Abstract
The ataxia-telangiectasia-mutated (ATM) kinase plays a central role in responses to various forms of DNA damage and has been suggested to facilitate human immunodeficiency virus type 1 (HIV-1) integration. Here, we describe a series of experiences that indicate that ATM can enhance HIV-1 replication by stimulating the action of the Rev viral posttranscriptional regulator. The Rev-dependent stimulation of viral late gene expression was observed with ATM-overexpressing cells, a result confirmed with a Rev-dependent reporter construct. Both parameters were also enhanced upon treatment of HeLa cells with caffeine, a xanthine that, in this cellular context, stimulates ATM activity. As well, decreased levels of virions with reduced infectivity were released by ATM knockdown cells. Notably, ATM overexpression did not stimulate the HIV-1 late gene expression within the context of Rev-independent constructs or the Rex-dependent production of capsid from human T-cell leukemia virus type 1 proviral constructs. Altogether, these results indicate that ATM can positively influence HIV-1 Rev function.
The temporally regulated nuclear export of viral mRNAs is an essential aspect of retroviral replication, for which these viruses have evolved cis-acting elements that specifically interact with cellular RNA export pathways and, in some cases, trans-acting viral regulatory proteins that facilitate these interactions (11). The nuclear export of the unspliced RNA (encoding Gag and Pol) from the simple retrovirus Mason-Pfizer monkey virus is mediated by the direct recognition of a cis-acting element termed the constitutive transport element (CTE) by the TAP/NXF1 RNA export factor, a protein also responsible for the nucleocytoplasmic transport of most cellular mRNAs (17, 28). In human immunodeficiency virus (HIV)-infected cells, three main categories of viral mRNAs are found: multiply spliced, singly spliced, and unspliced. The first class is readily exported to the cytoplasm and codes for the viral early proteins Tat, Rev, and Nef. In contrast, the transport of singly spliced and unspliced HIV RNAs, which notably encode the viral structural and enzymatic components, requires that their cis-acting Rev-responsive element (RRE) be recognized by the Rev posttranscriptional regulator. Rev in turn binds the cellular protein Crm1, a member of the karyopherin family, which acts as an RNA export factor (16, 27), allowing expression of the late viral proteins Gag, Pol, Env, Vif, Vpr, and Vpu and making the viral genomic RNA available for packaging. Several cellular factors, such as the DEAD box RNA helicase family members DDX1 and DDX3, have been proposed to regulate HIV type 1 (HIV-1) Rev function, albeit by mechanisms that are yet to be fully characterized (15, 33, 38). Here, we report the unexpected finding of a stimulating effect of the ataxia-telangiectasia-mutated (ATM) protein, a DNA damage sensor (31), on HIV-1 Rev function.
ATM, a phosphatidylinositol-3-OH-kinase-like serine/threonine kinase, and the related ATR (ATM- and Rad3-related) kinase regulate cellular responses to DNA damage by controlling cell cycle arrest and DNA repair pathways. Accordingly, a possible role for these proteins within the context of retroviral integration has been sought actively (34). Even though conflicting results were obtained (4, 12, 14, 22), indicating that neither protein is absolutely indispensable for HIV integration, recent data obtained with a small-molecule inhibitor of ATM kinase activity lend credence to a model in which ATM promotes productive retroviral integration by preventing integration-induced cell death (22). Here, we describe a set of experiments initially triggered by our finding that ATM overexpression could stimulate HIV-1 replication independently of integration.
MATERIALS AND METHODS
Cell culture.
293T and P4.2 (10) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37°C. Caffeine (Sigma) was dissolved in culture medium at a stock concentration of 100 mM just prior to use. Hydroxyurea (Sigma) was dissolved in water at 1 M.
DNA constructs, virus production, infections, transductions, and titrations.
The wild-type X4 HIV-1 proviral DNA clone (R9) and the vesicular stomatitis virus (VSV) G-pseudotyped HIV-1-based vector system were previously described (26, 41). We used the second-generation packaging construct pCMVΔR8.91 (where CMV is cytomegalovirus) (41), the envelope plasmid pMDG, and the green fluorescent protein (GFP)-expressing vector pWPTS-GFP. To examine Rev function, we used the Rev-dependent luciferase-based reporter pDM628; the Rev-expressing plasmid pcRev (15); the codon-optimized HIV-1 gag-pol construct pSYNGP (21); the HIV-1 gag-pol construct containing the Rev response element, pGPV-RRE; and the HIV-1 gag-pol construct containing four tandem copies of the Mason-Pfizer monkey virus CTE (4×CTE), pGPV-4×CTE (32, 36). The human T-cell leukemia virus type 1 (HTLV-1) molecular clones K30p (39) and λHTLV-1C (25) were previously described, as were pcDNA3 Flag-ATM (8), Tat101-FLAG (5), pHIV-1-LTR-Luc (where Luc is luciferase), and pcDNA3-FLAG (3). HIV-1 and retroviral vector particles were produced by transient transfection of 293T cells with Fugene 6 (Roche). Titrations were performed using a multinucleate activation of galactosidase indicator assay with CD4+ long terminal repeat (LTR)-β-galactosidase (β-Gal) HeLa-derived P4.2 cells (10). HIV-1 viral production was estimated by an HIV-1 p24 antigen capture assay (SAIC—Frederick, NCI—-Frederick). Reverse transcriptase (RT) activity was monitored as previously described (1).
RNA interference.
Oligonucleotides with the following sense and antisense sequences were used for the cloning of small-hairpin RNA (shRNA)-encoding sequences in lentiviral vector: MRE11, 5′-GATCCCCGGCACTGAGAAACATGCAATTCAAGAGATTGCATGTTTCTCAGTGCCTTTTTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAGGCACTGAGAAACATGCAATCTCTTGAATTGCATGTTTCTCAGTGCCGGG-3′ (antisense). The oligonucleotides described above were annealed and subcloned into the BglII-HindIII site of pSUPER (7). To construct pLVshRNA against MRE11, the BamHI-SalI fragment of pSUPER-MRE11i (where MRE11i represents MRE11 knockdown cells) plasmid was subcloned into the BamHI-SalI site of pRDI292 (6).
Western blotting.
Cells were lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 4 mM EDTA, 1% Nonidet P-40 (NP-40), 0.1% sodium dodecyl sulfate, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride. Supernatants from these lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblot analysis using anti-ATM (NB100-104; GeneTex), anti-MRE11 (GeneTex), anti-HIV-1 p24 rabbit polyclonal antibody (no. 5824), anti-HTLV-1 p24 mouse monoclonal antibody (a kind gift from Genoveffa Franchini), anti-HIV-1 Nef rabbit serum (2), anti-HIV-1 p17 MA (ABI), anti-Chk2 (NT; ProSci), or anti-phospho-Chk2 (Thr68) (Cell Signaling) antibody.
Luciferase assay.
Plasmids were transfected into 293T cells (2 × 104 cells) by use of the Fugene 6 transfection reagent (Roche). Luciferase assays were performed 24 h after transfection by use of luciferase assay reagent (Promega) as previously described (3). All transfections utilized equal total amounts of plasmid DNA quantities, owing to the addition of empty vector into the transfection mixture. Results were obtained through three independent transfections.
Northern blotting.
Cytoplasmic RNA was obtained using an RNeasy kit (QIAGEN), DNase treatment, and DNA-free removal reagent (Ambion). RNA (30 μg) was denatured, electrophoresed through a 0.8% agarose-formaldehyde gel, transferred to a Hybond-N+ nylon membrane (Amersham Bioscience), and hybridized at 68°C with an SP6 RNA polymerase-generated 32P-riboprobe complementary to HIV-1 nef from pG-N3X by use of a MAXIscript in vitro transcription kit (Ambion). After UV cross-linking at 150 mJ with a UV cross-linker (Bio-Rad), the filter was washed twice in low-stringency wash solution (NorternMax kit; Ambion) at room temperature and twice in high-stringency wash solution (NorternMax kit; Ambion) at 68°C and exposed to X-ray film.
RESULTS
ATM stimulates expression of HIV-1 late genes.
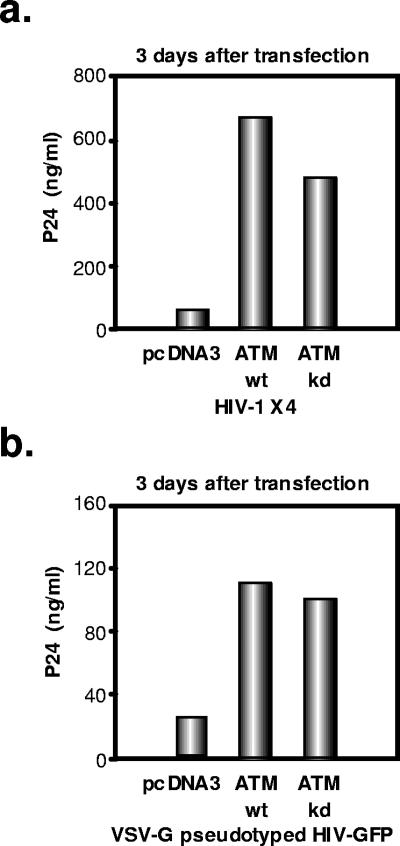
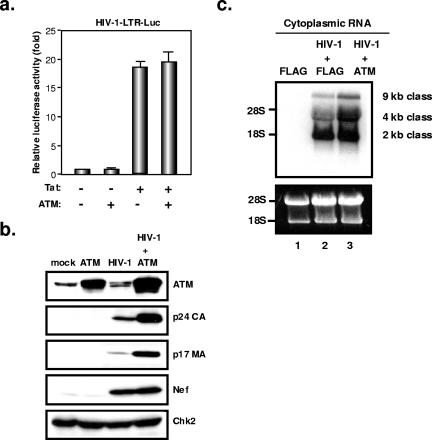
Using RNA interference, we previously failed to observe an influence of ATM on the early steps of HIV-1 infection (4). However, in parallel, we probed possible alterations of viral behavior in cells overexpressing the kinase. For this, we transfected 293T cells with either an HIV-1 molecular clone (R9, an X4 virus) or an HIV-derived lentiviral vector packaging construct (ΔR8.91), together with ATM-expressing or control plasmids, and we measured viral production in the supernatant by p24-specific enzyme-linked immunosorbent assay (ELISA). ATM overexpression, whether in wild-type or kinase-defective form, significantly enhanced HIV-1 production from either type of virus-derived construct (Fig. 1a and b). One possible explanation was that ATM stimulated transcription from the viral promoter. However, overexpression of the cellular protein affected neither basal level nor Tat-induced transcription from the HIV-1 LTR within the context of a transfected LTR-luciferase plasmid (Fig. 2a). To confirm this result, we performed Western blot analyses of cytoplasmic extracts of cells transfected with an HIV-1 proviral construct, with or without an ATM-expressing plasmid. ATM overexpression had no influence on the levels of the viral early protein Nef, confirming that it did not affect viral transcription. In contrast, it significantly increased levels of the late viral structural capsid (p24 CA) and matrix (p17 MA) (Fig. 2b) proteins, as well as envelope protein (data not shown).
FIG. 1.
Overexpression of ATM enhances HIV-1 replication in a kinase-independent manner. (a) 293T cells (2 × 105 cells) were cotransfected with an HIV-1 molecular clone (1 μg) and wild-type (wt) or kinase-defective (kd) ATM-expressing plasmid or the empty vector pcDNA3-FLAG (2 μg). Three days after transfection, p24 levels in the culture supernatant were measured by p24 ELISA. (b) Overexpression of ATM enhances VSV G-pseudotyped HIV-1 production. 293T cells (2 × 105 cells) were cotransfected with HIV-1 packaging construct pCMV R8.91 (2 μg), HIV-GFP (WPTS-GFP) (1 μg), VSV G-envelope-expressing plasmid (pMDG) (1 μg), and wt or kd ATM-expressing plasmids or the empty vector pcDNA3-FLAG (2 μg). Three days after transfection, p24 levels in the culture supernatant were measured by ELISA.
FIG. 2.
Overexpression of ATM enhances HIV-1 replication at a posttranscriptional level. (a) Overexpression of ATM does not affect Tat-mediated transcription from HIV-1 LTR. 293T cells (2 × 104 cells) were cotransfected with the HIV-1-LTR-luciferase (HIV-1-LTR-Luc) reporter gene (100 ng), Tat-expressing plasmid (Tat101-FLAG) (100 ng), and/or ATM-expressing plasmid (200 ng). Twenty-four hours after transfection, luciferase activity in the cellular lysates was measured. Results from three independent experiments are shown, with error bars indicative of the standard deviations from the means. (b) Overexpression of ATM increases production of late viral proteins. 293T cells (2 × 105 cells) were cotransfected with an HIV-1 molecular clone (1 μg) and an ATM-expressing or a control plasmid (2 μg). Three days after transfection, Western blotting of the cellular lysate was performed with anti-ATM, anti-p24 CA, anti-p17 MA, anti-Nef, or anti-Chk2 antibody. (c) Northern blot analysis of cytoplasmic RNA from 293T cells cotransfected with an HIV-1 molecular clone with or without an ATM-expressing plasmid. The loading control was rRNA (18S and 28S rRNA) stained with ethidium bromide.
To further examine the level of ATM action, we performed Northern blot analyses of RNA purified from the cytoplasm of 293T cells transfected with the HIV-1 R9 proviral construct and control or ATM-expressing plasmids (Fig. 2c). Levels of fully spliced (2-kb) viral RNAs were similar whether or not ATM was overexpressed. In contrast, the kinase strikingly increased the amounts of singly spliced (4-kb) and full-length (9-kb) RNAs.
ATM overexpression enhances Rev function.
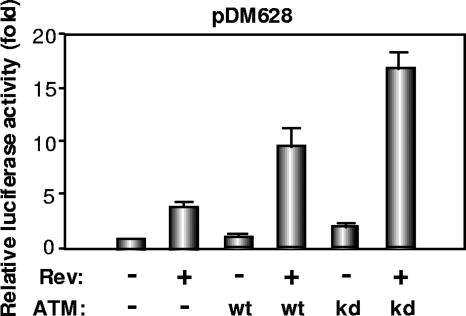
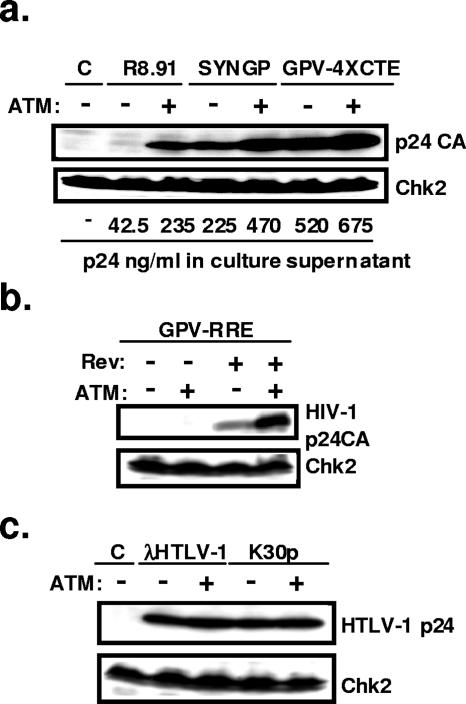
A selective increase in the levels of incompletely spliced HIV-1 RNA and of viral late proteins was consistent with either an inhibition of splicing or a stimulation of Rev function. To probe this issue, we first used the luciferase-based reporter plasmid pDM628 (15) (Fig. 3). As previously reported, luciferase production in 293T cells transfected with this plasmid was markedly stimulated by Rev, which induced a four- to fivefold increase in reporter signal. While ATM alone had no effect, it synergized with Rev, whether in wild-type or kinase-defective form, with luciferase levels increased by 10-fold, when both proteins were expressed. To further examine a possible influence of ATM on HIV RNA transport, we then compared three Gag-Pol expression systems: the Rev-dependent HIV-1 gag-pol expressing construct, pCMVΔR8.91 (41); a codon-optimized Rev-independent HIV-1 gag-pol vector, pSYNGP (21); and a Rev/Crm1-independent HIV-1 gag-pol construct containing four tandem copies of the Mason-Pfizer monkey virus CTE, pGPV-4×CTE (32, 36). Cotransfection of an ATM vector increased p24 CA protein production from the Rev-expressing HIV-1 packaging construct CMVΔR8.91 approximately sixfold. In contrast, it had only a modest effect (twofold) on the Rev-independent codon-optimized HIV-1 gag-pol construct SYNGP and did not significantly stimulate expression from the CTE-containing GPV-4×CTE plasmid (Fig. 4a). In line with these results, ATM overexpression also increased p24 CA production from the HIV-derived RRE-containing HIV-1 gag-pol construct GPV-RRE, but it did not alleviate its Rev dependence (Fig. 4b). Finally, ATM overexpression did not induce a general enhancement of Crm1-mediated nuclear export, because it did not stimulate capsid production from two HTLV-1 molecular clones (25, 39) (Fig. 4c), in which the Rex-mediated export of late viral mRNAs also proceeds through the Crm1 pathway (18).
FIG. 3.
Overexpression of ATM enhances Rev function. 293T cells (2 × 104 cells) were cotransfected with the Rev-dependent luciferase-based reporter gene pDM628 (100 ng), the Rev-expressing plasmid pcRev (100 ng), and/or wild-type (wt) or kinase-defective (kd) ATM-expressing plasmids (200 ng). Twenty-four hours after transfection, luciferase activity in the cellular lysates was measured. Results are from three independent experiments.
FIG. 4.
ATM effect is specific to Rev-dependent gene expression. (a) 293T cells (2 × 105 cells) were cotransfected with the Rev/RRE-dependent gag-pol construct pCMV R8.91, the Rev-independent codon-optimized HIV-1 gag-pol construct pSYNGP, or an HIV-1 gag-pol construct containing four tandem copies of the Mason-Pfizer monkey virus CTE, pGPV-4×CTE (2 μg), as well as ATM-expressing or empty vectors (2 μg). Three days after transfection, Western blotting of the cellular lysate was performed with anti-p24 CA or anti-Chk2 antibody, and p24 levels in the supernatant were measured by ELISA. (b) Overexpression of ATM increases HIV-1 p24 only in the presence of Rev. 293T cells were cotransfected with an RRE-containing HIV-1 gag-pol construct (pGPV-RRE) (2 μg), a Rev-expressing plasmid (1 μg), and/or an ATM-expressing plasmid (2 μg). Western blotting was performed as described for panel a. (c) Overexpression of ATM does not affect HTLV-1 Rex function. 293T cells were cotransfected with the HTLV-1 molecular clone λHTLV-1C or K30p (2 μg) with or without an ATM-expressing plasmid (2 μg). Three days after transfection, Western blotting of the cellular lysate was performed with anti-HTLV-1 p24 or anti-Chk2 antibody. C, control.
Reduced HIV replication in ATM knockdown cells.
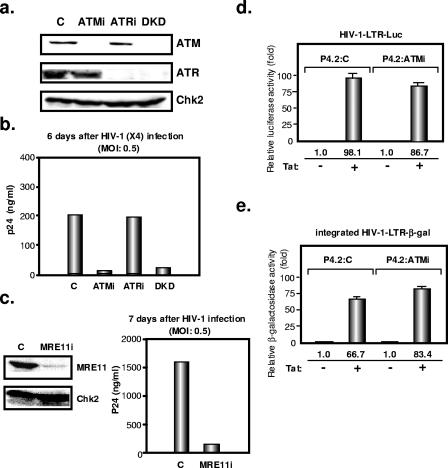
We then measured HIV-1 replication in CD4+ HeLa cells in which ATM and/or ATR expression was down-regulated by RNA interference (4) (Fig. 5a). The viability and growth kinetics of these cells were comparable to those of control cells (not illustrated). After infection with an X4 strain, p24 viral capsid levels were strongly reduced in the supernatants of ATM knockdown and ATM-ATR double-knockdown cells but not in that of ATR knockdown cells (Fig. 5b). We found a similar degree of HIV-1 inhibition in ATM knockdown SupT1 T-lymphoid cells (data not shown). ATM function requires the formation of a complex with MRE11 (9, 23). We thus examined HIV-1 replication in CD4+ HeLa cells in which MRE11 expression was down-regulated by RNA interference. Consistent with the results obtained with ATM knockdown cells, p24 production was significantly reduced in the MRE11 knockdown cells (Fig. 5c). Analyses of single-round infections in control, ATM knockdown, and/or ATR knockdown CD4+ HeLa cells confirmed that they were all equally susceptible to infection with wild-type HIV-1 (as determined by a multinucleate activation of galactosidase indicator assay) or to transduction with a VSV G-pseudotyped HIV-derived lentiviral vector, as previously described (data not shown) (4). ATM knockdown thus does not interfere with the early steps of infection, including integration, in this cell culture system. As well, HIV-1 Tat-mediated transcription was normal in ATM knockdown HeLa cells, as assessed by transient transfection of a Tat-responsive HIV-1-LTR-driven luciferase reporter plasmid (3) and by measuring the virus-induced transactivation of β-Gal activity from a stably integrated HIV-1-LTR-β-Gal construct (10) (Fig. 5d and e). Taken together, these results pointed to a defect in the late steps of viral replication in ATM knockdown cells. To test this hypothesis, we infected these and control cells with wild-type VSV G-pseudotyped HIV-1 at a multiplicity of infection (MOI) of 0.5 and measured the release of RT activity or p24 antigen in the supernatant. Forty-eight hours postinfection with wild-type HIV-1, that is, at a time corresponding to one or at most two rounds of replication, RT activity was decreased by about 30% in the supernatant of ATM knockdown cells (Fig. 6a). To further investigate this result, we transduced ATM knockdown and control cells with VSV G-pseudotyped Env-defective HIV-1 (capable of only one single round of infection) at an MOI of 5 and measured p24 production in the supernatant 55 h later. Virus production from the ATM knockdown cells was approximately 20% lower than from control cells (Fig. 6b). We then measured the infectivity of wild-type HIV-1 virions produced from ATM knockdown cells and found it to be reduced by about 50% compared with that of the control (Fig. 6c). While these defects did not appear individually spectacular, their cumulated impact resulted in a significant suppression of HIV-1 replication over multiple rounds, as reflected by a growth curve in which viral production was monitored over 6 days (Fig. 6d). However, the Rev-dependent luciferase-based pDM628 reporter plasmid failed to detect a significant defect of Rev function in this setting (not illustrated).
FIG. 5.
Reduced HIV replication in ATM knockdown cells. (a) Inhibition of endogenous ATM and ATR protein expression by shRNA-producing lentiviral vectors. Results are shown for Western blotting of cellular lysates with anti-ATM, anti-ATR, and anti-Chk2 antibodies in ATM knockdown (ATMi), ATR knockdown (ATRi), and double-knockdown (DKD) P4.2 cells as well as in P4.2 cells transduced with a control (C) lentiviral vector. (b) Each line was infected with HIV-1 at an MOI of 0.5. HIV-1 replication was assayed by p24 ELISA with the culture supernatants 6 days later. (c) Inhibition of endogenous MRE11 protein expression by shRNA-producing lentiviral vector. MRE11 knockdown (MRE11i) P4.2 cells were infected with HIV-1 at an MOI of 0.5. HIV-1 replication was assayed by p24 ELISA with the culture supernatants 7 days later. (d) HIV-1 Tat-mediated transcription in ATM knockdown HeLa cells. Tat-expressing plasmid (100 ng) and HIV-1-LTR-luciferase (HIV-1-LTR-Luc) reporter plasmid (100 ng) were cotransfected into control P4.2 cells (P4.2:C) or ATM knockdown P4.2 cells (P4.2:ATMi) (2 × 104 cells). A luciferase assay was performed 24 h later. Results are from three independent transfections. (e) Tat-expressing plasmid (100 ng) was transfected into P4.2 cells in triplicate. β-Gal activity of LTR-LacZ-containing cellular lysates was measured at an optical density at 570 nm 24 h later.
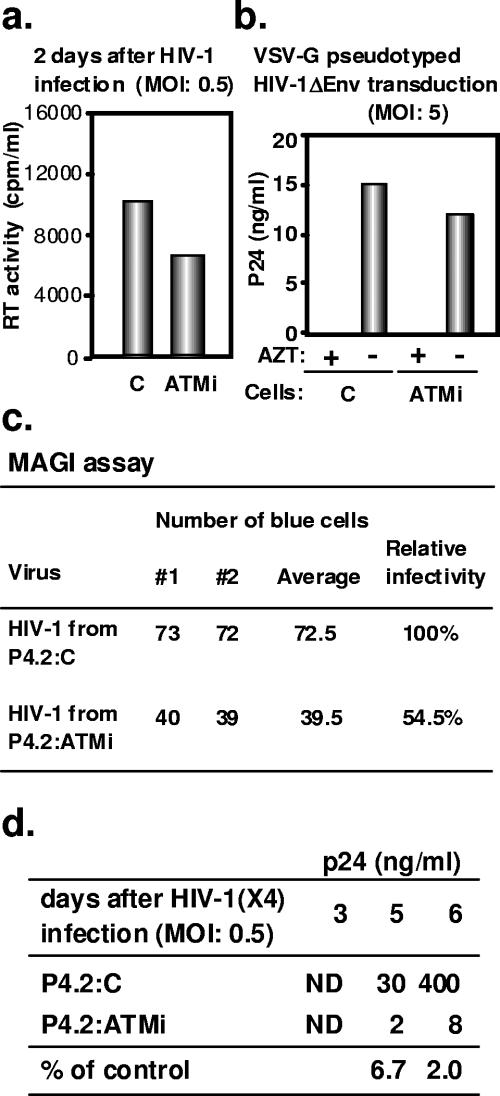
FIG. 6.
Decreased production of HIV-1 virions with reduced infectivity from ATM knockdown cells. (a) RT activity in the supernatants of control (C) or ATM knockdown (ATMi) P4.2 cells was measured 2 days after infection with HIV-1 at an MOI of 0.5. (b) Virion particle production in the supernatants of control or ATM knockdown P4.2 cells 55 h after transduction with VSV G-pesudotyped HIV-1 at an MOI of 5. Cells treated with 50 μM zidovudine (AZT) served as a negative control. Results are indicative of duplicate measurements that gave quasi-identical numbers. (c) ATM knockdown P4.2 cells (P4.2:ATMi) or control P4.2 cells (P4.2:C) were infected with wild-type HIV-1 (X4) at an MOI of 0.5. Normalized amounts of virions (as assessed by RT activity) harvested 6 days later from the supernatant of these cells were used to infect P4.2 cells. Virion infectivity was determined by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of the LTR-LacZ-containing target cells. Results from two independent experiments are shown. MAGI, multinucleate activation of galactosidase indicator. (d) p24 production in the supernatants of control and ATM knockdown P4.2 cells was measured by ELISA at indicated days after infection with HIV-1 at an MOI of 0.5. Percentages at bottom represent the ratio between ATM knockdown and control cell values. ND, under detection level.
Effect of caffeine on the late steps of HIV replication.
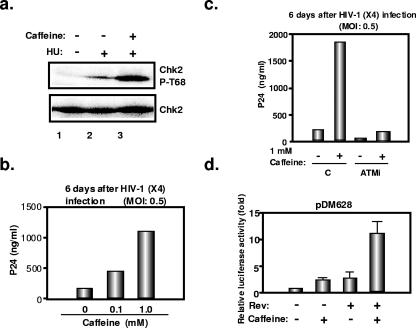
In an effort to examine further the role of ATM in HIV-1 replication, we used caffeine. This xanthine is commonly used as an inhibitor of ATM and ATR. However, with CD4+ HeLa cells, we unexpectedly found that it instead enhanced ATM kinase activity, as reflected by an increased accumulation of phosphorylated Chk2 in response to the DNA damaging agent hydroxyurea (Fig. 7a). Consistent with our failure to observe an influence of ATM overexpression or knockdown on the early steps of infection, including integration, in this system, caffeine had no influence on these events (not illustrated). In contrast, it increased the levels of p24 capsid antigen measured in the supernatant of CD4+ HeLa cells following HIV-1 infection (Fig. 7b). Caffeine also exerted a positive effect on this parameter with ATM knockdown cells, but in that case p24 levels remained only about 10% of those obtained from the supernatant of caffeine-treated ATM-positive cells (Fig. 7c). Finally, caffeine synergized with Rev to stimulate expression from the Rev-dependent pDM628 plasmid (Fig. 7d).
FIG. 7.
Effect of caffeine on the late steps of HIV-1 replication. (a) Caffeine induces hyperphosphorylation of the ATM kinase substrate Chk2, following hydroxyurea treatment. P4.2 cells were pretreated with 4 mM caffeine for 1 h and then treated with 5 mM hydroxyurea (HU) in the presence or absence of 4 mM caffeine for 3 h. Western blotting of the cellular lysates was performed with anti-Chk2 antibody or anti-phospho-Chk2 (Thr68) antibody (P-T68). (b) Caffeine enhances HIV-1 replication in P4.2 cells. P4.2 cells (2 × 105 cells) were pretreated with the indicated concentration of caffeine for 1 h and infected with HIV-1 (X4) at an MOI of 0.5. p24 levels in the supernatants were measured by ELISA 6 days later. (c) ATM knockdown (ATMi) or control (C) P4.2 cells (2 × 105 cells) were pretreated with 1 mM caffeine for 1 h and infected with HIV-1 (X4) at an MOI of 0.5 in the presence or absence of caffeine. p24 levels in the supernatants were measured at 6 days. (d) Caffeine enhances Rev function in P4.2 cells. P4.2 cells (2 × 104 cells) treated with 1 mM caffeine were transfected with the Rev-dependent luciferase-based reporter gene pDM628 (100 ng) with or without the Rev-expressing plasmid pcRev (100 ng). Twenty-four hours later, luciferase activity in the cellular lysates was determined. Results are from three independent experiments.
DISCUSSION
A large number of studies have examined a possible role of the DNA damage control protein ATM in HIV integration. Results have been conflicting, most likely because of cell-type-specific differences and possibly the vector systems used by various investigators. Daniel et al. found only a marginal impact of ATM on retroviral transduction by use of ATM knockout fibroblasts (12). Similarly, we observed that the transduction efficiency of HIV-based vector was decreased by 20% at most in ATM knockout fibroblasts (4). In addition, HIV-1 could integrate normally in both ATM knockdown HeLa cells (4) and SupT1 cells (data not shown) in spite of very effective downregulation of ATM. Also using RNA interference, DeHart et al. reported that ATM was not required for transduction with a lentiviral vector (14). Moreover, we and others observed that HIV-based vector could normally integrate into M059J cells, which lack DNA-PKcs and express very low levels of ATM (4, 20). Finally, ATM knockout mice are as sensitive to Friend leukemia virus-induced leukemogenesis as wild-type mice, indicating that ATM is not essential for Friend leukemia virus integration (19). While these results establish that integration of retroviruses in general and HIV-1 in particular does not absolutely require ATM, data obtained from primary T lymphocytes with a small-molecule inhibitor suggest that the kinase can play a positive role in HIV-1 replication by minimizing retrovirus-induced cell death (22). Here, we describe a series of experiments revealing that ATM can also stimulate a late phase of viral replication.
We first observed that ATM overexpression enhanced the production of late viral proteins within the context of HIV-1 provirus-derived constructs. This correlated with a preferential increase in the levels of incompletely spliced and unspliced viral mRNAs, pointing to a stimulation of Rev function. Confirming this hypothesis, ATM overexpression augmented Rev-induced expression from an HIV intron-containing reporter system. This effect was also detected when CD4+ HeLa cells were treated with caffeine, which in these cells stimulates ATM activity. In ATM knockdown CD4+ HeLa cells, HIV-1 replication was slowed down, correlating with slightly decreased levels of particle release and of virion infectivity. While it is conceivable that the latter phenotype resulted from an impairment of Rev function, this could not be confirmed with the Rev-dependent reporter system. It may be that, in these cells, residual ATM expression or the production of a compensating factor had an attenuating effect.
Remarkably, ATM had no significant influence on HIV-1 late gene expression when nuclear export of the corresponding viral RNAs was artificially targeted to the TAP1-dependent pathway via the Mason-Pfizer monkey virus CTE sequence. It also did not stimulate viral capsid production from two HTLV-1 proviral clones, an event regulated by the Rex-mediated connection of HTLV-1 unspliced mRNA to the Crm1-dependent RNA export pathway (18). Therefore, ATM is not a general modulator of the Crm1 pathway but rather acts specifically on Rev-dependent RNAs. Interestingly, retroviral mRNA export is linked with the trafficking of Gag to cellular membranes and efficient HIV assembly (32). In this regard, even though ATM is located predominantly in the nucleus, a significant proportion of the protein can be detected in cytoplasmic vesicles (24, 35), suggesting that ATM might influence such a trafficking step. It has also been demonstrated that several cellular cofactors, such as the DEAD box RNA helicase family members DDX1 and DDX3, regulate Rev function (15, 33, 38). As well, IκB has been shown to inhibit HIV-1 Rev function (37), pointing to a posttranscriptional role for the NF-κB/IκB pathway in HIV-1 replication. Interestingly, the activation of NF-κB following DNA damage is abrogated in ATM-deficient cells (29), indicating a link between NF-κB and the ATM signaling pathway. However, IκB inhibition was found to be equally active on HTLV-1 Rex and HIV-1 Rev functions (37), which suggests that it does not underlie the hereby-described ATM effect, since the latter is restricted to Rev (the present work).
Caffeine is a reported inhibitor of ATM and ATR both in vitro and in vivo (30, 40). However, we found that this xanthine stimulated ATM activity in CD4+ HeLa P4.2 cells and that, in this setting, it also enhanced HIV replication. With other cell lines, such as the human T-lymphoid SupT1 and Jurkat cells, caffeine partly inhibited viral replication, which correlated with a decrease of Tat function (Y. Ariumi and D. Trono, unpublished data). In none of these settings, however, did we observe an effect of caffeine on the integration of HIV and derived vectors, which contrasts with results with human peripheral blood mononuclear cells recently described by Daniel et al. (13).
The precise mechanism by which ATM stimulates HIV-1 late gene expression is unclear. The wild-type levels of activity of a catalytic site mutant for this type of effect suggest that ATM does not act by phosphorylating a substrate important for Rev function, be it Rev or one of its cellular cofactors. Of note, the ATM cofactor MRE11, the knockdown of which we found to decrease HIV-1 replication, binds with both wild-type and kinase-defective ATM (23). It could be that ATM, which has been shown capable of interacting with many proteins, stabilizes or retargets within the cell a Rev cofactor or, on the contrary, that it sequesters an inhibitor of the nuclear export of Rev-dependent RNAs, thus facilitating the action of the viral protein. Irrespectively, these data justify investigating the antiviral effect of ATM inhibitors not only on integration but also on the late steps of HIV replication.
Acknowledgments
We thank Priscilla Turelli, Bastien Mangeat, Sandrine Vianin, Romaine Stalder, and Elisabeth Buhlmann for discussion and help with the experiments and Michael H. Malim, Jianhua Fang, Roger J. Pomerantz, Kyriacos A. Mitrophanous, Monsef Benkirane, Thomas J. Kindt, Masakazu Hatanaka, Hiroyuki Sakai, Genoveffa Franchini, Amalio Telenti, Richard Iggo, and Michael B. Kastan for the gift of reagents.
This work was supported by the Swiss National Science Foundation.
REFERENCES
- 1.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C., L. Krause, Y.-L. Chen, and D. Trono. 1996. Mutational analysis of HIV-1 Nef: identification of two mutants that are temperature-sensitive for CD4 downregulation. Virology 217:293-300. [DOI] [PubMed] [Google Scholar]
- 3.Ariumi, Y., A. Kaida, M. Hatanaka, and K. Shimotohno. 2001. Functional cross-talk of HIV-1 Tat with p53 through its C-terminal domain. Biochem. Biophys. Res. Commun. 287:556-561. [DOI] [PubMed] [Google Scholar]
- 4.Ariumi, Y., P. Turelli, M. Masutani, and D. Trono. 2005. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J. Virol. 79:2973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bres, V., R. E. Kiernan, L. K. Linares, C. Chable-Bessia, O. Plechakova, C. Treand, S. Emiliani, J. M. Peloponese, K. T. Jeang, O. Coux, M. Scheffner, and M. Benkirane. 2003. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat. Cell Biol. 5:754-761. [DOI] [PubMed] [Google Scholar]
- 6.Bridge, A. J., S. Pebernard, A. Ducraux, A.-L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Canman, C. E., D.-S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Apella, M. B. Kastan, and J. D. Siliciano. 1988. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 9.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 11.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419-424. [DOI] [PubMed] [Google Scholar]
- 12.Daniel, R., G. Kao, K. Taganov, J. G. Greger, O. Favorova, G. Merkel, T. J. Yen, R. A. Katz, and A. M. Skalka. 2003. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc. Natl. Acad. Sci. USA 100:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel, R., E. Marusich, E. Argyris, R. Y. Zhao, A. M. Skalka, and R. J. Pomerantz. 2005. Caffeine inhibits human immunodeficiency virus type 1 transduction of nondividing cells. J. Virol. 79:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeHart, J. L., J. L. Andersen, E. S. Zimmerman, O. Ardon, D. S. An, J. Blackett, B. Kim, and V. Planelles. 2005. The ataxia telangiectasia-mutated and Rad3-related protein is dispensable for retroviral integration. J. Virol. 79:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang, J., S. Kubota, B. Yang, N. Zhou, H. Zhang, R. Godbout, and R. J. Pomerantz. 2004. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology 330:471-480. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 17.Grüter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from nucleus. Mol. Cell 1:649-659. [DOI] [PubMed] [Google Scholar]
- 18.Hakata, Y., T. Umemoto, S. Matsushita, and H. Shida. 1998. Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type 1. J. Virol. 72:6602-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa, M., S. Yamaguchi, S. Aizawa, H. Ikeda, K. Tatsumi, Y. Noda, K. Hirokawa, and M. Kitagawa. 2005. Resistance against Friend leukemia virus-induced leukemogenesis in DNA-dependent protein kinase (DNA-PK)-deficient scid mice associated with defective viral integration at the Spi-1 and Fli-1 site. Leuk. Res. 29:933-942. [DOI] [PubMed] [Google Scholar]
- 20.Kilzer, J. M., T. Stracker, B. Beitzel, K. Meek, M. Weitzman, and F. D. Bushman. 2003. Roles of host cell factors in circularization of retroviral DNA. Virology 314:460-467. [DOI] [PubMed] [Google Scholar]
- 21.Kotsopoulou, E., V. N. Kim, A. J. Kingsman, S. M. Kingsman, and K. A. Mitrophanous. 2000. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 74:4839-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau, A., K. M. Swinbank, P. S. Ahmed, D. L. Taylor, S. P. Jackson, G. C. M. Smith, and M. J. O'Connor. 2005. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat. Cell Biol. 7:493-500. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J.-H., and T. T. Paull. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304:93-96. [DOI] [PubMed] [Google Scholar]
- 24.Lim, D.-S., D. G. Kirsch, C. E. Canman, J.-H. Ahn, Y. Ziv, L. S. Newman, R. B. Darnell, Y. Shiloh, and M. B. Kastan. 1998. ATM binds to β-adaptin in cytoplasmic vesicles. Proc. Natl. Acad. Sci. USA 95:10146-10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori, K., H. Sabe, H. Siomi, T. Iino, A. Tanaka, K. Takeuchi, K. Hirayoshi, and M. Hatanaka. 1987. Expression of a provirus of human T cell leukemia virus type I by DNA transfection. J. Gen. Virol. 68:499-506. [DOI] [PubMed] [Google Scholar]
- 26.Naldini, L., U. Blömer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 27.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 28.Pasquinelli, A. E., R. K. Ernst, E. Lund, C. Grimm, M. L. Zapp, D. Rekosh, M.-L. Hammarskjöld, and J. E. Dahlberg. 1997. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 16:7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piret, B., S. Schoonbroodt, and J. Piette. 1999. The ATM protein is required for sustained activation of NF-κB following DNA damage. Oncogene 18:2261-2271. [DOI] [PubMed] [Google Scholar]
- 30.Sarkaria, J. N., E. C. Busby, R. S. Tibbetts, P. Roos, Y. Taya, L. M. Karnitz, and R. T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375-4382. [PubMed] [Google Scholar]
- 31.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 32.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trono, D., and D. Baltimore. 1990. A human cell factor is essential for HIV-1 Rev action. EMBO J. 9:4155-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turlure, F., E. Devroe, P. A. Silver, and A. Engelman. 2004. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9:3187-3208. [DOI] [PubMed] [Google Scholar]
- 35.Watters, D., P. Kedar, K. Spring, J. Bjorkman, P. Chen, M. Gatei, G. Birrell, B. Garrone, P. Srinivasa, D. I. Crane, and M. F. Lavin. 1999. Localization of a portion of extranuclear ATM to peroxisomes. J. Biol. Chem. 274:34277-34282. [DOI] [PubMed] [Google Scholar]
- 36.Wodrich, H., A. Schambach, and H. G. Kräusslich. 2000. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 28:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, B.-Y., C. Woffendin, C. S. Duckett, T. Ohno, and G. J. Nabel. 1995. Regulation of human retroviral latency by the NF-κB/IκB family: inhibition of human immunodeficiency virus replication by IκB through a Rev-dependent mechanism. Proc. Natl. Acad. Sci. USA 92:1480-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yedavalli, V. S. R. K., C. Neuveut, Y. Chi, L. Kleiman, and K.-T. Jeang. 2004. Requirement of DDX3 dead box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381-392. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, T. M., M. A. Robinson, F. S. Bowers, and T. J. Kindt. 1995. Characterization of an infectious molecular clone of human T-cell leukemia virus type I. J. Virol. 69:2024-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, B.-B. S., P. Chaturvedi, K. Spring, S. P. Scott, R. A. Johanson, R. Mishra, M. R. Mattern, J. D. Winkler, and K. K. Khanna. 2000. Caffeine abolishes the mammalian G2/M DNA damage checkpoint by inhibiting ataxia-telangiectasia-mutated kinase activity. J. Biol. Chem. 275:10342-10348. [DOI] [PubMed] [Google Scholar]
- 41.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]