Abstract
Coronaviruses are positive-strand, RNA-dependent RNA polymerase-utilizing viruses that require a polymerase template switch, characterized as discontinuous transcription, to place a 5′-terminal genomic leader onto subgenomic mRNAs (sgmRNAs). The usually precise switch is thought to occur during the synthesis of negative-strand templates for sgmRNA production and to be directed by heptameric core donor sequences within the genome that match an acceptor core (UCUAAAC in the case of bovine coronavirus) near the 3′ end of the 5′-terminal genomic leader. Here it is shown that a 22-nucleotide (nt) donor sequence engineered into a packageable bovine coronavirus defective interfering (DI) RNA and made to match a sequence within the 65-nt virus genomic leader caused a template switch yielding an sgmRNA with only a 33-nt minileader. By changing the donor sequence, acceptor sites between genomic nt 33 and 97 (identical between the DI RNA and the viral genome) could be used to generate sgmRNAs detectable by Northern analysis (∼2 to 32 molecules per cell) by 24 h postinfection. Whether the switch was intramolecular only was not determined since a potentially distinguishing acceptor region in the DI RNA rapidly conformed to that in the helper virus genome through a previously described template switch known as leader switching. These results show that crossover acceptor sites for discontinuous transcription (i) need not include the UCUAAAC core and (ii) rest within a surprisingly wide 5′-proximal “hotspot.” Overlap of this hotspot with that for leader switching and with elements required for RNA replication suggests that it is part of a larger 5′-proximal multifunctional structure.
Among the known positive-strand RNA viruses that use an RNA-dependent RNA polymerase (RdRp) for replication, only members of the Coronaviridae (11) and Arteriviridae (45) families in the order Nidovirales (12) require a 3′-coterminal nested set of subgenomic mRNAs (sgmRNAs) on which a common leader sequence, encoded only at the 5′ end of the genome, is placed. Curiously, not all families in the Nidovirales order have the same requirement. In the Toroviridae, for example, only one of the three sgmRNA species possesses a leader in common with the genome (51), and in the Roniviridae, no common leader on sgmRNAs is found (9). Thus, precisely how nidoviruses differ with regard to the mechanisms of sgmRNA synthesis remains to be determined. Attachment of the leader to sgmRNAs involves a mechanism of high-frequency RdRp template switching (i.e., discontinuous transcription) that is not fully understood but may be a target for designed molecular intervention against such infections as those caused by the severe acute respiratory syndrome coronavirus (17).
Results from several recent studies have generally supported a model for coronavirus discontinuous transcription (34, 36) in which positive-strand-to-positive-strand RdRp template switching occurs during the generation of sgmRNA-length negative-strand templates (14, 16, 41, 42) that are then used reiteratively in double-stranded transcriptive intermediates (33, 35, 37, 40) for the synthesis of positive-strand leader-containing sgmRNAs. Consistent with this model is the observation that sgmRNAs do not replicate upon transfection into helper virus-infected cells (7, 24, 26), suggesting that the sgmRNA-length negative strands may not be synthesized from sgmRNA templates but rather are dependent on the viral genome as a template for their synthesis (33, 46, 58). In this model for discontinuous transcription, the donor sites for template switching are internal in the viral genome, and the acceptor site, an acceptor “hotspot,” is near the 3′ end of the 5′-proximal leader on the genome. Since the infectious coronaviral genome (6, 39, 48, 54) is initially the only acceptor template available, the template switch is necessarily intramolecular, as depicted previously (34, 36, 46, 58). The template switch, furthermore, shares features with similarity-assisted copy choice recombination (5, 19, 22, 27) in that it occurs during RNA synthesis and (usually) requires a matching of bases between the heptameric (or hexameric) core donor and acceptor sites (2, 29, 46, 49, 58) (alternatively called intergenic sequences [ISs] or, when they include some flanking sequences, transcription regulating sequences [2] or transcription activating sequences [50]). This model has also been applied to discontinuous transcription in arteriviruses, and it was with the equine arteritis virus reverse genetics system that a conclusive demonstration for the requirement of matching bases between the donor and acceptor core sequences, which are hexameric in this case, was first made (30, 50). Whether mechanistic differences exist between coronaviruses and arteriviruses with regard to discontinuous transcription remains to be determined.
In the context of this model for coronavirus discontinuous transcription, it remains to be determined which conditions in the microenvironment of the RdRp during negative-strand synthesis lead to high-frequency template switching. For example, how is it that some, but not all, canonical core sequences in the viral genome induce a template switch (29, 46), and conversely, how is it that some template switches are induced by noncanonical sequences (18, 29, 57, 58)? How do the signals leading to usually precise leader-body junctions on sgmRNAs (58) differ from those leading to unusually truncated (mini) leaders (13, 20, 55) or extended (i.e., by UCUAA repeats) leader-body junctions (55)? Is template switching for synthesis of the negative-strand sgmRNA templates restricted to an intramolecular pathway, or can it be intermolecular such that a separate genome or perhaps preexisting sgmRNAs are used as donor or acceptor molecules?
Here we describe experiments with a cloned bovine coronavirus (BCoV) defective interfering RNA (DI RNA) replicon and its trans-acting helper virus in which we sought to explain synthesis from the DI RNA template of an sgmRNA species possessing only the 5′-terminal 33 nucleotides (nt) of the 65-nt leader. The results were interpreted in the context of the model for template switching during negative-strand synthesis, wherein the donor sites are internal on the DI RNA and the acceptor sites are in the 5′-proximal region of the DI RNA (if in cis) or the viral genome (if in trans). Inasmuch as the DI RNA and the viral genome are identical for the 5′-proximal 495 nt, or rapidly become so in the 5′-proximal 93-nt region through the previously described template switching process known as leader switching (8, 25), it was not possible from the experiments presented here to determine whether the switch was in cis only or in trans as well. The properties of the DI RNA donor molecule were manipulated (all within one or the other of two internal 22-nt donor regions), but the acceptor windows on the DI RNA and the viral genome were left unmodified. The results of this study demonstrate that (i) sequences flanking the 5′-proximal heptameric genomic core for distances of 31 nt upstream and 27 nt downstream of the UCUAAAC core can be made to substitute for the core acceptor site in the induction of template switching for discontinuous transcription and (ii) the acceptor window (nt 33 to 97), which can be characterized as an acceptor “hotspot,” is surprisingly wide and overlaps with that (nt 65 to 93) previously tentatively identified for leader switching (8) and other cis elements required for replication (4, 7, 31, 32). Additionally, it was found that for two DI RNA mutants, helper virus-derived sgmRNA 7 was the source of the leader on some of the DI RNA-derived sgmRNA molecules, but the RdRp template switching pathways giving rise to these mutants appear to have been infrequent crossover events within a 114-nt stretch of homologous sequence between DI RNA and sgmRNA 7 rather than due to involvement of the above-mentioned acceptor hotspot.
MATERIALS AND METHODS
Viruses and cells.
A DI RNA-free stock of the Mebus strain of BCoV (GenBank accession number U00235), at 4.5 × 108 PFU/ml, was used as a helper virus on human adenocarcinoma (HRT-18) cells as previously described (7). In some experiments, the porcine hemagglutinating encephalomyelitis virus (HEV; GenBank accession number AF523845) was used as a helper virus, as previously described (53).
Synthetic oligonucleotides.
The oligonucleotides used for this study are described in Table 1.
TABLE 1.
Oligonucleotides used for this study
| Oligonucleotidea | Polarityb | Sequence (5′→3′) | nt binding regionc |
|---|---|---|---|
| 5′gD(+) | − | GAGAGAGGCATCCGCCAAGGCATATTTG | 1854-1885 in Wt12.7 |
| 5′GpD4(+) | − | CGATTCGGGTCGGCCATCTT | 1865-1893 in Wt12.7 |
| 5′RAAP(−)d | + | GGCCACGCGTCGACTAGTACGGGGGGGGGG | 3′ poly(C) on cDNA |
| DrepNIS(−) | + | GGATCCCCGACTGCCATCAACCCAAAAGGGTTCTGG | 29,324-29,353 in BCoV |
| DrepNIS(+) | − | GGTACCTGGTTTGAACATTTCTAGATTGGTCGGACTG | 29,481-29,511 in BCoV |
| Eco9(−) | + | GGCCCCAATCAGAATTTTGGTGGTGGAG | 1396-1423 in Wt12.7 |
| Leader11(−) | + | GATTGTGAGCG | 1-11 in BCoV |
| Leader16(−) | + | GATTGTGAGCGATTTG | 1-16 in BCoV |
| M1(−) | + | CACGGTTTTAGTAGGTTTGCATATTATAATTTACCATCTGGAATATTCTTTAAGC | 1721-1776 in WtX |
| M11(−) | + | GGTGTTGAATGAGACACTGATCTCTTGTTAGATCTTGTTCTATAGG | 1641-1686 in WtX |
| M14(−) | + | GGTGTTGAATGAGATAAACTTTGCATGGATCCGGC | 1641-1686 in WtX |
| M15(−) | + | GGTGTTGAATGAGATTTGTAATCTAAACTTTATAAAGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M16(−) | + | GGTGTTGAATGAGAGATTGTGAGCGACCTAGGCCGAGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M17(−) | + | GGTGTTGAATGAGATTTATAAAAACATCCACTAAATGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M18(−) | + | GAGTGCATGGATCCGCGAGTTCTATAGG | 1659-1686 in WtX |
| M21(−) | + | GGTGTTGAATGAGATAAACGGACGATGGATCCGGCTG | 1641-1677 in WtX |
| M22(−) | + | GAGTGCATGGATCCGGCAGTTCTATAGG | 1659-1686 in WtX |
| M23(−) | + | GAGTGCATGGATCCGGGAGTTCTATAGG | 1659-1686 in WtX |
| M24(−) | + | GGTGTTGAATGAGAGATTGTGAGCGATTTGGGTGCGGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M25(−) | + | GGTGTTGAATGAGACTATGCTTGTGGGCGTAGATTTGTTCTATAGG | 1641-1686 in WtX |
| M26(−) | + | GGTGTTGAATGAGACAGCCAGGGACGTGTTGTATCCGTTCTATAGG | 1641-1686 in WtX |
| M27(−) | + | GGTGTTGAATGAGAGTGATGGTGGATTGCCGCCGACGTTCTATAGG | 1641-1677 in WtX |
| M28(−) | + | GGTGTTGAATGAGATAAACGGAGCATGGATCCGGCTG | 1641-1677 in WtX |
| M29(−) | + | GGTGTTGAATGAGATAAACGGACCATGGATCCGGCTG | 1641-1686 in WtX |
| M32(−) | + | CATAATGAAGGTGATTGTGAGCGATTTGCGTGCGTGCATCCCGCTGTTCTATAGG | 1632-1686 in WtX |
| M33(−) | + | CATAATGAAGGTGATTGTGAGCGATTTGCGTGCGTGCATCGGCGAGTTCTATAGG | 1632-1686 in WtX |
| M34(−) | + | GGTGTTGAATGAGAAAAAACATCCACTCCCTGTATTGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M35(−) | + | GGTGTTGAATGAGACATCCACTCCCTGTATTCTATGGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M36(−) | + | CATAATGAAGGTGATTGTGAGCGATTTGCGTGCGTGCATCCCGCAGTTCTATAGG | 1632-1686 in WtX |
| M37(−) | + | CATAATGAAGGTGATTGTGAGCGATTTGCGTGCGTGCATCCCGGAGTTCTATAGG | 1632-1686 in WtX |
| M41(−) | + | GGTGTTGAATGAGAAAACATCCACTCCCTGTATTCTGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M42(−) | + | GGTGTTGAATGAGAAACATCCACTCCCTGTATTCTAGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| M43(−) | + | GGTGTTGAATGAGAAACATCCACTCCCTGTATTCTAGTTCTATAGGTATTGAC | 1641-1693 in WtX |
| WtNM1(−) | + | GTTGAGAAATAATATTTGCGTGCGTGCATCCCGCTTTACTCCTGGTAAG | 1,718-1767 in WtN |
| WtNM2(−) | + | CCGCATTGTTGAGAAATAATCACTGATCTCTTGTTAGATCTTTTACTCCTGGTAAGC | 1711-1768 in WtN |
The positive and negative symbols in the oligonucleotide names indicate the polarities of the nucleic acids to which the oligonucleotides anneal. M oligonucleotides named with a plus sign in the text have a sequence complementary to a minus-sense oligonucleotide of the same name. For example, M(+) is an oligonucleotide with sequence complementary to M(−).
Polarity of the oligonucleotide relative to the positive-strand viral genome.
If the probe binds to a negative-strand sequence, then the numbers given correspond to the complementary sequence. For the sequence of the entire BCoV genome, numbers given are for the BCoV Mebus strain. Wt12.7 and WtX are the same length, so common nucleotides are in the same positions in both.
Supplied with the RACE kit (Invitrogen).
Plasmid constructs.
The construction of pDrepIS12.7gD (referred to herein as pWt12.7), from which T7 RNA polymerase-generated Wt12.7 DI RNA (Fig. 1A) was made, has been described previously (29). To construct mutant 1 of pWt12.7 (called pWtX herein; Fig. 1A), from which WtX DI RNA was made, an overlap PCR mutagenesis procedure was used as previously described (29), but with oligonucleotides Eco9(−) and M1(+) and pWt12.7 DNA in the first PCR, oligonucleotides M1(−) and 5′gD(+) and pWt12.7 DNA in the second PCR, and oligonucleotides Eco9(−) and 5′gD(+) and the products of the first two reactions in a third PCR to make a 498-nt product that was cloned into the TOPO XL vector (Invitrogen). From this, a 415-nt fragment obtained by digestion with SpeI and KpnI was cloned into SpeI- and KpnI-linearized pWt12.7. Mutants of pWtX, named pM11, pM14-18, pM21-29, pM32-37, and pM41-43, were similarly constructed, except that the template used in the first two reactions was pWtX DNA and the corresponding oligonucleotides used in the first and second reactions were as described in Table 1 [for example, oligonucleotides M14(−) and M14(+), the complement of M14(−), were used to make pM14].
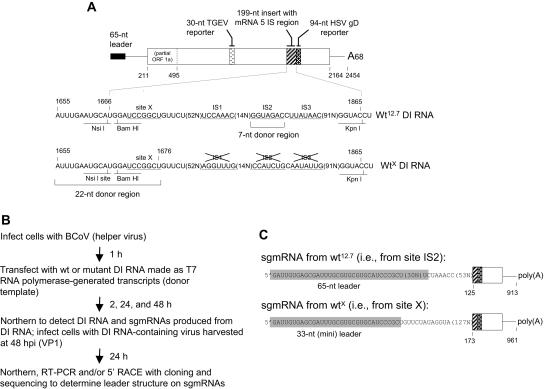
FIG. 1.
Scheme for measuring high-frequency template switching. (A) Structure of Wt12.7 DI RNA, generated from MluI-linearized pWt12.7 by T7 RNA polymerase, and its mutant derivative, WtX. The wt BCoV DI RNA is composed of the two ends of the viral genome joined at nt 494 and 29,392 and, for the formation of pWt12.7, has placed within it an in-frame 30-nt reporter sequence from the porcine transmissible gastroenteritis coronavirus N gene, a 199-nt insert containing the 191-nt BCoV intergenic sequence (IS) region from upstream of the open reading frame for sgmRNA 5 (genomic nt 27,967 to 28,157), and a 94-nt insert containing a 92-nt reporter sequence from the HSV gD gene (29). Template switch donor sites X, IS1, IS2, and IS3 are underlined. In WtX DI RNA, sites IS1, IS2, and IS3 have been made nonfunctional as donor sites. The 22-nt donor region within which all other mutations of pWtX were made for this study is shown. (B) Overall experimental scheme. (C) Structure of the 33-nt minileader on sgmRNA compared to the wt 65-nt BCoV leader.
To construct pDrepISNgD (referred to herein as pWtN), a 200-nt reverse transcription-PCR (RT-PCR) product obtained with the KpnI site-containing primer DrepNIS(+), the BamHI site-containing primer DrepNIS(−), and BCoV genomic RNA was cloned into the TOPO XL vector. From this, a 198-nt fragment obtained by digestion with BamHI and KpnI was cloned into BamHI- and KpnI-linearized pWt12.7. To construct mutant 1 of pWtN (named pWtN-M1), overlap PCR mutagenesis was done, wherein oligonucleotides Eco9(−) and M11-NIS(+) and pWtN DNA were used in the first PCR, oligonucleotides M11-NIS(−) and 5′gD(+) and pWtN DNA were used in the second PCR, and oligonucleotides Eco9(−) and 5′gD(+) and the products of the first two reactions were used in a third PCR to make a 498-nt product that was cloned into the TOPO XL vector. From this, a 199-nt fragment obtained by digestion with BamHI and KpnI was cloned into BamHI- and KpnI-linearized pWt12.7.
Mutated regions in all constructs were confirmed by DNA sequencing.
Northern assay for DI RNA and DI RNA-encoded sgmRNA.
A Northern assay for detecting reporter-containing DI RNAs and sgmRNAs was performed as described previously (7, 29), except that Trizol (Invitrogen) was used for RNA extraction. Briefly, 5 μg of MluI-linearized plasmid DNA, blunt ended with mung bean nuclease, was transcribed with 4 U of T7 RNA polymerase (Promega) in a 100-μl reaction mixture to produce uncapped DI RNA. The reaction mixture was treated with 5 U of RNase-free DNase (Promega), and the RNA was chromatographed through a Biospin 6 column (Bio-Rad). Cells in 35-mm dishes at 50 to 80% confluence (approximately 4 × 106 cells) were infected with BCoV at a multiplicity of infection of 5 PFU per cell and then transfected with 200 ng of RNA by the use of Lipofectin (Bethesda Research Laboratories). For passage of progeny virus, supernatant fluids were harvested at 48 h postinfection (hpi), and 500 μl was used to directly infect freshly confluent cells in a 35-mm dish (8 × 106 cells), from which RNAs were extracted at 24 hpi. RNAs were transferred from the gel to a Nytran membrane by vacuum blotting, and the UV-irradiated blots were probed with oligonucleotide 5′GpD4(+) 5′ end labeled with 32P to specific activities of 2.8 × 104 to 1.5 × 105 cpm/pmol, as determined with a Packard Instant Imager autoradiography system. Probed blots were read by the same system for quantification and exposed to Kodak XAR-5 film for 6 h to 48 h at −80°C for imaging.
Leader-junction sequence analyses of progeny sgmRNAs and their negative-strand counterparts.
Leader-body junctions of progeny sgmRNAs resulting from transfections with WtX (initially), M11, M14, M15, M17, and M34 DI RNAs were determined by sequencing cloned RT-PCR products made with oligonucleotide 5′gD(+) (for RT and PCR) and oligonucleotide leader16(−) (for PCR). Junctions on the sgmRNA negative strands were determined in the same way except that oligonucleotide leader16(−) was used for the RT step. Although no sgmRNAs were apparent by Northern analyses following transfections with M18, M22, M23, M25 to M29, M35, and M41 to M43, attempts to obtain RT-PCR products from these by using the same protocols were made. RT-PCRs were carried out essentially as previously described (29). Briefly, cDNAs were prepared from 2.5 μg of cytoplasmic RNAs (one-fourth of the total harvest from a 35-mm dish) in a 20-μl reaction mixture with reverse transcriptase, and 5 μl of this mixture was used in a 25-μl PCR mix that was heated to 94°C for 2 min and then subjected to 25 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. To obtain 5′-terminal leader and leader-junction sequences of sgmRNAs apparent by Northern analyses from transfections with WtX and M32 DI RNAs and from transfections with M16, M18, M22 to M24, M33, M36, and M37 DI RNAs for which there were no apparent sgmRNAs by Northern analyses, 5′ rapid amplification of cDNA ends (RACE) (5′ RACE kit; Invitrogen) was used. For this assay, oligonucleotide 5′gD(+) was used for the RT reaction and oligonucleotides 5′gD(+) and 5′RAAP(−) were used for PCR under the conditions described above, except that 35 cycles were used. RT-PCR and 5′ RACE products were separated by electrophoresis in the presence of ethidium bromide in native agarose gels at the indicated concentrations. Material from the resolved bands was obtained by punching (with suction) with a micropipette, and the PCR fragments were cloned into the TOPO XL vector (Invitrogen). For WtX and its derivatives, plasmid DNAs from three to six isolated colonies made from the positive-strand sgmRNA templates and three to six colonies from the sgmRNA negative strands were used for automated DNA sequencing of the cloned inserts. For pWtN and its derivatives, RT-PCRs and cloning were carried out in the same way, except that oligonucleotide leader11(−) replaced oligonucleotide leader16(−) and three colonies each from clones made from positive-strand sgmRNAs were sequenced.
RESULTS
Discovery of a donor site yielding a subgenomic mRNA with only a 33-nt leader (termed a minileader).
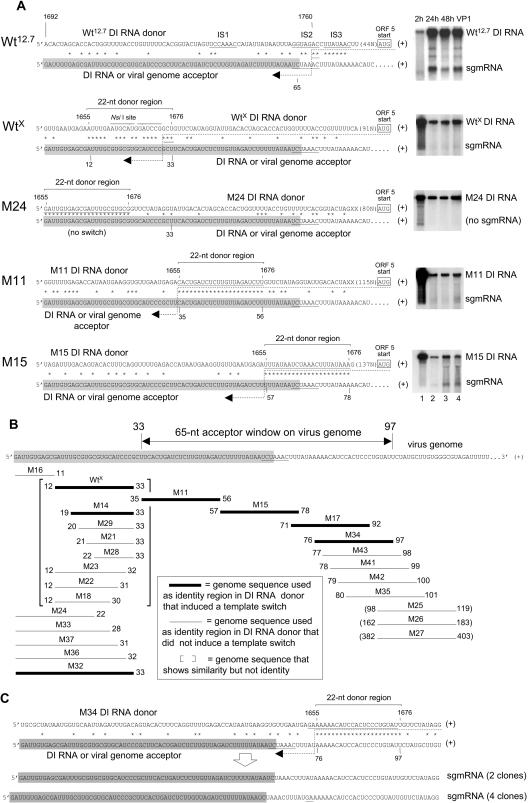
A 2.2-kilobase helper virus-dependent BCoV DI RNA replicon (made as a T7 RNA polymerase-generated transcript from pWt12.7) (Fig. 1A) was used (29) (see the experimental scheme outlined in Fig. 1B) to examine how BCoV mRNA 5-like transcripts are made during virus infection from the noncanonical intergenic donor core sequences GGUAGAC (mostly) and UUAUAAC (rarely) (named IS2 and IS3, respectively, in Fig. 1A) and not from a nearby upstream canonical core sequence, UCCAAAC (IS1 in Fig. 1A) (15, 16). The expression pattern from Wt12.7 DI RNA (29) (Fig. 2A and Table 2) was found to mimic that observed from the virus genome (i.e., the same choice of the noncanonical over the canonical core was made to produce an sgmRNA with a full-length [65-nt] leader [Fig. 1C]), although the rate of accumulation was approximately 18-fold less, as judged from the molar ratios of sgmRNA to the genome at 24 hpi (0.2 for Wt12.7 DI RNA versus 3.5 for the viral genome). The sgmRNAs were detectable by Northern analysis with an end-labeled reporter-specific probe, and the yields were ∼16 molecules of sgmRNA per cell from Wt12.7 DI RNA and ∼70 molecules per cell from the viral genome at 24 hpi, as calculated from the specific activity of the probe and the number of cells in the dish, as previously described (16, 42). With mutant 1 of pWt12.7, in which the natural sites IS1, IS2, and IS3 had been changed to nonfunctional noncanonical core sequences, herein called pWtX (Fig. 1A), and with the same experimental protocol (Fig. 1B), an artifactual heptameric core-like motif identified as site X was found to be used as the high-frequency template switch site for the synthesis of all sgmRNAs identified by the sequencing of 12 clones from RT-PCR products (6 from positive-strand sgmRNA and 6 from negative-strand sgmRNA) and 6 clones from 5′ RACE products (3 from positive-strand sgmRNA and 3 from negative-strand sgmRNA) (Fig. 1A and C and 2A, B, and D, lane 3). Site X begins 5 nt downstream of the NsiI ligation site used in the construction of pWt12.7. Here and throughout this report, a template switch is reported as a high-frequency discontinuous transcription event if (i) an sgmRNA product from the switch was detectable by Northern analysis of RNAs extracted at 24 h postinfection with the DI RNA-containing virus (i.e., VP1) and (ii) RT-PCR or 5′ RACE yielded a discrete product whose sequence, as determined by sequencing molecular clones, revealed an sgmRNA structure with a fused 5′-terminal leader sequence. sgmRNAs containing no leader would have been revealed by sequencing a cloned 5′ RACE product. Note that no special selection pressures were applied before RNA extraction to observe the leader-containing sgmRNA molecules. The striking feature of transcripts generated from site X was that a leader sequence of only 33 nt (a minileader), and not the 65-nt wild-type (wt) leader, was present (Fig. 1C and 2A to C). Although the rate of accumulation of sgmRNA from WtX DI RNA was twofold less than that from Wt12.7 DI RNA (i.e., the sgmRNA to DI RNA molar ratio at 24 hpi in VP1 was 0.1 for WtX versus 0.2 for Wt12.7 [Table 2]), sgmRNA was still faintly observable by Northern analysis at 24 hpi (Fig. 2A), and the number of sgmRNA molecules per cell was ∼4.
FIG. 2.
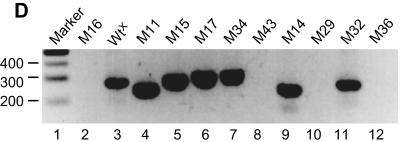
Template switching from internal DI RNA donor sites to a 5′-proximal genomic acceptor window. (A) RdRp template switch sites and Northern RNA blotting results for Wt12.7 DI RNA and selected WtX-derived DI RNA mutants. DI RNAs were transfected as in vitro T7 RNA polymerase-generated transcripts into BCoV helper virus-infected cells and analyzed as described in Fig. 1B. Results of Northern analyses in which the probe was specific for the HSV gD reporter sequence are shown. The molar ratios of sgmRNA to DI RNA found at 24 hpi with first-passage virus (VP1) for selected mutants are shown in Table 2. Leader-body junctions of the sgmRNAs were amplified by RT-PCR, and the products were cloned and sequenced. The upper sequence in each panel represents the positive-strand DI RNA donor, and the lower sequence represents the positive-strand DI RNA or helper virus genome acceptor. The leader sequence on the DI RNA or the viral genome is highlighted. Asterisks identify nt matches between donor and acceptor templates. The dashed lines represents the nascent negative-strand RNA that serves as a primer for reinitiation of RNA synthesis following the RdRp template switch. For the Wt12.7 DI RNA donor, the template switch occurs within site IS2 on either side of nt 1760 or 1761 (note underlined asterisks), and for WtX DI RNA, it occurs within site X on either side of nt 1674 to 1676. Note that the template switch could occur anywhere within the solid row of asterisks identifying the donor and acceptor regions. (B) Mapping of the 5′-proximal acceptor window on the genome. The 22-nt regions of matching sequence between the internal DI RNA sites and 5′-proximal genomic sequences are illustrated with bars. Thin bars indicate no template switch (i.e., no sgmRNAs were generated in VP1 RNA at 24 hpi), whereas thick bars indicate that a template switch occurred (i.e., sgmRNAs were generated). Bars within brackets illustrating WtX and mutants M14, M18, M21 to 23, and M28 represent regions of similarity, not identity; all other bars represent regions of identity. The 65-nt genomic acceptor window is noted. (C) Base insertions at a template switch site. The sequence of four cDNA clones from sgmRNAs of mutant M34 identifying a GA insertion (underlined) at the presumed crossover site is shown. (D) Electrophoresis of 5′ RACE and RT-PCR products in a gel of 2.5% agarose. Lane 1, marker DNA; lanes 2, 3, 11, and 12, 5′ RACE products; lanes 4 to 10, RT-PCR products.
TABLE 2.
Numbers of molecules per cell at 24 hpi and leader types on sgmRNAs
| Genomea | No. of viral genomesb | No. of DI RNA genomesb | No. of sgmRNA moleculesb | Molar ratio of sgmRNA to genome | Type of leader on sgmRNA |
|---|---|---|---|---|---|
| Viral | 20 | 70 (sgmRNA 5) | 3.5 | 65-nt wt leader (from 12.7 IS2 site)c | |
| Wt12.7 DI RNA | 97 | 16 | 0.2 | 65-nt wt leader (from 12.7 IS2 site)c | |
| M24 DI RNA | 155 | 0 | 0 | No sgmRNA by Northern analysis and no 5′ RACE product | |
| WtX DI RNA | 44 | 4 | 0.1 | 33-nt minileaderc,e | |
| M11 DI RNA | 130 | 14 | 0.1 | 56-nt minileaderc | |
| M15 DI RNA | 137 | 32 | 0.2 | 65-nt wt leaderc | |
| M17 DI RNA | 38 | 7 | 0.2 | 65-nt wt leaderc | |
| M34 DI RNA | 149 | 18 | 0.1 | 65-nt wt leaderc | |
| M43 DI RNA | 260 | 0 | 0 | No sgmRNA by Northern analysis and no RT-PCR product | |
| Viral | 20 | 2,000 (sgmRNA 7) | 100 | 65-nt wt leader (from N IS site)c | |
| WtN DI RNA | 17 | 13 | 0.8 | 65-nt wt leader (from N IS site)c | |
| WtN-M1 DI RNA | 30 | 4 | 0.1 | 34-nt minileaderd | |
| 65-nt wt leader (from sgmRNA 7)c | |||||
| WtN-M2 DI RNA | 75 | 2 | 0.03 | 60-nt minileaderd | |
| 65-nt wt leader (from sgmRNA 7)c |
Genomes were either viral or mutant DI RNA generated from plasmid DNA.
Determined from Northern analyses and the specific activity of the probe at 24 hpi with virus alone or with first-passage virus (VP1) following transfection with DI RNA transcripts. For the virus genome and sgmRNAs 5 and 7, the Northern probe detecting an N gene-specific sequence was used, and for DI RNA and DI RNA-derived sgmRNAs, the Northern probe detecting an HSV gD reporter-specific sequence was used.
Determined by sequencing cloned RT-PCR products using leader16(−) as one of the primers.
Determined by sequencing cloned RT-PCR products using leader11(−) as one of the primers.
Determined by sequencing cloned 5′ RACE products.
By aligning the internal WtX presumed donor site (i.e., DI RNA nt 1665 to 1676) with the 5′-proximal DI RNA or viral genomic presumed acceptor site (i.e., genomic nt 12 to 33) (the 5′-proximal DI RNA and viral genomic sequences are identical for 495 nt [7]), a 22-nt region within which 17 nt are identical was revealed (Fig. 2A). This degree of matching suggested that a sequence similarity-induced RdRp template switch had taken place. Furthermore, the switch could have occurred on either side of genomic nt 31, 32, or 33 (note the underlined asterisks in the WtX sequence in Fig. 2A). In conformity with the model of template switching during negative-strand synthesis, Fig. 2A (and others that follow) is drawn with DI RNA as the donor template, wherein the dashed arrow represents the nascent negative strand serving as a primer for the reinitiation of RNA synthesis following the template switch, and the RdRp crossover could be anywhere within the region identified by the contiguous sequence of asterisks.
To map the parts of the 22-nt DI RNA donor region (i.e., nt 1655 to 1676) that contributed to the switch, sequence matches were decreased stepwise by single nucleotides from either end, and the constructs were tested for sgmRNA synthesis from site X (results are summarized within brackets in Fig. 2B). The template switch still took place when the 5′-terminal identity was decreased by 5 nt, as in M14 (as revealed by the presence of an sgmRNA in Northern analysis [not shown], by an RT-PCR product [Fig. 2D, lane 9], and by sequence analysis [Fig. 2B]), but not by 6 nt, as in M29 (in this case, there was no sgmRNA by Northern analysis [not shown] and no RT-PCR product [Fig. 2D, lane 10]). When the identity was decreased by 1, 2, or 3 nt from the 3′ end, none of the resulting mutants, M23, M22, and M18, respectively, induced the switch. Thus, the minimum number of matching bases required for template switching was 12 within the 22-nt donor region, and their importance was position dependent. For the switch, the upstream limit in the virus genome was nt 33.
The genomic acceptor window for the high-frequency template switch is 65 nt wide and maps between nt 33 and 97.
To test whether the intraleader RdRp acceptor site on the genome could be moved farther upstream and thus generate sgmRNAs with a still shorter or even no leader, DI RNA donor mutants were made in which sequences identical to the 5′ end of the genome for distances of 11, 22, 28, 31, 32, and 33 nt, represented by mutants M16, M24, M33, M37, M36, and M32, respectively, were placed into the donor region and examined (Fig. 2B). Of these, only M32, with a match of 33 nt, directed the RdRp switch and generated an sgmRNA, as revealed by Northern analysis (not shown), and a 5′ RACE product (Fig. 2D, lane 11) with a 33-nt minileader (Fig. 2B). Thus, nt 33 was the upstream limit for the similarity-assisted high-frequency template switch induced under these experimental conditions.
To test whether an acceptor site further downstream but still within the leader could be induced, a 22-nt sequence matching genomic leader nt 35 to 56 was used in the DI RNA donor region to form mutant M11. From this, sgmRNA with a 3′-terminally truncated minileader of 56 nt was obtained, identifying another optional intraleader acceptor site for the template switch (Fig. 2A, B, and D, lane 4). Sites extending further downstream to as far as nt 403 were then tested with mutants M15, M17, M25 to M27, M34, M35, and M41 to M43 (summarized in Fig. 2B, with representative RT-PCR results shown in Fig. 2D). From these, sgmRNAs, as revealed by Northern analyses (Fig. 2A and data not shown), and RT-PCR products (Fig. 2D, lanes 5 to 7) were found only for mutants M15, M17, and M34, and sequence analyses of cloned RT-PCR products revealed fusion sites at genome nt positions 78, 92, and 97, respectively (Fig. 2B). No fusion sites mapping further downstream were found, however, indicating that nt 97 is at or near the downstream limit. Note that in four of the six clones of RT-PCR products from mutant M34 sgmRNA, nucleotides GA, appearing to be template insertions, were found at the 5′ end of the matching sequence, lending support to the idea that the leader fusion event occurs during RNA synthesis (Fig. 2C).
These data therefore establish that the genomic acceptor sites at which the RdRp can be induced to switch by matching sequences of 22 nt is limited upstream by nt 33 and downstream by nt 97. Thus, there exists an acceptor window of 65 nt in width near the 5′ end of the DI RNA or viral genome at which 22-nt matching donor sequences in the DI RNA can induce a high-frequency template switch.
The 22-nt donor region in an alternate sequence context also functions to generate sgmRNA with a minileader.
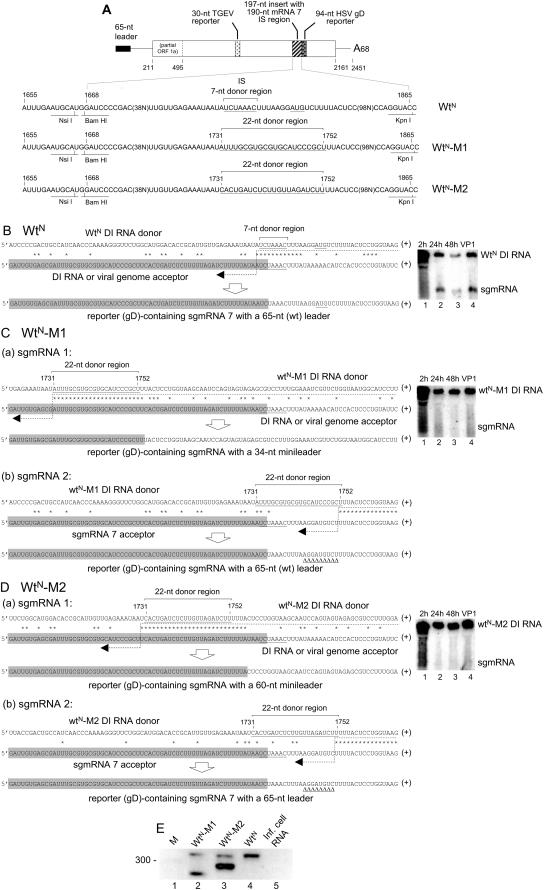
To test whether use of the wide acceptor window to generate sgmRNAs with minileaders is a function of an artifactual microenvironment surrounding donor site X, a different sequence context was tested. For this, a 197-nt insert containing the 190-nt intergenic sequence region encompassing the N gene (gene 7)-associated UCUAAAC core sequence (i.e., BCoV genomic nt 29,324 through 29,513) was used to replace the 191-nt intergenic sequence region encompassing the 12.7-kDa protein gene (gene 5)-associated core sequences IS1 to IS3 (i.e., BCoV genomic nt 27,967 through 28,457) in pWt12.7 (Fig. 1A) to form pWtN (Fig. 3A). When T7 RNA polymerase-generated WtN DI RNA was transfected into helper virus-infected cells and VP1 RNA was tested at 24 hpi, both sgmRNA and an RT-PCR product were found, and the sequence of the cloned product revealed a full-length leader of 65 nt and a leader-body junction sequence identical to that of viral sgmRNA 7 (Fig. 3B and E, lane 4), as anticipated from the results of earlier experiments in which an N gene intergenic region of only 18 nt in a similar DI RNA construct generated sgmRNAs with mRNA 7-like junctions (21). pWtN was then mutated to replace a 22-nt segment containing the UCUAAAC core sequence with the 22-nt donor sequence representing leader nt 12 to 33, the region responsible for the generation of sgmRNA with the 33-nt minileader in pWtX described above, to form mutant pWtN-M1 (Fig. 3A). When WtN-M1 DI RNA was transfected into helper virus-infected cells and the RNA at 24 hpi from VP1 infection was examined by quantitative Northern analysis, approximately eightfold fewer sgmRNA molecules had accumulated than for WtN, as determined from the molar ratio of sgmRNA to DI RNA (Fig. 3C and Table 2). RT-PCR carried out 24 h after infection with VP1, however, revealed two distinct PCR bands (Fig. 3E, lane 2), and the sequences of the cloned products in these bands revealed that they represented two populations of sgmRNAs (Fig. 3C, panels a and b). The shorter of the two RT-PCR products was from WtN-M1 sgmRNA 1 (Fig. 3C, panel a), which had resulted from a template switch within leader region nt 12 to 34. The resulting minileader in this case, however, was 34 nt, not 33 nt, because of the additional matching base at leader nt 34. The longer of the two PCR products was from WtN-M1 sgmRNA 2 (Fig. 3C, panel b), which had derived its leader from viral sgmRNA 7 (described below). Thus, an sgmRNA with a 34-nt minileader was generated by a 22-nt homologous donor region placed in an alternate site in the DI RNA genome.
FIG.3.
Template switching from an alternate internal DI RNA donor sequence to 5′-proximal genomic acceptor sites and involvement of viral sgmRNA 7 in formation of novel sgmRNAs. (A) Structure of WtN DI RNA and mutant derivatives WtN-M1 and WtN-M2. In WtN, the 197-nt insert contains the 190-nt sgmRNA 7 IS region of BCoV (genomic nt 29,324 to 29,513). (B) RdRp template switch site and Northern analysis results for WtN DI RNA containing the intergenic sequence region for sgmRNA 7. Note that the sgmRNA contained a full-length leader. (C) RdRp template switch sites for WtN-M1 DI RNA that yielded sgmRNAs of two types. (a) The first type resulted from an acceptor site within the DI RNA or viral genomic leader and yielded an sgmRNA with a 34-nt minileader. The structure of the sgmRNA is shown. (b) The second type resulted from a template switch in trans involving the 5′-proximal region of helper virus sgmRNA 7. The structure of the novel sgmRNA is shown, with the unique sgmRNA 7-specific nucleotides identified with triangles. (D) RdRp template switch sites for WtN-M2 DI RNA that yielded sgmRNAs of two types. (a) The first type resulted from an acceptor site within the DI RNA or viral genomic leader and yielded an sgmRNA with a 60-nt minileader. The structure of the sgmRNA is shown. (b) The second type resulted from a template switch in trans involving the 5′-proximal region of helper virus sgmRNA 7. The structure of the novel sgmRNA is shown, with the unique sgmRNA 7-specific nucleotides identified with triangles. (E) Electrophoresis of RT-PCR products in a gel of 3.0% agarose.
To determine whether an sgmRNA with a 56-nt minileader could also be induced from within the alternate donor sequence context, a WtN-M2 DI RNA was made in which the 22-nt donor sequence representing leader nt 35 to 56 was placed into the alternate 22-nt donor region (Fig. 3A). As with WtN-M1 DI RNA, a faint band of sgmRNA was observed from VP1 RNA by Northern analysis (Fig. 3D), and from this sgmRNA, two distinct RT-PCR products were obtained (Fig. 3E, lane 3). The shorter product represented an sgmRNA species containing the predicted minileader (Fig. 3D, panel a), except that in this case, the minileader was 60 nt, not 56 nt, as with M11 (described above; Fig. 2B), because of an additional four common 3′-proximal nt between the donor and acceptor regions. The longer of the two RT-PCR products, as with the longer of the two products from WtN-M1, appeared to have derived its leader from viral sgmRNA 7 (described below). Thus, an alternate sequence context in the DI RNA genome, i.e., one that comes from the intergenic sequence region just upstream of gene 7, functioned as a donor site for the synthesis of sgmRNA with a minileader. These results show that the template switch to acceptor sites within the 65-nt 5′-proximal genomic window was directed, at least in part, by the 22-nt donor sequence and not entirely by a larger sequence context in the donor molecule.
Subgenomic mRNA 7 served as the source of leaders for rare novel sgmRNA species.
Sequence analyses of the longer of the two RT-PCR products from WtN-M1- and WtN-M2-derived sgmRNAs (Fig. 3E, lanes 2 and 3, respectively) interestingly showed that the leaders on these molecules had resulted from template switching events involving either helper virus-derived sgmRNA 7 or its negative-strand counterpart (Fig. 3C, panel b, and D, panel b, respectively). The sequences of the cloned RT-PCR products on both the positive and negative strands demonstrated that the template switches had occurred at some position downstream of nt 83 on sgmRNA 7 in the case of WtN-M1 and downstream of nt 82 in the case of WtN-M2. If sgmRNA-length negative strands were not synthesized from sgmRNA templates, as suggested by the failure of sgmRNAs to replicate following transfection (7), a template switch during negative-strand synthesis would necessarily have used the DI RNA genome as a donor, as depicted in Fig. 3C, panel b, and D, panel b. On the other hand, if template switching had occurred during positive-strand synthesis by the use of virus-derived sgmRNA 7 negative strands as donors, then the arrows would have to be drawn in the opposite direction from those shown in Fig. 3C, panel b, and D, panel b. Whether the template switch took place during negative-strand or positive-strand RNA synthesis in these cases remains to be determined. Based on the distances over which the template switch would have occurred in these cases (i.e., 114 nt and 115 nt, representing the distances between nt 83 or 82 and the herpes simplex virus [HSV] gD reporter sequence, respectively), this crossover event may be more typical of random homologous recombination (22) than of the hotspot-related template switching described above. Thus, leader acquisition by this method on nascent sgmRNAs may not be a common event during virus replication. These results are reminiscent of those of a previously described study (29) in which a single sgmRNA species appeared to have resulted from a template switch between Wt12.7 DI RNA and viral sgmRNA 5.
DISCUSSION
In the experiments presented here, we sought to explain the origin of an unexpected 3′-truncated leader, a minileader of 33 nt rather than the full-length 65 nt, on sgmRNA generated from a DI RNA replicon in bovine coronavirus-infected cells. For reasons stated in the introduction, the results were interpreted in accordance with a model of positive-strand-to-positive-strand RdRp template switching, during which time the negative-strand templates for sgmRNA synthesis are made (33, 36). The internal sites on the DI RNA were therefore viewed as the donor sites, and the 5′-proximal region between nt 33 and 97 on either the DI RNA or the viral genome was viewed as the acceptor region. The properties of the DI RNA donor molecule were manipulated (within one or the other of two internal 22-nt donor regions), but the potential acceptor sites within the acceptor window were unmodified wild-type structures.
We view the significance of our results as being twofold, as follows. (i) They show that sequences flanking the 5′-proximal heptameric genomic core for distances of 31 nt upstream and 27 nt downstream from the UCUAAAC core can substitute for the core acceptor site in the induction of template switching for discontinuous transcription. Although these sites were used with far lower apparent efficiencies than the wt core acceptor site (sgmRNA to genomic molar ratios ranged from 0.03:1 to 0.8:1 for DI RNA [generating ∼2 to 32 molecules per cell by 24 hpi] versus 3.5:1 for mRNA 5 and 100:1 for mRNA 7 in the virus genome [generating ∼70 and ∼2,000 molecules per cell by 24 hpi, respectively]) (Table 2; calculated from data obtained here and in reference 16), special selection pressures were still not needed in order to visualize the sgmRNA products of this high-frequency template switch by Northern, RT-PCR, and 5′ RACE analyses soon (24 h) after infection. It is possible that smaller amounts of sgmRNAs with atypical leaders accumulated because of lower stabilities, but no comparative measurement of stabilities was made to evaluate this possibility. However, there was no discernible difference between in vitro translation rates for 33-nt minileader- and 65-nt wt leader-containing uncapped transcripts (data not shown), suggesting that the stabilities of the two transcripts were similar within the cell. Therefore, these results characterize a large but discrete acceptor “hotspot” for coronavirus discontinuous transcription. They also show that not only is a canonical core donor site dispensable for discontinuous transcription (18, 29, 46, 57, 58), but a canonical core acceptor site is dispensable as well. This particular feature may reveal a difference compared to arterivirus discontinuous transcription, which seems to require strict identity between donor and acceptor core sequences (30, 50). The results suggest, furthermore, that chance base matches between noncanonical donor sites and noncanonical acceptor sites within the window may explain the origins of truncated (13, 20, 55) and extended (i.e., by multiple junctional UCUAA elements or use of the downstream 8-nt UUUAUAAA palindromic sequence) (55, 56) leaders, and they may also explain a 5′-proximal enhancer-like function for sgmRNA synthesis (52). The results also suggest that flanking sequences of >4 nt upstream and 4 nt downstream of the acceptor core, although 4-nt distances proved useful for predicting the likelihood of a template switch (46, 58), might also be involved in regulating template switching rates.
(ii) The results show that the 5′-proximal genomic acceptor window for discontinuous transcription is surprisingly wide and overlaps the tentative window previously identified for leader switching in BCoV (nt 65 to 93) (8). This overlap therefore raises the question, again, of which mechanistic features might be shared between these two remarkable high-frequency RdRp template switching phenomena. Because of the different positional requirements for the template switch during discontinuous transcription (employing internal donor sites and a 5′-proximal genomic acceptor region) versus leader switching (employing only genomic 5′-proximal donor and acceptor sequences), it is quite possible that the two processes differ mechanistically, at least in part. Whereas template switching during discontinuous transcription appears restricted to negative-strand synthesis, template switching during leader switching might occur during either negative- or positive-strand synthesis or both.
The question of whether template switching during discontinuous transcription can occur in trans (25), as happens during leader switching, has been examined previously (3, 56), and the conclusion was that it can. In these analyses, however, it was not reported whether leader switching had occurred simultaneously, as might be expected based on other studies with the same experimental system (23), leaving open the possibility that the observed discontinuous transcription-related template switch in trans in reality reflected a cis event. It is clear from our experiments that template switching during discontinuous transcription does occur in cis (56; data not shown). This was demonstrated by using BCoV WtX DI RNA with porcine HEV as the helper virus. HEV is another group 2 coronavirus whose leader differs from the BCoV leader at nt positions 10, 13, and 27 (53). Interestingly, in addition to minileaders of 33 nt derived in cis from BCoV DI RNA on sgmRNAs at 24 hpi with VP1, 33-nt minileaders of HEV origin were also found. The HEV minileaders could have been derived in trans or, alternatively, in cis from DI RNA molecules that had already undergone a leader switch (53; data not shown). If it is confirmed that template switching during discontinuous transcription does occur in trans, it would suggest that a variant of the model described by Enjuanes and coworkers (46, 58), itself a modification of the model of Sawicki and Sawicki (34, 36), should be considered. Whereas template switching in the model of Enjuanes and coworkers occurs in cis within a 3′-to-5′-traversing transcription complex containing the acceptor-bearing genomic 5′ terminus (Fig. 4B), for template switching in trans the transcription complex might carry within it the acceptor-bearing 5′ terminus of another DI RNA or viral genome, as depicted in Fig. 4C.
FIG. 4.
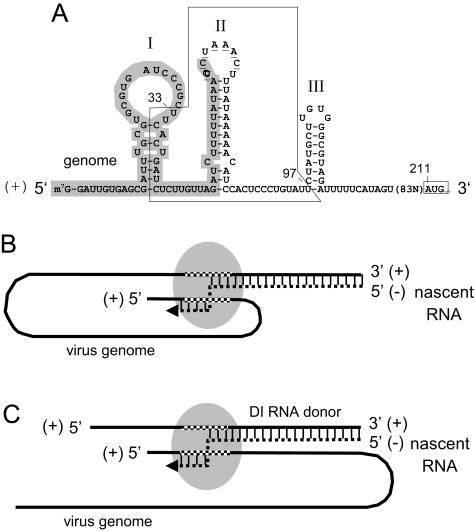
Model for higher-order structure of the 5′-proximal genomic acceptor region and proposed pathways for the switch in cis and in trans. (A) Mapped genomic acceptor window (boxed) for high-frequency template switching in the context of stem-loops I, II, and III in the BCoV genome. The 65-nt leader sequence is shaded, nucleotides making up the leader-associated intergenic core sequence (UCUAAAC) in the loop of stem-loop II are underlined, and the start codon for ORF 1 is boxed. (B) Model for positive-strand-to-positive-strand template switching in cis during synthesis of negative-strand templates for sgmRNA production, as illustrated by Enjuanes et al. (46, 58). The matching donor and acceptor regions are checkered. Base-paired regions between the positive-strand genome and negative-strand nascent RNA are indicated by vertical lines. The transcription complex is depicted by the shaded oval. (C) Model for positive-strand-to-positive-strand template switching in trans.
What mechanistic features define the wide acceptor hotspot observed here remain to be determined, but precedents of template-switching hotspots in other positive-strand RNA viruses (summarized in reference 27) might suggest some. (i) A/U richness may contribute to, but cannot be the sole explanation for, high-frequency acceptor activity, since the window, showing an A+U content of 72%, is not uniquely A/U-rich. Scanning of the BCoV genome with Vector NTI software (Invitrogen) revealed seven other windows in the genome of 71 nt in width with A/U richness exceeding 70% (data not shown), and none have been revealed as hotspots for template switching. (ii) Higher-order protein-binding promoter and enhancer RNA structures might contribute, and in this case, stem-loops II and possibly III (Fig. 4A) are candidates. So far, the coronaviral nucleocapsid (N) protein, recently shown to be required for genome replication (1, 38), is the only viral protein shown to bind 5′ untranslated region (UTR) structures. These structures include the UC(U/C)AAAC core (28), which in BCoV maps to the loop of stem-loop II (7) (Fig. 4A), and the positive-strand form of stem-loop III (S. Raman and D. Brian, unpublished). Whether N facilitates template switching remains to be determined; it is certainly not required for sgmRNA synthesis, as green fluorescent protein-expressing autoreplicons of human coronavirus 229E lack the N gene (47). Since coronavirus ORF 1 gene products have predicted or demonstrated functions in RNA binding (10) or RdRp, helicase, exonuclease, endonuclease, or methyl transferase activity (44), they too are potential components of a template switching apparatus and must be examined further for such activity. Cellular proteins that bind coronaviral 5′ UTR structures (43) might also play a role.
The fact that the window, as identified here, overlaps that for leader switching (8) and partially overlaps 5′ UTR cis-acting elements for BCoV genome replication (4, 7, 31, 32) suggests that it is part of a larger multifunctional structure involved in both genome replication and discontinuous transcription. The details of this structure remain to be characterized.
Acknowledgments
We thank Cary Brown, Shamila Raman, Gwyn Williams, and Kim Nixon for many valuable discussions and Ruey-Yi Chang (Dong-Hua University, Taiwan), Cheng Kao (Texas A&M University), and John Taylor (Fox Chase Institute) for helpful comments on an early version of the manuscript.
This work was supported by Public Health Service grant AI14367 from the National Institute of Allergy and Infectious Diseases and by funds from the University of Tennessee, College of Veterinary Medicine, Center of Excellence Program for Livestock Diseases and Human Health. H.-Y.W. was supported by a stipend from the Center of Excellence Program. A.O. was supported by a stipend grant from the Turkiye Bilim Teknik ve Arastirma Kurumu (TUBITAK), Government of Turkey.
REFERENCES
- 1.Almazan, F., C. Galan, and L. Enjuanes. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 78:12683-12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, S. C., and M. M. Lai. 1990. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 9:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brian, D. A., and W. J. M. Spaan. 1997. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brian, D. A., D. E. Dennis, and J. S. Guy. 1980. Genome of porcine transmissible gastroenteritis virus. J. Virol. 34:410-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, R. Y., M. A. Hofmann, P. B. Sethna, and D. A. Brian. 1994. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 68:8223-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, R. Y., R. Krishnan, and D. A. Brian. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 70:2720-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowley, J. A., C. M. Dimmock, and P. J. Walker. 2002. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83:927-935. [DOI] [PubMed] [Google Scholar]
- 10.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enjuanes, L., D. Brian, D. Cavanagh, K. Holmes, M. M. C. Lai, H. Laude, P. Masters, P. Rottier, S. G. Siddell, W. J. M. Spaan, F. Taguchi, and P. Talbot. 2000. Coronaviridae, p. 835-849. In C. M. F. M. H. V. van Regenmortel, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 12.Enjuanes, L., W. Spaan, E. Snijder, and D. Cavanagh. 2000. Nidovirales, p. 827-834. In C. M. F. M. H. V. van Regenmortel, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 13.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, M. A., and D. A. Brian. 1991. The 5′ end of coronavirus minus-strand RNAs contains a short poly(U) tract. J. Virol. 65:6331-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann, M. A., R. Y. Chang, S. Ku, and D. A. Brian. 1993. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology 196:163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann, M. A., P. B. Sethna, and D. A. Brian. 1990. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 64:4108-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes, K. V., and L. Enjuanes. 2003. Virology. The SARS coronavirus: a postgenomic era. Science 300:1377-1378. [DOI] [PubMed] [Google Scholar]
- 18.Hussain, S., J. Pan, Y. Chen, Y. Yang, J. Xu, Y. Peng, Y. Wu, Z. Li, Y. Zhu, P. Tien, and D. Guo. 2005. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 79:5288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis, T. C., and K. Kirkegaard. 1991. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 7:186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keck, J. G., S. A. Stohlman, L. H. Soe, S. Makino, and M. M. Lai. 1987. Multiple recombination sites at the 5′-end of murine coronavirus RNA. Virology 156:331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, M. M. 1992. Genetic recombination in RNA viruses. Curr. Top. Microbiol. Immunol. 176:21-32. [DOI] [PubMed] [Google Scholar]
- 23.Liao, C. L., and M. M. Lai. 1992. RNA recombination in a coronavirus: recombination between viral genomic RNA and transfected RNA fragments. J. Virol. 66:6117-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino, S., and M. Joo. 1993. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J. Virol. 67:3304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino, S., and M. M. Lai. 1989. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J. Virol. 63:5285-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masters, P. S., C. A. Koetzner, C. A. Kerr, and Y. Heo. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, G. W., S. A. Stohlman, and S. M. Tahara. 2000. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 81:181-188. [DOI] [PubMed] [Google Scholar]
- 29.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raman, S., and D. A. Brian. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 79:12434-12446. [DOI] [PMC free article] [PubMed]
- 32.Raman, S., P. Bouma, G. D. Williams, and D. A. Brian. 2003. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 77:6720-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawicki, D., T. Wang, and S. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385-396. [DOI] [PubMed] [Google Scholar]
- 34.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287:31-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 37.Schaad, M. C., and R. S. Baric. 1994. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J. Virol. 68:8169-8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schelle, B., N. Karl, B. Ludewig, S. G. Siddell, and V. Thiel. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79:6620-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schochetman, G., R. H. Stevens, and R. W. Simpson. 1977. Presence of infectious polyadenylated RNA in coronavirus avian bronchitis virus. Virology 77:772-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethna, P. B., and D. A. Brian. 1997. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J. Virol. 71:7744-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, S. T., and M. M. C. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 46.Sola, I., J. L. Moreno, S. Zuniga, S. Alonso, and L. Enjuanes. 2005. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 79:2506-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 75:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel, V., and S. G. Siddell. 2005. Reverse genetics of coronaviruses using vaccinia virus vectors. Curr. Top. Microbiol. Immunol. 287:199-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Most, R. G., R. J. de Groot, and W. J. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Y., and X. Zhang. 2000. The leader RNA of coronavirus mouse hepatitis virus contains an enhancer-like element for subgenomic mRNA transcription. J. Virol. 74:10571-10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, H. Y., J. S. Guy, D. Yoo, R. Vlasak, E. Urbach, and D. A. Brian. 2003. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology 315:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, X., and M. M. Lai. 1994. Unusual heterogeneity of leader-mRNA fusion in a murine coronavirus: implications for the mechanism of RNA transcription and recombination. J. Virol. 68:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, X., C. L. Liao, and M. M. Lai. 1994. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J. Virol. 68:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, X., and R. Liu. 2000. Identification of a noncanonical signal for transcription of a novel subgenomic mRNA of mouse hepatitis virus: implication for the mechanism of coronavirus RNA transcription. Virology 278:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuniga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]