Abstract
Using Meta-BASIC, a highly sensitive method for detection of distant similarity between proteins, we have identified another potential PD-(D/E)XK endonuclease in human herpesvirus 1 (HHV-1) encoded by the UL24 gene. The universal presence of UL24 in completed herpesviral genomes of three major subfamilies, Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae, suggests a fundamental role for this predicted PD-(D/E)XK endonuclease activity in the viral life cycle.
Herpesviridae encompass a diverse family of animal viruses that possess a large linear double-stranded DNA genome (120 up to 230 kbp), share a common virion morphology, and retain the capacity to enter a latent phase of infection in particular host cells (24). Historically herpesvirus species, including eight known to infect humans (HHV-1 to HHV-8), have been classified according to these biological characteristics into three major subfamilies: Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. Recent analysis of herpesviral gene content and sequence similarity substantiates this classification scheme and defines a subset of common genes (43 core sequences) thought to be involved in fundamental viral processes such as capsid assembly and egress from the nucleus, as well as DNA replication, processing, and packing into assembled virions (8). Of the core herpesviral genes, only one (UL24 in HHV-1) remains unassigned to any broad functional category. Using Meta-BASIC (http://basic.bioinfo.pl) (10), a highly sensitive fold recognition method that applies a comparison of sequence profiles enriched by predicted secondary structure, we predict that the UL24 gene encodes a novel PD-(D/E)XK endonuclease belonging to a large superfamily of restriction endonuclease-like fold proteins.
A consensus UL24 family (PFAM [3] accession no. PF01646) sequence maps with an above-threshold Meta-BASIC score (Z-score, 12.22) to a domain of unknown function, DUF91 (PFAM accession no. PF01939), recently identified as possessing a restriction endonuclease-like fold (17). In addition, for the majority of UL24 family members Meta-BASIC provides reliable hits (Z-scores above 12) to several PD-(D/E)XK endonuclease families such as DUF91, DUF911 (PFAM accession no. PF06023) (17), or SfsA (PFAM accession no. PF03749) (18). Further analysis performed using a Meta Server (http://bioinfo.pl/meta) (9) that combines several top-of-the-line fold recognition methods revealed additional weak hits (for both family consensus and HHV-1 UL24 sequences) to several structures with a restriction endonuclease-like fold: Holliday junction-resolving enzymes (PDB 1ob8, PDB 1hhl and PDB 1ipi), DNA mismatch repair protein (PDB 1azo), and DNA restriction endonuclease NaeI (PDB 1ev7). The correct but highly nontrivial fold assignment includes good mapping of predicted (mainly with PSI-PRED) (16) and observed secondary structure elements, general conservation of hydrophobicity patterns, and absolute conservation of the signature PD-(D/E)XK endonuclease motifs critical for function (Fig. 1).
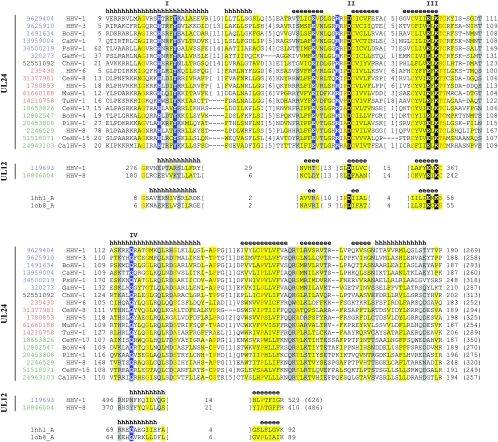
FIG. 1.
Multiple sequence alignment for selected UL24 and UL12 family representatives and Holliday junction-resolving structures. UL24 and UL12 family sequences are labeled according to NCBI gene identification (gi) number followed by an abbreviation of the species name according to the Universal Virus Database of the International Committee on Taxonomy of Viruses (http://www.ncbi.nlm.nih.gov/ICTVdb): BoHV-1, Bovine herpesvirus 1; BoHV-4, Bovine herpesvirus 4; CalHV-3, Callitrichine herpesvirus 3; CaHV-1, Canine herpesvirus 1; CeHV-15, Cercopithecine herpesvirus 15; CeHV-17, Cercopithecine herpesvirus 17; CeHV-8, Cercopithecine herpesvirus 8; ChHV-1 Chelonid herpesvirus 1; GaHV-1, Gallid herpesvirus 1; HHV-1 Human herpesvirus 1; HHV-3, Human herpesvirus 3; HHV-5, Human herpesvirus 5, HHV-6, Human herpesvirus 6; HHV-8, Human herpesvirus 8; MuHV-1, Murid herpesvirus 1; PlHV-1, Porcine lymphotropic herpesvirus 1; PsHV-1, Psittacid herpesvirus 1; and TuHV-1, Tupaiid herpesvirus 1. Labels are colored according to taxonomy: alphaherpesviruses in blue, betaherpesviruses in red, gammaherpesviruses in green, and unclassified herpesvirus in black. The residue numbers are indicated before and after each sequence, with total sequence lengths following in parentheses. The numbers of excluded residues are specified in square brackets. Residue conservation in the UL24 family (90% identity threshold) is denoted by the following scheme: uncharged at dominantly hydrophobic positions, highlighted in yellow; charged or polar at dominantly hydrophilic positions, highlighted in gray; and small residues, letters in red. Conserved PD-(D/E)XK signature amino acids are highlighted in black, while additional UL24 family invariant residues are highlighted in blue. Restriction endonuclease-like motifs are labeled above the corresponding residue columns. Locations of predicted (gi 9629404 and gi 119693) and observed (PDB 1hh1) secondary structure elements (e, β-strand; h, α-helix) are marked above the corresponding sequences, respectively. The UL24 family multiple-sequence alignment was generated using the PCMA (22) program, while structurally conserved elements of restriction endonuclease-like fold from Holliday junction resolvases (PDB 1ob8 and PDB 1hh1) and UL12 representatives were added manually, taking into account mapping of secondary structures, hydrophobic profiles, and conservation of presumably catalytic residues. UL24 family sequences were collected using exhaustive transitive PSI-BLAST (1) searches performed against the NCBI nonredundant protein database (posted 15 June 2005, 2,506,589 sequences) until profiles convergence (inclusion threshold, 0.01) and were filtered by identity.
Existing classification of PD-(D/E)XK endonuclease structures groups a number of families, including several restriction endonucleases (EcoRI, EcoRII, BamHI, BglI, Cfr10I, NaeI, etc.), DNA repair enzymes (MutH and Vsr), Holliday junction resolvases (Hjc and Hje), and other nucleotide-cleaving enzymes, into a large and diverse superfamily of restriction endonuclease-like fold proteins (20). These enzymes contribute to important biological functions such as protecting against foreign DNA (restriction endonucleases), repairing damaged DNA (MutH and Vsr), resolving Holliday junctions (endonuclease I, Hjc, Hje, and XPF/Rad1/Mus81-dependent nuclease), or recombining DNA (lambda exonuclease and TnsA) (4, 5, 23). The common core structure (αβββαβ topology) includes a four-stranded, mixed β-sheet flanked by an α-helix on both sides. The PD-(D/E)XK signature (2, 6, 17) provides active site residues responsible for cleaving a variety of nucleic acid substrates.
UL24 homologs encompass an N-terminal restriction endonuclease-like domain (ααβββαβββα topology, with the common core elements underlined) followed by a low-complexity, highly basic region that is probably unstructured. UL24 family proteins include all restriction endonuclease-like core elements (with a short α-helical insertion between the N-terminal core α-helix and first β-strand) in addition to three C-terminal elements (two β-strands and α-helix) that probably extend the core of the fold. UL24 sequences retain absolute conservation of the PD-(D/E)XK signature (motifs II and III, Fig. 1). Several additional UL24 invariant residues (H and Y in motif I, Q in motif IV, and E and R preceding motif II; highlighted in blue in Fig. 1) map near the presumed catalytic residues and are poised to supplement the active site architecture, perhaps contributing to the substrate specificity of the enzyme family.
Assignment of UL24 to the PD-(D/E)XK endonuclease superfamily provides new insight into its functional role in herpesviral replication. Initially, the UL24 gene was identified in the HSV-1 genome as an open reading frame overlapping the gene for thymidine kinase (tk) (11). Disruption of conserved UL24 sequence elements [including the first D of the signature PD-(D/E)XK motif] was shown to correlate with a small syncytial plaque phenotype observed in mutants initially designed to study tk expression (15). Subsequent analysis of UL24 mutants with wild-type tk activity substantiated the small-plaque phenotype and showed less efficient replication in the eye (10- to 30-fold reduction) as well as severe impairment of replication in ganglia (14). These phenotypes support a general and nonessential role for UL24 activity in mediating viral replication and perhaps membrane fusion events.
Interestingly, another PD-(D/E)XK nuclease (alkaline nuclease) is encoded in the HHV-1 genome by the UL12 gene (7). Like the UL24 gene product, UL12 is not essential for herpesviral replication (26), although its mutation decreases viral DNA synthesis and processing. UL12 exonuclease activity mediates strand exchange (in vitro) in DNA recombination events thought to be an integral part of herpesviral replication (25). Although UL12 and UL24 do not share any significant sequence similarity [apart from the PD-(D/E)XK signature], these two enzymes might perform redundant nucleotide cleavage activities essential for low levels of herpesviral replication.
The universal presence of UL24 in completed avian-mammalian and reptilian herpesviruses (although not detected in amphibian-fish and invertebrate herpesviruses) indicates a fundamental role of this protein in the viral life cycle. Identification of UL24 as a potential PD-(D/E)XK endonuclease suggests that this role might involve cleaving nucleic acid substrate. Consistent with this hypothesized activity, UL24 protein localizes to the nuclei of infected cells (12, 21). Accordingly, UL24 could participate in homologous recombination of viral DNA, a process intimately associated with replication and thought to drive herpesviral evolution (25). Alternatively, UL24 could mediate linear-to-circular genome transitions that emerge in herpesviral latency (13) or might recognize and resolve specific genome structures that result from concurrent replication and recombination. Finally, UL24 could act on host genetic material, perhaps to trigger cellular DNA damage response machinery shown to accumulate at viral replication centers (19). Ultimately, experimental investigations should address the predicted UL24 PD-(D/E)XK endonuclease activity and clarify potential substrates, allowing further insight into the fundamental role of this protein in herpesviral latency and replication.
Acknowledgments
This work was supported in part by the NIH grant GM67165 to N.V.G. and by MNI and the European Commission with grants LSHG-CT-2003-503265 and LSHG-CT-2004-503567 to L.R.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Survey and summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhagwat, A. S., and M. Lieb. 2002. Cooperation and competition in mismatch repair: very short-patch repair and methyl-directed mismatch repair in Escherichia coli. Mol. Microbiol. 44:1421-1428. [DOI] [PubMed] [Google Scholar]
- 5.Bujnicki, J. M. 2003. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the “midnight zone” of homology. Curr. Protein Pept. Sci. 4:327-337. [DOI] [PubMed] [Google Scholar]
- 6.Bujnicki, J. M., and L. Rychlewski. 2001. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 3:69-72. [PubMed] [Google Scholar]
- 7.Bujnicki, J. M., and L. Rychlewski. 2001. The herpesvirus alkaline exonuclease belongs to the restriction endonuclease PD-(D/E)XK superfamily: insight from molecular modeling and phylogenetic analysis. Virus Genes 22:219-230. [DOI] [PubMed] [Google Scholar]
- 8.Davison, A. J., D. J. Dargan, and N. D. Stow. 2002. Fundamental and accessory systems in herpesviruses. Antivir. Res. 56:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Ginalski, K., A. Elofsson, D. Fischer, and L. Rychlewski. 2003. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19:1015-1018. [DOI] [PubMed] [Google Scholar]
- 10.Ginalski, K., M. von Grotthuss, N. V. Grishin, and L. Rychlewski. 2004. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 32:W576-W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gompels, U., and A. Minson. 1986. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology 153:230-247. [DOI] [PubMed] [Google Scholar]
- 12.Hong-Yan, Z., T. Murata, F. Goshima, H. Takakuwa, T. Koshizuka, Y. Yamauchi, and Y. Nishiyama. 2001. Identification and characterization of the UL24 gene product of herpes simplex virus type 2. Virus Genes 22:321-327. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, J. G., S. L. Martin, and D. M. Coen. 1989. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J. Virol. 63:1839-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 17.Kinch, L. N., K. Ginalski, L. Rychlewski, and N. V. Grishin. 2005. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 33:3598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosinski, J., M. Feder, and J. M. Bujnicki. 12 July 2005, posting date. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics 6:172. [Online.] doi: 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 102:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, A., and D. M. Coen. 2002. Identification, localization, and regulation of expression of the UL24 protein of herpes simplex virus type 1. J. Virol. 76:10821-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei, J., R. Sadreyev, and N. V. Grishin. 2003. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics 19:427-428. [DOI] [PubMed] [Google Scholar]
- 23.Sharples, G. J. 2001. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 39:823-834. [DOI] [PubMed] [Google Scholar]
- 24.Subak-Sharpe, J. H., and D. J. Dargan. 1998. HSV molecular biology: general aspects of herpes simplex virus molecular biology. Virus Genes 16:239-251. [DOI] [PubMed] [Google Scholar]
- 25.Thiry, E., F. Meurens, B. Muylkens, M. McVoy, S. Gogev, J. Thiry, A. Vanderplasschen, A. Epstein, G. Keil, and F. Schynts. 2005. Recombination in alphaherpesviruses. Rev. Med. Virol. 15:89-103. [DOI] [PubMed] [Google Scholar]
- 26.Weller, S. K., M. R. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 71:2941-2952. [DOI] [PubMed] [Google Scholar]