0.96 (0.96) | ND |
| 5 | 1.46 (1.46) | ND |
| Sag-2 | ||
| 1 | ND | ND |
| 2 | ND | ND |
| 3 | 1.74 (1.74) | 0.00 (0) |
| 4 | 0.44 (0.44) | 0.00 (0) |
| 5 | ND | ND |
ND, not detectable.
DISCUSSION
L. monocytogenes is capable of adhesion and biofilm formation on a number of abiotic surfaces (29). By using polystyrene as a model hydrophobic surface, we obtained two Tn917-LTV3 insertion mutants unable to grow after initial adhesion (Fig. 1 and 2). The inability of the mutants to grow in a biofilm (attached state) despite their ability to grow in a planktonic state suggests that the physiological requirements of growth under these conditions are distinct.
Characterization of Sag-1 and Sag-2 identified insertions in the relA and hpt genes, respectively. Unlike the wild-type strain, both mutants were unable to mount a stringent response and accumulate (p)ppGpp in response to amino acid starvation (Fig. 5). The stringent response is well characterized in E. coli and is a pleiotropic physiological response in which (p)ppGpp indirectly reduces protein synthesis by the repression of stable RNA synthesis (6). Gram-negative bacteria possess two (p)ppGpp synthetase activities. The relA gene encodes (p)ppGpp synthetase, while spoT encodes a (p)ppGpp 3′-pyrophosphohydrolase that also possesses (p)ppGpp synthetase activity (44). Gram-positive bacteria appear to express only one protein, termed RelA, which contains both (p)ppGpp activities (31, 43). The observation that low levels of pppGpp were detectable in Sag-1 following stress induction (Fig. 5) suggests that the Tn917-LTV3 insertion at amino acid position 259 in the relA gene in Sag-1 does not abolish all (p)ppGpp synthetase activity. The most likely explanation for this is that a truncated RelA protein with residual (p)ppGpp synthetase activity is still being expressed in Sag-1. This is in keeping with previous findings in B. subtilis, where a mutation which resulted in a truncated RelA protein consisting of the first 240 amino acids showed low-level (p)ppGpp synthetase activity (43). However, the possibility that another enzyme can synthesize low-level (p)ppGpp cannot at this stage be ruled out. The role of (p)ppGpp in the growth of adhered L. monocytogenes is novel and intriguing. The observation that transcription of the relA gene is induced following adhesion suggests that the ability to mount a stringent response and undergo physiological adaptation to nutrient deprivation after attachment is essential for the subsequent growth of the adhered bacteria. This is in keeping with the results of other studies indicating that bacteria undergo a number of physiological changes during the early stages of surface-attached culture (10).
Other stress response proteins have been shown to be important in surface-attached growth and subsequent biofilm formation, including the gram-negative sigma factor ςS. An rpoS mutant of E. coli was reduced in biofilm cell density by 50% in comparison to the wild type, and the arrangement of cells within the biofilm was altered, suggesting that ςS is important for biofilm physiology (1). Mutants unable to synthesize (p)ppGpp fail to induce the RpoS regulon upon entering stationary phase, and it has been shown that (p)ppGpp is required for the synthesis of ςS under stringent conditions, with (p)ppGpp being required for the induction of ςS-dependent promoters (16, 25). It is not known whether an equivalent interaction takes place in gram-positive bacteria between the functional ςS analogue, ςB, and (p)ppGpp.
The Tn917-LTV3 insertion in Sag-2 was in the promoter region of the hpt gene (Fig. 3). The Hgprt enzyme is a 6-oxopurine phosphoribosyltransferase that converts the purine base (guanine) into the corresponding nucleotide (GMP) and is involved in the purine salvage pathway (30, 33). Salvage pathways perform a number of functions in bacteria. First, they scavenge exogenous preformed bases for nucleotide synthesis. Second, they act to reutilize bases generated endogenously as a result of nucleotide turnover. Third, they provide a route for the catabolism of the pentose moieties of exogenous nucleosides (46). In the presence of a functional de novo pathway for purine biosynthesis, Hgprt is not essential for growth, and Sag-2 was capable of growing in defined medium lacking exogenous purines (42). The lack of detectable (p)ppGpp in Sag-2 following amino acid starvation indicates that Sag-2 cannot undergo a stringent response, and this probably accounts for its inability to grow after adhesion. The absence of (p)ppGpp in Sag-2 is curious and suggests that a functional Hgprt is important in the synthesis of (p)ppGpp. One interpretation is that the RelA protein has a high Km for GDP/GTP and that Hgprt is needed to maintain the intracellular GDP/GTP at levels sufficient for RelA activity. The observation that Hgprt is inhibited by (p)ppGpp (20) would support this notion.
Both Sag-1and Sag-2 were avirulent in the murine model of infection, although the hemolytic activity and secreted protein profiles of both mutants were unaffected in vitro (data not shown). Sag-1 and Sag-2 were either absent or barely detectable in the livers and spleens of infected animals (Table 1), suggesting that both mutants were rapidly cleared by the host. It has been shown that the accumulation of (p)ppGpp in response to amino acid depletion is necessary for entry of intracellular Legionella pneumophila into a virulent state that permits escape from the dying infected amoeba (18). Virulence genes of L. monocytogenes are regulated by PrfA (8, 38), and intracellular growth and invasion by L. monocytogenes is well characterized (7). However, there is little data on the physiological adaptation of L. monocytogenes to the nutrient stresses associated with intracellular growth. It has been shown that mutations in genes encoding enzymes for the biosynthesis of nucleotides and for the uptake of amino acids reduce virulence (23), as do mutations in the clpE gene, which is essential for cell division (32). In addition, it has been shown that the ClpP serine protease of L. monocytogenes is essential for intracellular invasion and virulence of L. monocytogenes (14). The ClpP protein is a subunit of a larger Clp ATP-dependent protease that acts as a molecular chaperone, assisting the proper folding, refolding, and assembly of proteins. In B. subtilis it has been shown that, under stringent conditions, the ClpP protease is upregulated following amino acid depletion and the initiation of a stringent response (43). As such, the inability of Sag-1 and Sag-2 to mount a stringent response could lower the levels of the ClpP protease, the consequence of which would be a reduction in virulence.
In summary, we have demonstrated that the ability to mount a stringent response and synthesize (p)ppGpp appears to be important in the growth of bacteria after adhesion and in the virulence of L. monocytogenes in mice. Since the growth of adhered bacteria is an early step in the development of a biofilm, it is likely that Sag-1 and Sag-2 will also be defective in biofilm formation. The next stage is to understand how (p)ppGpp regulation is achieved in L. monocytogenes and to determine the molecular basis for the role of this molecule in biofilm formation and in infection.
Acknowledgments
Work in the laboratory of I.S.R. is funded by the Lister Institute of Preventive Medicine, the BBSRC, and the Wellcome Trust. C.M.T. was the recipient of an MAFF studentship. M.B. was a recipient of an EPSRC studentship.
We gratefully acknowledge the generous help and advice of Mike Cashel.
REFERENCES
- 1.Adams, J. L., and R. J. C. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4286–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. L. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, M. 1996. Studies on the adhesion of Listeria monocytogenes to leaf and model surfaces. Ph.D thesis. University of Manchester, Manchester, United Kingdom.
- 4.Camilli, A., D. A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows directs cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cashel, M. 1994. Detection of (p)ppGpp accumulation patterns in Escherichia coli mutants, p.341–356. In, C. W. Adolph (ed.), Methods in molecular genetics, vol. 3. Academic Press, Inc., San Diego, Calif.
- 6.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p.1458–1496. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.
- 7.Chakraborty, T. 1999. Molecular and cell biological aspects of infection by Listeria monocytogenes. Immunobiology 201:155–163. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty, T., M. Leimeisher-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Co-ordinate regulation of the virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W., R. T. Irvin, and K. J. Cheng. 1981. The bacterial glycocalyx in nature and disease. Annu. Rev. Microbiol. 35:299–324. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. [DOI] [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of bacterial biofilm. Science 280:295–298. [DOI] [PubMed] [Google Scholar]
- 12.Demuth, D. R., Y. Duan, W. Brookes, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403–413. [DOI] [PubMed] [Google Scholar]
- 13.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286–1294. [DOI] [PubMed] [Google Scholar]
- 15.Genevaux, P., S. Muller, and P. Bauda. 1996. A rapid screening procedure to identify mini-Tn10 insertion mutants in Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 142:27–30. [DOI] [PubMed] [Google Scholar]
- 16.Gentry, D. R., J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor ςS is positively regulated by ppGpp. J. Bacteriol. 175:7982–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong, K., and M. C. Herzberg. 1997. Streptococcus sanguis expresses a 150-kilodalton two domain adhesin: characterization of several independent adhesin epitopes. Infect. Immun. 65:3815–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721–731. [DOI] [PubMed] [Google Scholar]
- 19.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013–1024. [DOI] [PubMed] [Google Scholar]
- 20.Hochstadt-Ozer, J., and M. Cashel. 1972. The regulation of purine utilisation in bacteria. J. Biol. Chem. 21:7067–7072. [PubMed] [Google Scholar]
- 21.Jenkinson, H. F. 1995. Genetic analysis of adherence by oral streptococci. J. Ind. Microbiol. 15:186–192. [DOI] [PubMed] [Google Scholar]
- 22.Jones, C. E., G. Shama, D. Jones, I. S. Roberts, and P. W. Andrew. 1997. Physiological and biochemical studies on psychrotolerance in Listeria monocytogenes. J. Appl. Microbiol. 83:31–35. [DOI] [PubMed] [Google Scholar]
- 23.Klarsfield, A. D., P. L. Goossens, and P. Cossart. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE, and an arginine ABC transporter gene apr. J. Mol. Microbiol. 13:585–597. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn, M., T. Pfeuffer, L. Greiffenberg, and W. Goebel. 1999. Host cell signal transduction during Listeria monocytogenes infection. Arch. Biochem. Bipophys. 372:166–172. [DOI] [PubMed] [Google Scholar]
- 25.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of ςS. J. Biol. Chem. 275:14795–14798. [DOI] [PubMed] [Google Scholar]
- 26.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorber, B. 1990. Clinical listeriosis: implications for pathogenesis, p.41–49. In A. L. Miller, J. L. Smith, and G. A. Somkuti (ed.), Foodborne listeriosis. Society for Industrial Microbiology. Elsevier Science, Amsterdam, The Netherlands.
- 28.Low, J. C., and W. Donachie. 1997. A review of Listeria monocytogenes and listeriosis. Vet. J. 153:9–29. [DOI] [PubMed] [Google Scholar]
- 29.Mafu, A. A., D. Roy, J. Goulet, and L. Savoie. 1991. Characterization of physichochemical forces involved in adhesion of Listeria monocytogenes to surfaces. Appl. Environ. Microbiol. 57:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLauchlin, J. 1997. The pathogenicity of Listeria monocytogenes: a public health perspective. Rev. Med. Microbiol. 8:1–14. [Google Scholar]
- 31.Mechold, U., M. Cashel, K. Steiner, D. Gentry, and H. Malke. 1996. Functional analysis of a relA/spoT homolog from Streptococcus equisimilis. J. Bacteriol. 178:1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair, S., C. Frehel, L. Nguyen, V. Escuyer, and P. Berche. 1999. ClpE, a novel member of the HSP1000 family is involved in cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 31:185–196. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson, D., and A. A. Lauridsen. 1992. Isolation of purine auxotrophic mutants of Lactococcus lactis and characterization of the gene hpt encoding hypoxanthine guanine phosphoribosyltransferase. Mol. Gen. Genet. 235:359–364. [DOI] [PubMed] [Google Scholar]
- 34.Ogasawara, N., S. Nakai, and H. Yoshikawa. 1994. Syst. sequencing of the 180-kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1:1–14. [DOI] [PubMed] [Google Scholar]
- 35.O’Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas aeruginosa WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461. [DOI] [PubMed] [Google Scholar]
- 36.O’Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm deveploment. Mol. Microbiol. 30:295–304. [DOI] [PubMed] [Google Scholar]
- 37.Park, S. F., and G. S. A. B. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin treated cells. Gene 94:129–132. [DOI] [PubMed] [Google Scholar]
- 38.Portnoy, D. A., T. Chakrabarty, W. Goebel, and P. Cossart. 1992. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60:1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Stephens, J. C., I. S. Roberts, D. Jones, and P. W. Andrew. 1991. Effect of growth temperature on virulence of strains of Listeria monocytogenes in the mouse: evidence for dose dependence. J. Appl. Bacteriol. 70:239–244. [DOI] [PubMed] [Google Scholar]
- 41.Taylor, C. M. 2000. The molecular determination of the adhesion of Listeria monocytogenes to plant surfaces. Ph.D. thesis. University of Manchester, Manchester, United Kingdom.
- 42.Taylor, C. M., and I. S. Roberts. Unpublished results
- 43.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65–79. [DOI] [PubMed] [Google Scholar]
- 44.WHO Working Group. 1988. Foodborne listeriosis. Bull. W. H. O. 66:421–428. [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990. [PubMed] [Google Scholar]
- 46.Zalkin, H., and P. Nygaard. 1996. Biosynthesis of purine nucleotides, p.561–579. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.