Abstract
JC virus (JCV), a human polyomavirus, exhibits oncogenic activity in rodents and primates. The large tumor antigens (TAgs) of the polyomaviruses play key roles in viral replication and oncogenic transformation. Analyses of JCV TAg phosphorylation mutants indicated that the amino-terminal phosphorylation site at threonine 125 (T125) is critical to TAg replication function. This site is also conserved in the TAg splice variants T′135, T′136, and T′165. By constructing stable cell lines expressing JCV T125A and T125D mutants, we show that mutation of this phosphorylation site to alanine generates an unstable TAg; however, the stability of the three T′ proteins is unaffected. JCV T125A mutant proteins bind the retinoblastoma protein (RB) family members p107 and p130 with slightly reduced efficiencies and fail to induce the release of transcriptionally active E2F from RB-E2F complexes. On the other hand, cell lines expressing JCV T125D mutant proteins produce stable TAg and T′ proteins which bind p107 and p130 more efficiently than do the wild-type proteins. In addition, T125D mutant proteins efficiently induce the release of E2F from RB-E2F complexes. T125D mutant cell lines, unlike the T125A mutant lines, continue to grow under conditions of low serum concentration and anchorage independence. Finally, both T125A and T125D mutant viruses are replication defective. Phosphorylation of the T125 site is likely mediated by a cyclin-cyclin-dependent kinase, suggesting that JCV TAg and T′ protein functions that mediate viral replication and oncogenic transformation events are regulated in a cell cycle-dependent manner.
The human polyomavirus JC virus (JCV) may persist in the kidneys and brains of healthy individuals at subclinical levels but may cause the fatal demyelinating disease progressive multifocal leukoencephalopathy in immunocompromised patients. Members of the Polyomaviridae family replicate in actively dividing cells because they rely on the host cell's machinery to copy their DNAs and produce infectious virions (14). The best studied of these viruses, simian virus 40 (SV40), shares 69% sequence similarity with JCV. Both viruses produce a multifunctional T protein (TAg) that mediates viral DNA replication and promotes the transition of cells from G0 into S phase (6). Depending on the cell type, the latter event may either support a lytic infection of permissive cells or contribute to the transformation of nonpermissive cells.
Hypophosphorylated forms of retinoblastoma (RB) pocket proteins bind members of the E2F family of transcription factors, thereby providing a gatekeeper function that prevents unscheduled cell cycle progression. The RB-E2F complexes regulate genes that control entry into and progression through mitosis, and they coordinate these cell cycle programs via regulation of checkpoint controls, DNA damage responses, apoptosis, and differentiation (5). Upon receipt of mitogenic signals, cells express cyclin proteins that interact with specific cyclin-dependent kinases (Cdk) to promote cell cycle transitions. Cyclin-Cdk complexes phosphorylate pRb, p107, and p130, thereby causing the release of their E2F partners, which activate the expression of S-phase genes (21, 26). DNA tumor virus proteins, including TAg, override normal cell cycle regulation by binding to RB proteins and displacing the E2F proteins, resulting in far wider consequences than simply increasing cellular proliferation (5).
Polyomavirus TAgs have multiple functional domains that support their roles in promoter activation and viral DNA replication. SV40 TAg has two phosphorylation domains containing eight or more sites, clustered between serine 106 (S106) and threonine 124 (T124) at its amino terminus and S639 and T701 at its carboxy terminus (24). Mutations to the carboxy-terminal sites affect phosphorylation at the amino-terminal sites (23). Posttranslational modifications of SV40 TAg modulate its DNA binding (27), replication (20), and p53 binding (4) activities. Phosphopeptide mapping of JCV TAg also revealed two phosphorylation domains (31), and a mutation to T125 abolished viral DNA replication (32). To initiate viral DNA replication, monomeric TAg oligomerizes to form hexamer and double-hexamer complexes in the presence of ATP, and these TAg structures bind and unwind the viral origin of replication (reviewed in reference 1 and references therein). The formation of the first SV40 TAg hexamer is independent of the phosphorylation status of the critical T124 residue (1, 17). However, mutation of this residue to alanine (T124A) disrupts formation of the second SV40 TAg hexamer and origin unwinding without preventing specific DNA binding activity (1, 17-19, 38). Furthermore, mutation of T124 to aspartic acid (T124D) does not affect double-hexamer formation, but DNA binding of such as mutant is inefficient (12). These results suggest that T124 phosphorylation leads to changes in TAg-DNA interactions to facilitate structural changes required for hexamer formation, origin unwinding, and DNA replication (12).
The present report details our investigation of two mutants of JCV TAg altered at amino acid residue 125 and of the three TAg isoforms, T′135, T′136, and T′165. The mutation of T125 to alanine (T125A) introduces an unphosphorylatable residue at this position, while mutation to aspartic acid (T125D) yields a negatively charged carboxyl group that may mimic a permanent phosphate group. The effects of these alterations on the function and stability of the JCV early proteins are presented.
MATERIALS AND METHODS
Plasmids.
CMV-JCVE expresses the entire JCV early region via the cytomegalovirus (CMV) early promoter and was constructed by replacing the intron-spanning region (nucleotides [nt] 4413 to 4877) from CMV-JCT (2) with that from pM1TCR1A (8), using a PflMI-EcoNI restriction enzyme fragment. BS/JCV(T125A) was created by ligating the JCV(T125A) genome from the recombinant pJCV-T125A clone (32) into the pBluescript II SK(−) vector, using an EcoRI restriction site. BS/JCV(T125D) was created by using a PCR-based site-directed mutagenesis approach (Stratagene) with primers T125Dfwd (5′-CCCAACACTCTGACCCACCTAAAAAG-3′) and T125Drev (5′-CTTTTTAGGTGGGTCAGAGTGTTGGG-3′) (mutations are shown in bold and underlined). CMV-JCVE(T125A) and CMV-JCVE(T125D) were constructed by inserting PflMI-BglII fragments containing either the T125A mutation from BS/JCV(T125A) or the T125D mutation from BS/JCV(T125D), respectively, into CMV-JCVE. Each DNA construct was confirmed by sequence analysis at the nucleic acid facility of the Huck Institutes of the Life Sciences at Penn State University.
Luciferase reporter plasmids containing either four copies of the E2F-1 promoter element (pE2F1-luc) or four copies of the E2F-4 promoter element (pE2F4-luc) and an isogenic construct lacking E2F promoter elements (ΔE2F-luc) were kind gifts from J. R. Nevins (Duke University). A β-galactosidase (β-Gal) expression vector under the control of a CMV promoter (pCMV-βgal) was obtained from G. H. Perdew (Penn State University).
Cells.
Rat 2 cells expressing various JCV wild-type (WT) and mutant proteins, i.e., R2-JCVE, R2-T125A, R2-T125D, R2-T′135, R2-SV40E, and R2CT (G418-selected Rat 2 cells) cells, were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% bovine calf serum (BCS), 2 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin at 37°C under 10% CO2. PHFG cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin at 37°C under 10% CO2.
G418 selection.
The cell lines R2-T125A and R2-T125D were created by transfecting Rat 2 cells with 1 μg CMV-JCVE(T125A) or CMV-JCVE(T125D) DNA, using Lipofectamine 2000 reagent (Invitrogen). Transfected cells were propagated in DMEM containing 400 μg/ml Geneticin sulfate and supplemented with 10% BCS. Surviving cells exhibiting G418 resistance were isolated at 10 days posttransfection (p.t.) and subcloned by limiting cell dilution.
IP and WB analysis.
Protein-protein interactions were demonstrated by a coimmunoprecipitation assay consisting of immunoprecipitation (IP) and Western blotting (WB) steps. The cells were lysed in EBC buffer (50 mM Tris, pH 8.0, 120 mM NaCl, 0.5% NP-40) containing protease and phosphatase inhibitors (10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml E-64, 1 mM Pefabloc, 5 mM sodium fluoride, 1 mM sodium vanadate, 500 mM EDTA). The IP step involved adding specific anti-T or anti-RB antibodies to cell lysates prepared in EBC buffer and proceeding as described earlier (36). The WB step involved electrophoresing the complexes in sodium dodecyl sulfate (SDS)-polyacrylamide gels, transferring the separated proteins to membranes, adding anti-RB or anti-T antibodies to the membrane, and adding a secondary antibody to permit the visualization of protein bands (36). The unstable TAg in R2-T125A cells was stabilized by treating the cells with the proteasome inhibitor MG-132 (10 μM) and incubating them for 4 h at 37°C before preparing the cell lysate in EBC buffer containing protease inhibitors.
IP/WB analysis was conducted using the following primary antibodies: anti-T (PAb 962 [35], PAb 2001, and PAb 2023 [3]), anti-p107 (c-18; Santa Cruz), and anti-p130 (c-20; Santa Cruz). Anti-mouse immunoglobulin G (IgG)-alkaline phosphatase conjugate (A-3562; Sigma) was used as the secondary antibody.
Pulse-chase analysis.
R2-JCVE and R2-T125D (5 × 105) cells were seeded into 60-mm dishes containing DMEM and 10% FBS. Twelve hours later, the medium was changed to methionine-free DMEM supplemented with 5% BCS, and cells were incubated at 37°C in 10% CO2. After 1 h, the medium on each plate of cells was replaced with DMEM containing 375 μCi [35S]methionine and 5% FBS, and cells were pulse labeled for 1 h. The cells were then washed twice in phosphate-buffered saline (PBS), refed with DMEM supplemented with 10% FBS, and incubated at 37°C under 10% CO2. Cell lysates were prepared at 0, 3, 6, 12, 24, and 36 h, using EBC buffer containing protease and phosphatase inhibitors. TAg was immunoprecipitated with PAb 962 and electrophoresed in an SDS-15% polyacrylamide gel. The protein bands were detected by autoradiography, and band intensities were quantitated using a phosphorimager and ImageQuant 5.2 software (Molecular Dynamics). The intensity (in counts per minute [cpm]) at each time point was divided by the cpm measured at 3 h postlabeling and multiplied by 100, and this value was plotted against time.
RT-PCR analysis.
Total cellular RNAs were isolated from R2CT, R2-JCVE, and R2-T125A cells by using an RNeasy mini kit (Promega). After DNase treatment, the isolated RNAs were subjected to reverse transcription-PCR (RT-PCR) analysis, using the Access RT-PCR system (Promega) and primers T′#5 (nt 5019 to 4993; 5′-GCTAAAATGGACAAAGTGCTGAATAGG-3′) and T′#6 (nt 4249 to 4274; 5′-CAGGAAAGTCTTTAGGGTCTTCTACC-3′).
DpnI replication assay.
PHFG cells were seeded in 60-mm dishes and transfected the following day with JCV DNA (400 ng), using the Lipofectamine 2000 reagent according to the manufacturer's protocol. Low-molecular-weight DNA was isolated from the cells at 0, 7, 10, and 14 days p.t. by the Hirt extraction procedure (10). A portion of the DNA was cleaved with the restriction enzymes DpnI and EcoRI, and digestion products were separated by electrophoresis on a 0.8% agarose gel. DNA fragments were transferred to a nylon membrane using a Rapid Downward transfer system and an alkaline transfer protocol (Schleicher & Schuell and Amersham, respectively). The immobilized samples were hybridized with linear full-length JCV DNA labeled with [α-32P]dCTP (Oligo labeling kit; Pharmacia). Relative replication activities of different viral DNAs were determined by quantitating band intensities using ImageQuant 5.2 software (Molecular Dynamics).
Anchorage-independent growth (AIG) assay.
Parental and G418-resistant Rat 2 cells were suspended in DMEM containing 10% FBS plus 0.25% agarose and seeded in 60-mm dishes coated with a layer of 0.5% agarose medium. Three dishes were seeded with cells from each line (1 × 105 cells/dish). Fresh medium was added to the plates every 7 days, and after 3 weeks colonies with diameters exceeding 0.5 mm were counted in 20 to 30 randomly selected fields. The percentage of cells developing into colonies was then calculated (2).
Cell cycle analysis.
Cells (2.2 × 105) were seeded in 60-mm dishes in DMEM containing 0.01% BCS. After 72 h, the cells were stimulated with medium containing 10% BCS. Cells were removed from the dishes with trypsin at 0, 9, 12, and 15 h poststimulation and then pelleted. After being washed in PBS, cells were pelleted and resuspended in 300 μl PBS. Cells were fixed in 300 μl 70% ethanol, and after 2 h the cells were pelleted, washed in PBS, and pelleted once more. Cells were suspended in 500 μl propidium iodide (PI) buffer (PBS containing 0.1% Triton X-100, 1 μg/ml RNase, and 20 μg/ml propidium iodide). Samples were incubated at 37°C for 15 min and then subjected to flow cytometric analysis (XL-MCL Coulter machine at the Center for Quantitative Cell Analysis, Penn State University).
Immunofluorescence.
R2CT, R2-JCVE, R2-T125A, and R2-T125D cells were plated at a low density on glass coverslips. The cells were fixed with a 1:1 mixture of methanol and acetone for 10 min and then incubated for 45 min with 12 μl of anti-T antibodies (PAb 962, PAb 2001, and PAb 2023) diluted 1:20. After washing of the coverslips in PBS three times, the cells were incubated with fluorescein-conjugated mouse immunoglobulin G serum for 45 min. The cells were observed and photographed using an Olympus BX-60 epifluorescence upright microscope with a Hamamatsu Orca-100 camera (Center for Quantitative Cell Analysis, Penn State University).
RESULTS
Derivation of Rat 2 cell lines expressing T125A and T125D TAg mutants.
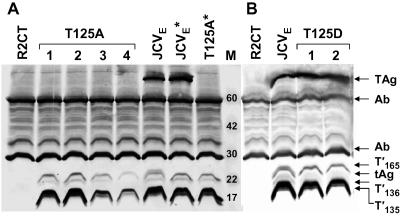
Mutation of T124 in SV40 TAg or T125 in JCV TAg to alanine results in a replication-defective virus (25, 31). These mutations affect not only TAg but also the SV40 and JCV TAg isoforms, 17KT, T′135, T′136, and T′165; small t antigen does not contain the mutation. To investigate the replication defect in the JCV T125A mutant, CMV-JCVE(T125A) DNA was transfected into Rat 2 cells to establish a G418-resistant cell line that expressed the mutated JCV early region. Independent clones were screened to identify cells expressing the five JCV early proteins. Nine cell lines containing JCV proteins were obtained, but surprisingly, none of them expressed detectable levels of TAg (data for four independent lines are shown in Fig. 1). The other four early proteins were detected, but the TAg isoforms, T′135, T′136, and T′165, which normally migrate as doublets in dividing cells due to phosphorylation (B. Bollag et al., unpublished data), were each represented by a single hypophosphorylated band that migrated at the same position as the phosphatase-treated protein bands derived from the WT JCVE control cells.
FIG. 1.
Rat 2 cells expressing JCVE proteins and T125A and T125D mutant proteins. Whole-cell extracts were prepared from subconfluent cultures of R2CT, R2-JCVE, R2-T125A (four independent clones), and R2-T125D (two clones) cells, and proteins were immunoprecipitated with PAb 962 antibody. Coimmunoprecipitated proteins were separated in two 15% SDS-polyacrylamide gels (A and B), transferred to nitrocellulose membranes, and probed with a mixture of anti-T antibodies (PAb 962, PAb 2003, and PAb 2023) to enhance detection. Anti-rabbit IgG alkaline phosphatase-conjugated antibody was used as the secondary antibody, and protein bands were visualized using Nitro Blue Tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) substrate buffer solution. The JCVE* and T125A* samples were treated with lambda protein phosphatase after the IP reaction. Protein molecular size standards (in kDa) are shown in lane M.
An aspartic acid residue was introduced at the T125 site to create the expression plasmid CMV-JCVE(T125D). Based on earlier studies (11), we predicted that this substitution would mimic the negative charge of a phosphorylated threonine residue in WT TAg. CMV-JCVE(T125D) DNA was transfected into Rat 2 cells to establish a G418-resistant cell line. Independent clones were screened to determine whether all JCV proteins were expressed at detectable levels. The introduction of a negative charge at amino acid 125 stabilized TAg expression, and the protein was readily detected in our IP/WB analysis (Fig. 1). The three mutant T′ proteins migrated as single bands and thus shared this property with the T125A mutant proteins.
Status of TAg in R2-T125A cells, as determined by mRNA and protein analyses.
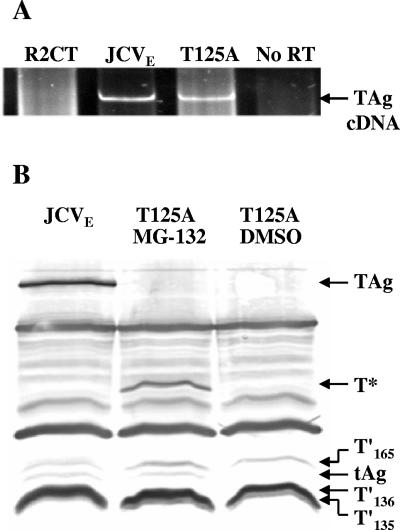
The lack of TAg expression in R2-T125A cells led us to speculate that either TAg mRNA was rapidly degraded or the translated protein was highly unstable in R2-T125A cells. To address these possibilities, steady-state levels of TAg mRNA were examined in R2-JCVE and R2-T125A cells by RT-PCR (Fig. 2A). TAg mRNA was readily detected in both lines, and the transcripts were the same size, suggesting that no major alterations to RNA processing had occurred. We then treated R2-T125A cells with the 26S proteasome inhibitor MG-132 and looked for TAg expression by IP/WB analysis (Fig. 2B). After MG-132 treatment, T125A mutant TAg was detected, although it migrated as a 42-kDa rather than an ∼92-kDa protein in the gel. The anti-T monoclonal antibody used in this experiment recognizes an epitope within the amino terminus of TAg. Therefore, the experiment was repeated using a second monoclonal antibody, PAb 901, which recognizes carboxy-terminal sequences. No TAg fragments were detected using this approach (data not shown).
FIG. 2.
Analyses of viral mRNAs and proteins in R2-T125A cells. (A) RT-PCR analysis of viral mRNAs in R2-T125A cells. Total cellular RNAs were collected from R2CT, R2-JCVE, and R2-T125A cells, treated with DNase, and subjected to RT-PCR analysis using primers T′#5 and T′#6. (B) IP/WB analysis of TAg in R2-T125A cells. Whole-cell extracts were prepared from subconfluent R2-JCVE and R2-T125A cells. The latter cells were treated with the proteasome inhibitor MG-132 (10 μM) for 4 h at 37°C before preparation of the cell lysate. Proteins were immunoprecipitated with PAb 962 antibody. Coimmunoprecipitated proteins were separated in a 15% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with PAb 962, PAb 2003, and PAb 2023 antibodies. Anti-rabbit IgG alkaline phosphatase-conjugated antibody was used as the secondary antibody, and protein bands were visualized using NBT/BCIP substrate buffer solution. T* indicates the position of a 42-kDa amino-terminal fragment of TAg.
The amino-terminal 42-kDa TAg fragment stabilized by MG-132 localizes to the cell nucleus.
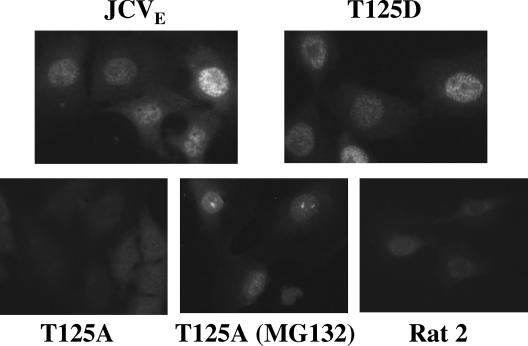
The nuclear localization signal (NLS) in SV40 TAg (amino acids 126 to 131) is positioned near the T124 residue, and mutation of this residue inhibits NLS function (11). A similar arrangement occurs in JCV TAg, but because the JCV T125A TAg mutant is unstable, we could not directly determine whether the mutation affected TAg subcellular localization. Therefore, we treated R2-T125A cells with MG-132 to yield a stable amino-terminal TAg fragment. Cells expressing WT and mutant JCV TAgs were examined by indirect immunofluorescence using anti-TAg antibodies (Fig. 3). No nuclear staining was observed in parental Rat 2 cells or in the untreated R2-T125A line. Nuclear fluorescence was detected in R2-JCVE and R2-T125D cells. Interestingly, punctate nuclear TAg staining was seen in R2-T125A cells treated with MG-132, indicating a functional NLS in the 42-kDa TAg fragment.
FIG. 3.
Cellular localization of T125A and T125D mutant TAgs. R2CT, R2-JCVE, R2-T125D, and R2-T125A (with or without MG-132 treatment) cells were plated at a low density on glass coverslips. The cells were fixed with a 1:1 mixture of methanol and acetone for 10 min and then incubated for 45 min with 12 μl of anti-T antibodies (PAb 962, PAb 2001, and PAb 2023) diluted 1:20. After the coverslips were washed three times with PBS, the cells were incubated with fluorescein-conjugated mouse immunoglobulin G serum for 45 min and observed under an Olympus BX-60 epifluorescence microscope. It should be noted that T′ proteins were not detected in this assay, although we have demonstrated localization of these proteins to the nucleus when fused to enhanced green fluorescent protein (Bollag et al., unpublished data).
Stabilities of WT and T125D mutant TAgs are similar.
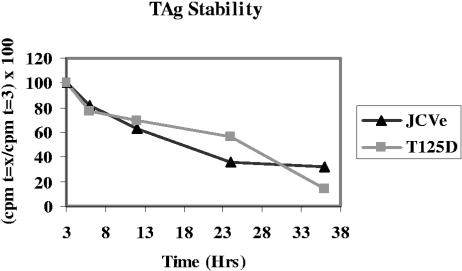
The T125A mutation resulted in a labile JCV TAg. To confirm this finding and to test if the negative charge at position 125 increased the stability of the T125D mutant TAg, R2-JCVE, R2-T125A, and R2-T125D cells were pulsed with [35S]methionine-containing medium for 1 h. After this time, the medium was replaced with unlabeled DMEM supplemented with 5% BCS. Cell lysates collected at regular intervals were immunoprecipitated with anti-TAg antibody, and TAg was electrophoresed in SDS-polyacrylamide gels. Band intensities were quantitated using a phosphorimager and ImageQuant 5.2 software, and the percent cpm remaining relative to that measured at 3 h postlabeling was plotted against time (Fig. 4). The T125A mutant TAg was not detected in these experiments (data not shown), whereas WT and T125D mutant TAgs exhibited similar stabilities.
FIG. 4.
Stability of WT and T125D mutant TAgs. R2-JCVE, R2-T125A, and R2-T125D cells were pulsed with [35S]methionine-containing medium for 1 h. Cell lysates were collected at regular intervals (at 0, 3, 6, 12, 24, and 36 h postlabeling), immunoprecipitated with PAb 962 antibody, and electrophoresed in SDS-15% polyacrylamide gels. TAg bands were detected by autoradiography, and band intensities were quantitated using a phosphorimager and ImageQuant 5.2 software. The percent radiolabeled protein relative to that measured at 3 h postlabeling (100%) was plotted against time. The T125A mutant TAg was not detected, whereas WT and T125D mutant TAgs exhibited similar stabilities. Results from one of three independent experiments are shown.
Interaction of T125A and T125D mutant proteins with p107 and p130.
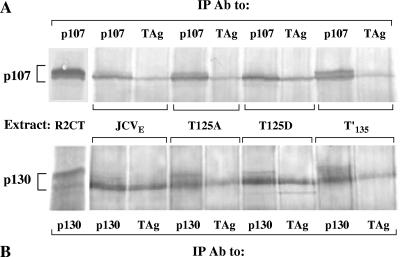
Recently, we showed that JCV T′ proteins interact with RB members and release E2F from RB-E2F complexes (S. K. Tyagarajan and R. J. Frisque, submitted for publication). To determine whether the T125 mutant proteins exhibited these properties, a co-IP/WB approach was taken, using extracts of the R2-T125A and R2-T125D cell lines (Fig. 5). Interactions between JCV proteins and cellular pRb were not detected in these experiments, presumably because of the low levels of pRb in rodent cells (39). Complex formation between JCV early proteins and p107 or p130 was observed in extracts of R2-JCVE, R2-T125A, R2-T125D, and R2-T′135 cells. In each extract, the viral proteins associated with only a fraction of the total pool of p107 or p130; slightly greater amounts of these cellular proteins were consistently found to coimmunoprecipitate with T125D mutant proteins. Furthermore, while the WT and mutant JCV proteins each interacted with only the faster migrating forms of p107 and p130, the total cellular pools of phosphorylated forms of p107 and p130 in the R2-JCVE, R2-T125A, R2-T125D, and R2-T′135 lines varied. The amounts of hyperphosphorylated p107 detected in R2-T125A and R2-T′135 cells were similar to that observed in the R2CT control line, whereas the levels of hyperphosphorylated p107 were greatly reduced in the R2-JCVE and R2-T125D cell lines (Fig. 5A). Several phosphorylated forms of p130 were apparent in the R2CT cells. The slowest migrating species of p130 was absent from the R2-JCVE and R2-T125D lines, but this band remained intact in R2-T125A and R2-T′135 cells (Fig. 5B).
FIG. 5.
T125A and T125D mutant proteins interact with p107 and p130. Whole-cell extracts were prepared from subconfluent cultures of R2CT, R2-JCVE, R2-T125A, R2-T125D, and R2-T′135 cells, and proteins were immunoprecipitated with either PAb 962, anti-p107, or anti-p130 antibody. Coimmunoprecipitated proteins were separated in a 6% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with either anti-p107 (A) or anti-p130 (B) antibody. Anti-rabbit IgG alkaline phosphatase-conjugated antibody was used as the secondary antibody, and protein bands were visualized using NBT/BCIP substrate buffer solution.
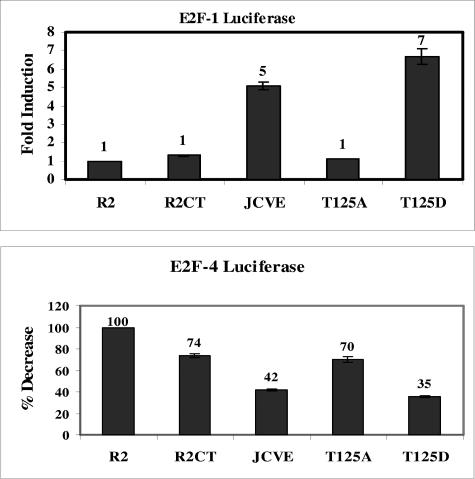
T125D mutant proteins induce the release of transcriptionally active E2F-1 and E2F-4.
Intact J and LXCXE domains of the JCV early proteins are essential for their interactions with the RB family members and their ability to effect the release of E2F from RB-E2F complexes (Tyagarajan and Frisque, submitted). Because mutations to the T125 residues of JCV early proteins alter their ability to bind p107 and p130 and their effect on cellular pools of these RB proteins, we asked whether phosphorylation at this position influenced the release of E2F-1 and E2F-4 from RB-E2F complexes in the cell. We tested this possibility by conducting luciferase reporter assays with the R2-JCVE, R2-T125A, and R2-T125D lines. Cells were cotransfected with a β-Gal-expressing vector (pCMV-βgal) and either an E2F-1 luciferase vector (pE2F1-luc) or an E2F-4 luciferase vector (pE2F4-luc), and the activity of the reporter gene was measured. Luciferase activities were normalized based on the β-Gal measurements, and changes in activity were expressed relative to activities measured in the parental Rat 2 cells (Fig. 6). Luciferase activity was nearly undetectable in cells transfected with the promoterless pΔE2F-Luc construct (data not shown). The induction of luciferase activity in R2-T125D cells transfected with the pE2F1-luc plasmid was consistently greater (sevenfold) than that measured in WT R2-JCVE cells (fivefold). Transfection of these two lines with the pE2F4-luc vector yielded similar levels of luciferase activity that were reduced relative to that measured in the parental Rat 2 control line. This observation supports the recent finding that E2F-4, a repressive transcription factor, is released from RB-E2F complexes by functional JCV early proteins (Tyagarajan and Frisque, submitted). Transfection of R2-T125A cells with either E2F-reporter construct yielded a luciferase activity that was similar to that observed in the parental Rat 2 cells.
FIG. 6.
E2F-1- and E2F-4-induced luciferase activity in R2-T125A and R2-T125D cell lines. Rat 2 (R2), R2CT, R2-JCVE, R2-T125A, and R2-T125D cells (2.5 × 105) were seeded into six-well (35-mm) plates. After 12 h, the cells were transfected with 750 ng pE2F1-Luc or pE2F4-Luc and 500 ng pCMV-βgal, using Lipofectamine 2000 reagent, and incubated at 37°C for 3 h. Cells were lysed 48 h later, and extracts were assayed for luciferase and β-Gal activities. The luciferase activity for each sample was normalized using the β-Gal readings and plotted as the degree of induction (E2F-1) or the percent decrease (E2F-4) relative to the activity measured in the parental Rat 2 cell line (value of 1.0 for E2F-1 or 100% for E2F-4). The E2F-1 and E2F-4 experiments were each repeated four times with duplicate samples. The error bars represent the ranges between duplicate samples in this representative experiment.
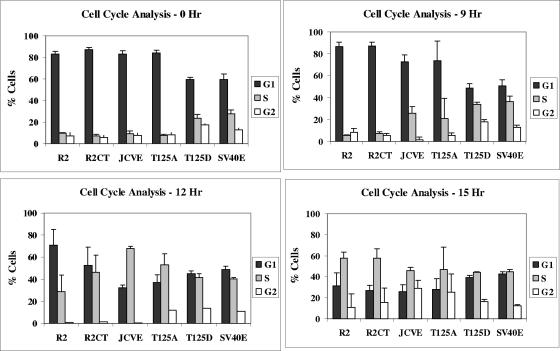
R2-T125D cell growth is not arrested by serum starvation.
Cells transformed by small DNA viruses often proliferate in the presence of reduced concentrations of growth factors in the culture medium (13). We tested the ability of our lines to be growth arrested under conditions of low serum and then to progress through the cell cycle upon serum stimulation. The R2-JCVE, R2-T125A, R2-T125D, R2-SV40E, and Rat 2 lines were first placed in medium containing 0.01% BCS for 72 h, followed by stimulation with medium containing 10% BCS. The progression of synchronized populations of cells through different cell cycle phases (G0/G1, S, and G2/M) was analyzed by fluorescence-activated cell sorting (FACS) analysis. Parental Rat 2 and R2CT (G418-selected Rat 2) cells were employed as negative controls to eliminate differences in cell cycle distribution that might arise from the G418 selection process. FACS analysis showed that following serum deprivation (0 h post-serum stimulation), >80% of the Rat 2, R2CT, R2-JCVE, and R2-T125A cells were arrested in G0/G1 phase, whereas the SV40 control line and R2-T125D cells continued to progress through the cell cycle (Fig. 7). By 9 h post-serum stimulation, R2-JCVE and R2-T125A cells had entered S phase. The two Rat 2 control lines were the last to leave G0/G1, and the R2CT cells appeared to enter S phase slightly ahead of the parental Rat 2 line. The patterns of cell cycle progression for SV40E and R2-T125D cells at 9, 12, and 15 h post-serum stimulation were not altered significantly from those seen at the 0-h time point.
FIG. 7.
Cell cycle analysis of R2-T125A and R2-T125D mutant cell lines. Rat 2 (R2), R2CT, R2-JCVE, R2-T125A, R2-T125D, and R2-SV40E cells (2.2 × 105) were seeded into six-well (35-mm) plates. After 3 h, the medium was replaced, and cells were incubated in DMEM containing 0.01% BCS to induce growth arrest. After 72 h, cells were then refed with DMEM containing 10% BCS and analyzed for cell cycle progression 0, 9, 12, and 15 h after serum stimulation. The percentages of cells in the G1, S, and G2/M phases for each cell line at each time point were determined by using an XL-MCL Coulter machine (Center for Quantitative Cell Analysis, Penn State University). Results from one of three independent experiments are shown.
AIG of cells expressing WT and mutant JCV early proteins.
Adherent cells exhibiting AIG are usually considered transformed. To test whether the mutations at residue 125 altered the ability of JCV tumor proteins to induce a transformed phenotype, we examined the ability of R2-T125A and R2-T125D cells to form colonies in soft agarose (Table 1). Parental Rat 2 cells and M1R2/7 cells, a JCV-transformed Rat 2 line isolated from a dense focus assay (2), were included as negative and positive controls, respectively. The M1R2/7 line, which was originally selected on the basis of its transformed phenotype, readily formed large colonies (10%), whereas G418-selected cells expressing a WT early region (R2-JCVE) formed colonies less efficiently (2%). The R2-T125A line, like the parental Rat 2 cells, failed to grow under these conditions; however, R2-T125D cells demonstrated efficient AIG (6%).
TABLE 1.
Anchorage-independent growth of G418-selected Rat 2 cell lines
AIG of G418-selected Rat 2 cells expressing WT (R2-JCVE), T125A mutant (R2-T125A), or T125D mutant (R2-T125D) protein was compared to that of parental Rat 2 cells and a JCV-transformed line (M1R2/7).
Colonies larger than 0.05 mm were counted in 20 randomly selected fields from two 60-mm dishes, and the percentage of cells developing into colonies was calculated.
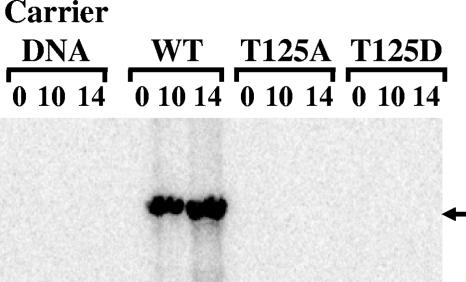
Threonine 125 plays an important role in viral replication.
Previous studies indicated that phosphorylation of T125 and T124 in the JCV and SV40 TAgs, respectively, is critical to viral DNA replication and viability (25, 31). However, the JCV T125D mutant created for the present study was active in a number of assays, perhaps because the negative charge at this position mimicked a phosphorylated T125 residue. Therefore, the DNA replication activity of the T125D mutant virus was compared to that of the T125A mutant and WT JCV. Viral DNAs were transfected into PHFG cells, and Hirt-extracted DNAs were collected at 0, 10, and 14 days p.t. Relative to WT JCV, both the T125A and T125D mutants were replication defective (Fig. 8).
FIG. 8.
DNA replication activities of the intact JCV WT and T125A and T125D mutant genomes in human cells. PHFG cells were seeded into 60-mm plates. After 12 h, the cells were transfected with WT JCV or T125A or T125D mutant viral DNA using the Lipofectamine 2000 reagent. After 3 h of incubation, DMEM containing 10% FBS was added to the transfected cells, and at days 0, 10, and 14 p.t., low-molecular-weight DNAs were collected by the Hirt procedure. Samples from two independent DpnI replication assays were analyzed. The band representing the replicated DNA is indicated (arrow).
DISCUSSION
The phosphorylation of amino- and carboxy-terminal clusters of serine and threonine residues in the SV40 and JCV TAgs influences multiple functions of these regulatory proteins (7, 23, 31, 32). JCV TAg's critical role in viral DNA replication has been established, and TAg isoforms (T′135, T′136, and T′165) contribute to replication efficiency (15, 22, 32, 33, 37). Genetic analyses of T′-deficient viruses showed 10- to 20-fold reductions in DNA replication (22, 37). In previous work, we demonstrated that a T125A TAg mutant was replication incompetent (32). The present study characterizes the activities of JCV tumor proteins mutated at residue 125.
An analysis of cell lines stably expressing T125A or T125D mutant proteins indicated that the T125 residue is essential to TAg's stability and to early viral protein function. Altered TAg stability could be due to the loss of a phosphorylated residue or to the substitution of an alanine residue that destabilizes the protein. The substitution of a second amino acid (aspartic acid) for T125 yielded a TAg that had a half-life similar to that of WT TAg, suggesting that the presence of a negatively charged residue at this position might be required for stability. We investigated the temperature-sensitive phenotype of TAg in R2-T125A cells by incubating cells at 32, 35, 40, and 42°C for 72 h prior to conducting IP/WB analysis. TAg expression was not detected in any of the samples (data not shown). In addition, pulse-chase experiments were carried out using pulses of short duration (5 or 15 min), but again the T125A mutant TAg was not detected under these conditions (data not shown).
We attempted to inhibit the degradation of the mutant JCV TAg by treating R2-T125A cells with the proteasome inhibitor MG-132, and in the process we recovered a 42-kDa amino-terminal TAg fragment in our gels. However, we were unable to detect the carboxy-terminal fragment with our IP/WB assay. It is possible that the degradation of TAg involves multiple cleavages of the protein's carboxy-terminal half by a cellular protease, followed by ubiquitination and destruction within the proteasome machinery. Activation of the cleavage site(s) would likely result from a structural change in TAg involving either the alanine substitution or the loss of the phosphorylated residue at position 125. The detection of all three T′ proteins with T125A mutations might indicate that this mutation either fails to induce structural changes that lead to protease activation or that T′ proteins lack the sequences recognized by the protease.
The JCV TAg and T′ proteins contain a functional NLS (amino acids 127 to 132) immediately adjacent to the T125 site (Bollag et al., unpublished). A similar arrangement occurs in the SV40 TAg and 17KT proteins, and substitution mutations at the T124 site result in defects in nuclear import (11). To determine if the T125A and T125D mutations altered TAg localization, immunofluorescence experiments were conducted. It should be noted that a positive nuclear signal in this assay establishes the presence of TAg only. While T′ proteins do localize to the nucleus, we are only able to visualize this event by using enhanced green fluorescent protein-T′ fusion proteins (Bollag et al., unpublished). Similar observations have been made for N-terminal fragments of SV40 TAg (T1-121 and T1-127) (M. J. Tevethia, personal communication). Nuclear TAg staining was readily observed in R2-T125D and R2-JCVE cells but not in R2-T125A cells. Treatment of the last cell line with MG-132 resulted in the appearance of TAg fluorescence, although it is unclear at present whether the atypical staining observed reflects altered nuclear localization of a partially degraded TAg molecule. Our results do indicate that, unlike the case for SV40 TAg, substitution of an aspartic acid for a threonine at residue 125 in JCV TAg does not interfere with nuclear import.
Aberrant proteins arise in cells, in part because of mutation or transcriptional or translational errors. The protein quality control system within the cell marks such proteins for degradation, usually via the ubiquitination system (28). Currently, it is unclear how cells contend with the presence of aberrant proteins in the nucleus. A recent report identified a protein quality control system within the nucleus of Saccharomyces cerevisiae (9). San1p, a ubiquitin ligase, is the defining member of this degradation system. In the previous study, San1p was found to differentiate between mutant and wild-type nuclear proteins, sending the mutant for rapid degradation. The RING domain of San1p is predicted to have parallels to other RING domain-containing ubiquitin ligases that function in the endoplasmic reticulum (9). These findings in yeast suggest that similar nuclear protein quality control machinery exists in higher eukaryotes and might influence the degradation of nuclear T125A mutant TAg.
We observed that T′ proteins expressed in R2-T125A cells migrated as single bands rather than doublets. T′ proteins typically migrate as hypophosphorylated and hyperphosphorylated bands in proliferating cells; in quiescent cells, only a faster migrating form is detected (Bollag et al., unpublished). Our results support the suggestion that the hyperphosphorylated band represents a T′ protein that is modified at T125. Furthermore, these results indicate that modification of this site might be influenced by the stage of the cell cycle through which the cell is passing. The latter suggestion is supported by reports indicating that both the T125 and T124 sites in JCV and SV40 early proteins, respectively, are part of a Cdk consensus sequence, and in the case of the SV40 protein, T125 is phosphorylated in vitro by a Cdk (16). At present, we do not know whether these differentially modified species of T′ proteins exhibit functional differences, such as the ability to bind or disrupt RB-E2F complexes.
We have demonstrated that WT JCV TAg and T′ proteins interact with pRb, p107, and p130 in vitro and in vivo (3; Bollag et al., unpublished) and degrade and/or dephosphorylate modified forms of p107 and p130 (Tyagarajan and Frisque, submitted). The present study suggests that phosphorylation of the T125 site also influences the ability of JCV early proteins to bind and alter the cellular pools of RB proteins. Relative to the JCV WT proteins, T125A mutant proteins bind p107 and p130 and reduce the levels of hyperphosphorylated RB species with slightly reduced efficiencies. On the other hand, these two activities are enhanced when measured in R2-T125D cells. Although we cannot rule out the possibility that reduced binding of RB proteins in the R2-T125A cells is due to the absence of a stable TAg, we have shown that binding of p107 and p130 in vivo is primarily a function of JCV T′ proteins, not TAg (Fig. 5; Bollag et al., unpublished). Thus, we speculate that the slight reduction in RB binding exhibited by the mutant T′ proteins is due to the T125A mutation and not to the loss of TAg. These observations lead us to speculate that early JCV proteins phosphorylated at the T125 residue have a greater influence on the regulation of the RB pathway than do unmodified species. Relevant to our hypothesis, Sullivan et al. (30) reported that only a subpopulation of SV40 TAg binds to RB proteins in cells.
Recently, we demonstrated that JCV T′ proteins effect the release of E2F-1 and E2F-4 from RB-E2F complexes in rat fibroblasts (Tyagarajan and Frisque, submitted). To test the prediction that differential binding of JCV WT and mutant proteins to p107 and p130 results in differential regulation of E2F-1 and E2F-4 members, we measured free, transcriptionally active E2F in R2-JCVE, R2-T125A, and R2-T125D cells. In agreement with our binding data, we measured minimal effects on E2F-1 and E2F-4 activation in R2-T125A cells but detected consistently higher levels of E2F release in R2-T125D cells. It appears that the T125A mutation has a greater negative impact on the activation of E2F and the disappearance of hyperphosphorylated RB species than it does on the binding of p107 and p130. In this respect, the activities of T125A mutants are more similar to those of J domain mutants than to those of LXCXE domain mutants (Tyagarajan and Frisque, submitted). The ability of JCV early proteins to effect the release of transcriptionally active E2F from E2F-RB complexes is expected to have significant consequences on cell growth. To investigate the impact of E2F release on cell cycle progression, cells expressing JCV WT or T125 mutant proteins were starved of serum for 72 h, refed with medium supplemented with 10% BCS, and then subjected to FACS analysis at several times post-serum stimulation. A high percentage of R2-T125D and R2-SV40E (Rat 2 cells expressing an intact SV40 early region) cells were distributed throughout S phase at the 0-h time point, indicating that these cells were not growth arrested even under stringent low-serum conditions. The remaining lines, including R2-JCVE, R2-T125A, and two lines of Rat 2 cells (parental Rat 2 and G418-selected Rat 2 cells [R2CT]), were growth arrested when incubated in 0.01% serum and progressed through each stage of the cell cycle after serum stimulation. These data suggest a possible correlation between cell cycle progression and the patterns of E2F-1 and E2F-4 release promoted by the WT and mutant JCV tumor proteins (Fig. 6); however, it will be necessary to look at additional cloned lines to confirm these results.
We believe that our failure to arrest the growth of R2-T125D cells in medium containing low serum concentrations reveals an important difference in the abilities of JCV WT and T125D mutant tumor proteins to influence cellular proliferation. Growth in low serum is one property exhibited by virally transformed cells. An even more stringent parameter associated with polyomavirus transformation of mammalian cells is the ability of those cells to exhibit AIG. The induction of AIG in rodent cells by SV40 TAg requires an intact RB-binding domain, but it is unclear whether a functional J domain is also required (29, 34). We found that cells producing the T125A mutant proteins failed to exhibit AIG. We have observed that JCV constructs expressing a subset of the early proteins fail to induce transformation, as measured by a dense focus assay (L. Kilpatrick and R. Frisque, unpublished data). Because TAg in R2-T125A cells is unstable, we predicted that these cells would not grow in soft agarose. On the other hand, cells expressing the JCV T125D mutant proteins grew more efficiently in soft agarose than did cells expressing the JCV WT proteins. Thus, in addition to demonstrating greater effects on RB protein function, the T125D mutant proteins exhibited a more robust effect on this transformation parameter than did the WT early proteins. It is likely that these two effects are related, that is, small differences in the efficiencies of the T125D mutant and WT proteins in binding RB family members and altering their phosphorylation status might translate into differences in their abilities to alter cell cycle progression and cellular growth parameters.
Phosphorylation of the T124 site in SV40 TAg is required for efficient viral DNA replication (25). Kim and coworkers (12) proposed that during replication initiation, TAg utilizes an unphosphorylated CDK/NLS motif to bind the core replication origin and form the first of two hexamer structures. The CDK consensus signal in this motif includes the T124 residue. Once T124 phosphorylation occurs in a cell cycle-dependent manner, the second hexamer is formed, creating the TAg helicase required for the elongation step of replication. The failure of both the T125A and T125D JCV mutants to replicate in human cells is consistent with the predictions of this model. As suggested by the model, some functions of TAg and TAg isoforms require that a specific residue alternate between a phosphorylated and an unphosphorylated state. In the present study, the mutant JCV TAgs with T125D and T125A mutations represent the continuously phosphorylated and dephosphorylated states, respectively. These two mutant TAgs, unlike WT proteins, are unable to switch to the alternate form, thus rendering both mutant viruses replication defective. In contrast, JCV TAg and T′ proteins may carry out some functions only when residue 125 is posttranslationally modified. In the current study, we found that T125D mutant proteins, which represent constitutively “phosphorylated” forms, interacted with RB-E2F complexes and transformed cells more efficiently than did WT JCV proteins, which exist as a mixture of phosphorylated and unphosphorylated species.
The polyomavirus TAgs are phosphorylated at multiple sites, in several combinations, and at different times during the cell cycle. With the discovery of TAg isoforms, an even greater number of tumor protein subpopulations are now known to exist in infected and transformed cells. The multitude of interactions occurring between these viral proteins and the cellular factors that drive the replication and cell cycle machinery most certainly contribute to the regulatory complexities involved in virus-host interactions.
Acknowledgments
We thank J. R. Nevins for the E2F-luciferase reporter constructs, G. H. Perdew for the β-Gal expression vector, and B. Bollag for assistance with the AIG assay and critical comments during manuscript preparation. We also thank Rujuta Bam for assistance with the replication assay.
This work was supported by a Public Health Service grant from the National Institutes of Neurological Disorders and Stroke (S11 NS41833).
REFERENCES
- 1.Barbaro, B. A., K. R. Sreekumar, D. R. Winters, A. E. Prack, and P. A. Bullock. 2000. Phosphorylation of simian virus 40 T antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollag, B., W. F. Chuke, and R. J. Frisque. 1989. Hybrid genomes of the polyomaviruses JC virus, BK virus, and simian virus 40: identification of sequences important for efficient transformation. J. Virol. 63:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollag, B., C. Prins, E. L. Snyder, and R. J. Frisque. 2000. Purified JC virus T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology 274:165-178. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, R. B., and E. G. Gurney. 1982. Time-dependent maturation of the simian virus 40 large T antigen-p53 complex studied by using monoclonal antibodies. J. Virol. 44:565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobrinik, D. 2005. Pocket proteins and cell cycle control. Oncogene 24:2796-2809. [DOI] [PubMed] [Google Scholar]
- 6.Dubbs, D. R., D. Trkula, and S. Kit. 1978. T antigen and initiation of cell DNA synthesis in a temperature-sensitive mouse line transformed by an SV40tsA mutant and in heterokaryons of the transformed cells and chick erythrocytes. Somatic Cell Genet. 4:95-110. [DOI] [PubMed] [Google Scholar]
- 7.Fanning, E. 1994. Control of SV40 DNA replication by protein phosphorylation: a model for cellular DNA replication? Trends Cell Biol. 4:250-255. [DOI] [PubMed] [Google Scholar]
- 8.Frisque, R. J. 1983. Regulatory sequences and virus-cell interactions of JC virus, p. 41-59. In D. L. Madden (ed.), Polyomaviruses and human neurological disease. Alan R. Liss, Inc., New York, N.Y. [PubMed]
- 9.Gardner, R. G., Z. W. Nelson, and D. E. Gottschling. 2005. Degradation-mediated protein quality control in the nucleus. Cell 120:803-815. [DOI] [PubMed] [Google Scholar]
- 10.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 11.Jans, D. A., M. J. Ackermann, J. R. Bischoff, D. H. Beach, and R. Peters. 1991. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J. Cell Biol. 115:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, R. J., S. Moine, D. K. Reese, and P. A. Bullock. 2002. Peptides containing cyclin/Cdk-nuclear localization signal motifs derived from viral initiator proteins bind to DNA when unphosphorylated. J. Virol. 76:11785-11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura, G., and A. Itagaki. 1975. Initiation and maintenance of cell transformation by simian virus 40: a viral genetic property. Proc. Natl. Acad. Sci. USA 72:673-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loche, M. P. 1979. Studies on polyoma virus DNA replication in synchronized C3H2K cells. J. Gen. Virol. 42:429-433. [DOI] [PubMed] [Google Scholar]
- 15.Lynch, K. J., and R. J. Frisque. 1991. Factors contributing to the restricted DNA replicating activity of JC virus. Virology 180:306-317. [DOI] [PubMed] [Google Scholar]
- 16.McVey, D., L. Brizuela, I. Mohr, D. R. Marshak, Y. Gluzman, and D. Beach. 1989. Phosphorylation of large tumour antigen by cdc2 stimulates SV40 DNA replication. Nature 341:503-507. [DOI] [PubMed] [Google Scholar]
- 17.McVey, D., S. Ray, Y. Gluzman, L. Berger, A. G. Wildeman, D. R. Marshak, and P. Tegtmeyer. 1993. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J. Virol. 67:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McVey, D., B. Woelker, and P. Tegtmeyer. 1996. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J. Virol. 70:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moarefi, I. F., D. Small, I. Gilbert, M. Hopfner, S. K. Randall, C. Schneider, A. A. Russo, U. Ramsperger, A. K. Arthur, and H. Stahl. 1993. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J. Virol. 67:4992-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohr, I. J., B. Stillman, and Y. Gluzman. 1987. Regulation of SV40 DNA replication by phosphorylation of T antigen. EMBO J. 6:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei, X. H., and Y. Xiong. 2005. Biochemical and cellular mechanisms of mammalian CDK inhibitors: a few unresolved issues. Oncogene 24:2787-2795. [DOI] [PubMed] [Google Scholar]
- 22.Prins, C., and R. J. Frisque. 2001. JC virus T′ proteins encoded by alternatively spliced early mRNAs enhance T antigen-mediated viral DNA replication in human cells. J. Neurovirol. 7:250-264. [DOI] [PubMed] [Google Scholar]
- 23.Scheidtmann, K. H., M. Buck, J. Schneider, D. Kalderon, E. Fanning, and A. E. Smith. 1991. Biochemical characterization of phosphorylation site mutants of simian virus 40 large T antigen: evidence for interaction between amino- and carboxy-terminal domains. J. Virol. 65:1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheidtmann, K. H., B. Echle, and G. Walter. 1982. Simian virus 40 large T antigen is phosphorylated at multiple sites clustered in two separate regions. J. Virol. 44:116-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider, J., and E. Fanning. 1988. Mutations in the phosphorylation sites of simian virus 40 (SV40) T antigen alter its origin DNA-binding specificity for sites I or II and affect SV40 DNA replication activity. J. Virol. 62:1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherr, C. J., and J. M. Roberts. 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18:2699-2711. [DOI] [PubMed] [Google Scholar]
- 27.Simmons, D. T., W. Chou, and K. Rodgers. 1986. Phosphorylation downregulates the DNA-binding activity of simian virus 40 T antigen. J. Virol. 60:888-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stirling, P. C., V. F. Lundin, and M. R. Leroux. 2003. Getting a grip on non-native proteins. EMBO Rep. 4:565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan, C. S., A. E. Baker, and J. M. Pipas. 2004. Simian virus 40 infection disrupts p130-E2F and p107-E2F complexes but does not perturb pRb-E2F complexes. Virology 320:218-228. [DOI] [PubMed] [Google Scholar]
- 31.Swenson, J. J., and R. J. Frisque. 1995. Biochemical characterization and localization of JC virus large T antigen phosphorylation domains. Virology 212:295-308. [DOI] [PubMed] [Google Scholar]
- 32.Swenson, J. J., P. W. Trowbridge, and R. J. Frisque. 1996. Replication activity of JC virus large T antigen phosphorylation and zinc finger domain mutants. J. Neurovirol. 2:78-86. [DOI] [PubMed] [Google Scholar]
- 33.Tavis, J. E., P. W. Trowbridge, and R. J. Frisque. 1994. Converting the JCV T antigen Rb binding domain to that of SV40 does not alter JCV's limited transforming activity but does eliminate viral viability. Virology 199:384-392. [DOI] [PubMed] [Google Scholar]
- 34.Tevethia, M. J., H. A. Lacko, T. D. Kierstead, and D. L. Thompson. 1997. Adding an Rb-binding site to an N-terminally truncated simian virus 40 T antigen restores growth to high cell density, and the T common region in trans provides anchorage-independent growth and rapid growth in low serum concentrations. J. Virol. 71:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tevethia, S. S., M. Epler, I. Georgoff, A. Teresky, M. Marlow, and A. J. Levine. 1992. Antibody response to human papovavirus JC (JCV) and simian virus 40 (SV40) T antigens in SV40 T antigen-transgenic mice. Virology 190:459-464. [DOI] [PubMed] [Google Scholar]
- 36.Trowbridge, P. W., and R. J. Frisque. 1993. Analysis of G418-selected Rat2 cells containing prototype, variant, mutant, and chimeric JC virus and SV40 genomes. Virology 196:458-474. [DOI] [PubMed] [Google Scholar]
- 37.Trowbridge, P. W., and R. J. Frisque. 1995. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J. Neurovirol. 1:195-206. [DOI] [PubMed] [Google Scholar]
- 38.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young, A. P., and G. D. Longmore. 2004. Differential regulation of apoptotic genes by Rb in human versus mouse cells. Oncogene 23:2587-2599. [DOI] [PubMed] [Google Scholar]