Abstract
Hepatitis C virus (HCV) RNA circulates in the blood of persistently infected patients in lipoviroparticles (LVPs), which are heterogeneous in density and associated with host lipoproteins and antibodies. The variability and lability of these virus-host complexes on fractionation has hindered our understanding of the structure of LVP and determination of the physicochemical properties of the HCV virion. In this study, HCV from an antibody-negative immunodeficient patient was analyzed using three fractionation techniques, NaBr gradients, isotonic iodixanol, and sucrose gradient centrifugation. Iodixanol gradients were shown to best preserve host lipoprotein-virus complexes, and all HCV RNA was found at densities below 1.13 g/ml, with the majority at low density, ≤1.08 g/ml. Immunoprecipitation with polyclonal antibodies against human ApoB and ApoE precipitated 91.8% and 95.0% of HCV with low density, respectively, suggesting that host lipoprotein is closely associated with HCV in a particle resembling VLDL. Immunoprecipitation with antibodies against glycoprotein E2 precipitated 25% of HCV with low density, providing evidence for the presence of E2 in LVPs. Treatment of serum with 0.5% deoxycholic acid in the absence of salt produced HCV with a density of 1.12 g/ml and a sedimentation coefficient of 215S. The diameters of these particles were calculated as 54 nm. Treatment of serum with 0.18% NP-40 produced HCV with a density of 1.18 g/ml, a sedimentation coefficient of 180S, and a diameter of 42 nm. Immunoprecipitation analysis showed that ApoB remained associated with HCV after treatment of serum with deoxycholic acid or NP-40, whereas ApoE was removed from HCV with these detergents.
Since the cloning of hepatitis C virus (HCV) by Choo et al. (11), the genome has become well characterized and the virus is classified as a separate genus, Hepacivirus, within the family Flaviviridae (43). An important long-term objective of HCV research is the identification of the HCV virion, the infectious particle, and determination of its size and structural composition. Currently, physicochemical data for the virus particle are inadequate due to the low titer of the virus in blood and the difficulty in isolating pure virus particles (50). Also, measurements of density and sedimentation properties produce a variety of fractions of HCV which are different in density and sedimentation rate. This may be due to differences in the structures of viruses, to the binding of HCV to host lipoproteins (low-density lipoprotein [LDL], very-low-density lipoprotein [VLDL], and high-density lipoprotein [HDL]), to specific anti-viral antibodies (immunoglobulin G [IgG] and IgM), or to rheumatoid factors and cryoglobulins. Furthermore, this heterogeneity could be the consequence of dissociative properties of the matrix of the gradients. Density heterogeneity is partly attributable to the association of circulating HCV RNA with host LDL/VLDL, as described by Thomssen et al. (52, 53), a finding subsequently confirmed by several groups (1, 4, 41, 59). This association may facilitate virus uptake via the LDL receptor (1, 19, 37, 56, 59), as uptake of serum-derived HCV by cultured cells correlates with the expression level of LDL receptors (4). In line with this, sera rich in low-density particles associated with host lipoproteins have been shown to be infectious in chimpanzees, whereas the infectivity of sera with higher-density particles was reduced (6).
HCV RNA in the blood may also be bound to immunoglobulin (12, 25, 27, 53). Immune complexes containing HCV and polyclonal IgG, as well as monoclonal IgM, have been described (2, 45). Some of this immunoglobulin may represent specific anti-viral antibody. However, in at least a proportion of cases, rheumatoid factors are involved, and the complexes tend to precipitate upon storage of serum in the cold. This phenomenon is particularly marked in patients with type II cryoglobulinemia (2). HCV RNA-containing particles associated with both host lipoproteins and immunoglobulins have been shown to differ in size, density, and cholesterol/triglyceride ratio from host LDL or VLDL and have been termed lipoviroparticles (LVPs) (4).
The low concentration of virus in serum, together with the association of HCV with lipoprotein and antibodies, has also been an obstacle to the determination of the physicochemical properties of the particles recovered (50). The favored methods for the analysis of the density heterogeneity of HCV have been sequential centrifugation in salt gradients and density gradient centrifugation using sucrose. With these techniques, HCV RNA has been recovered over a wide range of densities between 1.03 g/ml and 1.25 g/ml. In some sera, all HCV RNA found has been of low density (ρ [buoyant density] ≤ 1.06 g/ml) (25, 52), associated with LDL/VLDL. With other sera, the majority of the HCV RNA population has had a density between 1.06 g/ml and 1.17 g/ml (25, 28). This density range may contain putative free virus particles, as well as virus-HDL complexes, virus-antibody complexes, or lipid-virus-antibody complexes (1, 25, 38). There are also sera in which most of the HCV RNA has a density above 1.18 g/ml (27, 29). HCV particles with such a high density are hypothesized to be nucleocapsids or nucleocapsid-antibody complexes. The lability of host lipoproteins during fractionation by these techniques complicates the interpretation of these data and the cross-comparison of different studies. These issues have been addressed in the present report through the use of iodixanol (Optiprep; Axis-Shield, Upton Huntingdon, United Kingdom) density gradients. Iodixanol is isosmotic and isotonic with blood and was found here to be superior to sucrose and NaBr gradients in preserving the integrity of host lipoprotein-virus complexes. This technique has been applied to an HCV-positive serum from an immunosuppressed, immunodeficient patient without viral antibodies and with an exceptionally high virus load. HCV RNA-containing particles have also been recovered after treatment with deoxycholic acid and NP-40, to remove lipid, and characterized by rate-zonal centrifugation, to determine the sedimentation coefficient and particle size. A panel of antibodies to host lipoproteins and viral glycoproteins was used to identify host and viral proteins exposed on the surfaces of HCV particles from serum and thus to contribute knowledge about ultrastructural features of the HCV lipoviroparticle (40, 57).
MATERIALS AND METHODS
HCV samples from patients.
Patient S6 suffered from common variable immunodeficiency and was infected with the 1a genotype of HCV from contaminated intravenous immunoglobulin with the brand name Gammagard (39). The patient required orthotopic liver transplantation for severe HCV-related cirrhosis and liver failure. Posttransplantation, liver function deteriorated and the patient had a retransplantation (17). The transplanted liver (S6b) was removed at retransplantation and found to have a high titer of HCV, 5 × 109 IU per gram of liver (39). Serum from patient S6 collected at the time of the retransplantation also contained a high titer of HCV, 6 × 108 IU/ml, and was negative by recombinant immunoblot assay and also negative for antibodies against HCV core antigen. Serum S6 was not tested for antibodies against HCV glycoproteins E1 and E2, but HCV RNA could not be precipitated with anti-human IgG (data not shown).
Blood samples were also collected from three immunocompetent patients (S7 to S9) with chronic hepatitis C infection prior to antiviral treatment (Table 1). A sample of plasma from each immunocompetent patient was applied to an NaBr gradient within 4 hours of collection, having been held throughout at 37°C. A second sample of each serum was stored at +4°C overnight and then centrifuged at 10,000 × g for 10 min prior to application to an iodixanol density gradient. This centrifugation removes cryoglobulins prior to analysis by gradient centrifugation (2).
TABLE 1.
HCV genotype, RNA titre and disease characteristics of patients studied
| Patient | Sample | Genotype | HCV RNA titer (IU)a | Disease |
|---|---|---|---|---|
| S6 | Serum | 1a | 6.0 × 108 | Acute recurrent |
| S6 | Transplanted liver S6b | 1a | 5.0 × 109 | Acute recurrent |
| S7 | Plasma | 5a | 4.9 × 106 | Moderate HCV infection |
| S8 | Plasma | 3a | 2.1 × 105 | Mild HCV infection |
| S9 | Plasma | 3a | 1.7 × 106 | Cirrhotic liver |
HCV titers are listed in IU of HCV RNA per ml serum or plasma and as IU per gram of liver.
Collection of samples was made with informed consent from the patients themselves, and the research project has been approved by the Ethical Committee at Newcastle University.
Density gradients and sequential ultracentrifugation.
Iodixanol is a nonionic dimeric density gradient medium with a molecular mass of 1,550 Da (15). Isopycnic linear density gradients were prepared from 6% (wt/vol) (1.7 ml of 60% [wt/vol] iodixanol, 0.34 ml of 0.5 M Tris-HCl, pH 8.0, 0.34 ml of 0.1 M EDTA, pH 8.0, and 14.6 ml 0.25 M sucrose) and 56.4% (wt/vol) (16.0 ml of 60% iodixanol, 0.34 ml of 0.5 M Tris-HCl, pH 8.0, 0.34 ml of 0.1 M EDTA, pH 8.0, and 0.34 ml 0.25 M sucrose) iodixanol solutions in Ultra-Clear centrifuge tubes (Beckman, High Wycombe, United Kingdom) using a two-chamber gradient maker (Jencons, Leighton Buzzard, United Kingdom). The gradients were poured light end first using a peristaltic pump (21). A sample of 10 μl serum mixed with 200 μl of 10 mM Tris-HCl (pH 7.4), 250 mM sucrose, and 2 mM EDTA was applied to the top of 6 to 56% iodixanol gradients and centrifuged for 24 h in a Beckman L8-70 M ultracentrifuge at 90,000 × gav and +4°C in an SW40 rotor. The gradient was harvested by tube puncture using a Model 184 tube piercer (Isco, Lincoln, NE) and collected into 14 fractions of 1 ml each. The density of each fraction was determined using a digital refractometer (Atago, Japan).
Linear sucrose gradients were mixed from two solutions: 10% (wt/vol) (20 mM Tris-HCl, pH 7.4, 2 mM EDTA, 0.29 M sucrose) and 85.5% (wt/vol) (20 mM Tris-HCl, pH 7.4, 2 mM EDTA, 2.5 M sucrose). Sample preparation, centrifugation, and harvesting were performed as for isopycnic iodixanol gradients.
Sequential ultracentrifugation was performed with NaBr solutions at densities of 1.063 g/ml and 1.21 g/ml, respectively. A sample of 100 μl from patient S6 or 1 ml from patients S7 to S9 was mixed with NaBr solution with a density of 1.063 g/ml and centrifuged at 146,000 × gav (gav is the average centrifugal force) in a Beckman Type 50 Ti fixed-angle rotor (42). After sequential ultracentrifugation, three samples of 1 ml each were harvested from low-density (ρ < 1.063 g/ml), intermediate-density (1.063 g/ml < ρ < 1.21 g/ml), and high-density (ρ < 1.21 g/ml) fractions.
Rate-zonal centrifugation was performed in 4 to 24% iodixanol gradients using samples of HCV purified by isopycnic iodixanol density centrifugation. Samples were first concentrated to 200 μl using an Ultrafree centrifugal filter unit with a 10,000-Da nominal molecular mass limit (Millipore, Watford, United Kingdom) and then dialyzed at +4°C against 20 mM Tris-HCl, pH 7.4, containing 0.25 M sucrose in a Slide-A-Lyzer Mini dialysis unit (10,000-Da nominal molecular mass limit; Perbio Science, Tattenhall, United Kingdom). After dialysis, the sample had a density of 1.03 g/ml and was applied to the top of a 4% to 24% iodixanol gradient and centrifuged at 90,000 × gav for 2 or 4 h. Centrifugation was stopped without a brake, and the gradient was harvested to determine the position of HCV RNA.
Calculation of sedimentation coefficient and diameter.
The sedimentation coefficient (s20,w) was determined from data obtained after rate-zonal centrifugation according to the following formula (60):
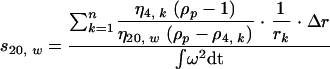 |
The s20,w was calculated as a sum over all fractions from the top fraction, k = 1, to the fraction n, where the particle is found after centrifugation for time t. η4,k/η20,w is the relative viscosity of iodixanol in fraction k at 4°C compared to water at 20°C and was determined with a viscometer. ρP is the buoyant density of the virus particle and was determined by isopycnic density gradient centrifugation using iodixanol. ρ4,k is the density of iodixanol at 4°C in fraction k. The time integral, ∫ω2 dt, includes acceleration and deceleration times. rk is the distance between fraction k and the center of rotation (31). The radius and diameter of the virus particle were calculated from the sedimentation coefficient according to Stokes' law: r2 = (9 · s20,w)/[20 · (ρ − ρ4,w)].
Precipitation of LDL/VLDL from serum with heparin/Mn2+ and lipoprotein antibody.
HCV-positive serum was centrifuged at 10,000 × g and +4°C for 30 min to remove aggregates. LDL/VLDL was then precipitated (34) by mixing 20 μl from the supernatant with an equal volume of 60 mM Tris-HCl (pH 8.0) containing 110 mM MnCl2, 154 mM NaCl, and 400 USP units/ml heparin (Sigma, Poole, United Kingdom). One hour of incubation at +4°C led to the appearance of a precipitate, which was recovered by centrifugation at 3,000 × g (30 min; +4°C). The precipitate was washed three times with a solution containing 50 mM Tris-HCl (pH 7.3), 55 mM MnCl2, 154 mM NaCl, and 200 USP units/ml heparin. The pellet was resolubilized in 150 μl of 50 mM Tris-HCl (pH 7.3) containing 150 mM NaCl and 100 mM EDTA.
LDL/VLDL was immunoprecipitated from 20 μl HCV-positive serum by the addition of an equal volume of rabbit anti-human ApoB antibody (Dako, Ely, United Kingdom) as described by Thomssen et al. (52). A multimeric complex (22) developed after overnight incubation at +4°C and was recovered by centrifugation at 5,000 × g (+4°C; 40 min). Normal rabbit IgG (Dako) was included as a control. The pellet after immunoprecipitation was washed twice with ice-cold phosphate-buffered saline and resuspended in either AVL lysis solution for RNA quantitation (QIAGEN, Crawley, United Kingdom) or sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (5) for protein analysis. LDL with a density between 1.018 g/ml and 1.063 g/ml was purified from normal human plasma by sequential flotation ultracentrifugation (34).
Treatment of serum with detergents and low or high pH.
Incubation of HCV serum with the anionic detergent deoxycholic acid was performed using a procedure modified from that of Prince et al. (41). Twenty-five microliters of serum was mixed with 200 μl of an ice-cold solution containing 10 mM Tris base (pH adjusted to 8.0 with 5 N NaOH), 2 mM EDTA, and 0.5% (wt/vol) deoxycholic acid (Ultra grade; Sigma). The sample was incubated for 30 min on ice prior to application to a precooled (0°C) 6% to 56% iodixanol density gradient made without the detergent. The gradient was spun at 90,000 × gav (24 h at 4°C).
Treatment of HCV serum with NP-40 was performed as described by Miyamoto et al. (36). Twenty-five microliters of serum was mixed with 200 μl of an ice-cold solution containing 10 mM Tris-HCl (pH 8.0), 2 mM EDTA, 100 mM NaCl, 0.2% (vol/vol) NP-40 (Serva, Heidelberg, Germany), and 200 units of RNasin (Promega, Southhampton, United Kingdom). Samples were held on ice for 12 min prior to analysis by isopycnic and rate-zonal centrifugations. Detergent-treated samples were analyzed by rate-zonal centrifugation in precooled 4% to 24% iodixanol gradients without detergent.
Treatment of HCV serum with low pH was performed using a method previously developed to test the low-pH sensitivity of hog cholera virus and bovine viral diarrhea virus (14). PIPPS [piperazine-N,N′-bis(3-propanesulfonic acid); Calbiochem, Nottingham, United Kingdom] was used as a biological buffer with pH 4.0. Ten microliters of serum was mixed with 200 μl of an ice-cold solution containing 20 mM PIPPS (pH 4.0 at 21°C), 250 mM sucrose, and 2 mM EDTA. The sample was applied to a 6% to 56% iodixanol gradient made with light and heavy solutions containing 10 mM PIPPS, pH 4.0.
RNA extraction and quantitation by real-time RT-PCR.
RNA was extracted from 100 μl of each gradient fraction using a QIAamp viral-RNA kit (QIAGEN, Crawley, United Kingdom). Reverse transcription (RT) and PCR were performed with two oligonucleotide primers corresponding to bases 29 to 46 (sense) and 251 to 268 (antisense) of the 5′ noncoding region of HCV as described previously (42). The 240-bp PCR product was analyzed on 2% agarose gels, and the intensities of bands were semiquantitated with an AlphaImager 2200 digital-image camera (Flowgen, Ashby de la Zouch, United Kingdom).
Real time quantitative RT-PCR was carried out as described previously (39) using primers and a probe annealing between positions 120 and 290 in the 5′ noncoding region of the HCV 1a genome. This assay was calibrated against the World Health Organization international standard for HCV 96/790 from the National Institute of Biological Standards and Controls (Hertfordshire, United Kingdom). The calibration curve was linear between 3 IU and 600,000 IU per tube.
Samples derived from heparin/Mn2+ precipitation experiments contained heparin, which is an inhibitor of PCR and is not removed by the QIAamp viral-RNA kit (QIAGEN, personal communication). Therefore, such samples were first mixed with 1 ml of 4.0 M guanidinium thiocyanate and layered onto a cushion of 4.0 ml 5.7 M CsCl in a thick-walled Ti 50 centrifuge tube (Beckman). After centrifugation at 146,000 × gav (18°C; 24 h), RNA in the pellet was resuspended in 100 μl 4.0 M guanidinium thiocyanate and repurified with a QIAamp viral-RNA kit.
Immunoprecipitation of HCV from iodixanol gradient fractions.
Immunoprecipitations were performed with fractions from an iodixanol density gradient in the range (i) less than 1.06 g/ml, (ii) 1.11 g/ml to 1.13 g/ml, or (iii) 1.18 g/ml. Each fraction was diluted to 2 ml with 10 mM Tris HCl, pH 8.0, containing 0.5% IgG-free bovine serum albumin (Sigma) and 0.25 M sucrose. Iodixanol was added to readjust the density to match that of the original gradient fraction. The polyclonal antibodies used in immunoprecipitations were rabbit anti-human ApoB and anti-human ApoE (both from Dako, Ely, United Kingdom), goat anti-HCV E2 (B65581G; Biødesign, Maine), and goat anti-HCV core protein (HG-22; Fitzgerald, MA). Mouse monoclonal antibodies (MAbs) used in immunoprecipitations were anti-HCV E1 (11B7D8 and 15G6B2; a gift from E. Depla, Innogenetics, Ghent, Belgium), anti-HCV E2 (AP33 and ALP98; a gift from A. Patel, Glasgow, United Kingdom), and anti-HCV core protein (Φ126; Biogenesis, Poole, United Kingdom). Human monoclonal antibodies used in immunoprecipitations were anti-HCV E2 (CBH-2, CBH-5, CBH-7, and CBH-4D) and anti-HCV E1 (H-111; a gift from S. Foung, Palo Alto, Calif.). Control reactions received either normal rabbit IgG (Dako), normal goat IgG (Sigma, Poole, United Kingdom), normal human IgG, or an isotype control monoclonal antibody against respiratory syncytial virus (2G12-2, isotype IgG1) (44). All antibodies were purified on protein G-Sepharose (Sigma). Aliquots from density gradient fractions were mixed with 6 μg monoclonal antibody or 12 μg polyclonal antibody in a final volume of 160 μl and then incubated at +4°C overnight. These samples did not contain detergent. Antibodies and HCV RNA-containing particles recognized by the antibodies were precipitated by the addition of 25 μl protein G-Sepharose. The Sepharose beads were incubated with the samples overnight at +4°C with rotation (11 rpm/min) and then pelleted by centrifugation at 3,000 × g (10 min; +4°C). The pellet was washed two times with 500 μl of 10 mM Tris HCl, pH 8.0, containing 0.25 M sucrose. RNA was extracted from the pellet and supernatant with a QIAamp viral-RNA kit, and the amount of HCV RNA in each compartment was determined by quantitative real-time PCR.
Immunoprecipitation of HCV structural proteins.
Antibodies which were evaluated for immunoprecipitation of HCV RNA-containing particles (see above) were also tested for the ability to precipitate S6b-derived HCV viral proteins. A liver macerate was prepared from the transplanted liver S6b using Dounce homogenization as previously described (39); 2.3 ml macerate with 5.5 × 108 IU of HCV RNA was applied to a self-forming iodixanol gradient to isolate lipoviroparticles with densities below 1.06 g/ml (39). Fractions from the top of the gradient were diluted in nondenaturing immunoprecipitation buffer (22) containing 50 mM Tris HCl, pH 7.4, 0.25 M sucrose, 1% NP-40, 2 mM EDTA, 6 μl/ml proteinase inhibitor cocktail (Sigma), and 0.1% IgG-free bovine serum albumin. Prior to immunoprecipitation, 25 μl of protein G-Sepharose beads was mixed with either 6 μg of monoclonal antibodies or 12 μg of polyclonal antibodies. Sepharose beads with bound antibody were washed with 0.2 M triethanolamine, pH 8.2, and then cross-linked with 40 mM dimethyl pimelimidate dihydrochloride (Sigma) in the same buffer for 1 h at 25°C (46). The beads were mixed with 1.8 ml lipoviroparticles in nondenaturing immunoprecipitation buffer and incubated with rotation at +4°C overnight. Bound protein was eluted by boiling it in SDS sample buffer (5) and analyzed by SDS-PAGE and Western blotting. Glycoprotein E1 was detected using antibody 11B7D8 at 1 μg/ml, and glycoprotein E2 was detected using antibody AP33 at 1 μg/ml. The secondary antibody was goat anti-mouse peroxidase conjugate (Novocastra Laboratories, Newcastle upon Tyne, United Kingdom) at 1/6,000, and Western blots were developed using ECL Plus (GE Healthcare, Buckinghamshire, United Kingdom). The amount of HCV glycoprotein precipitated was evaluated using a GS-800 calibrated densitometer with Quantity One software (Bio-Rad, Hemel Hempstead, United Kingdom).
RESULTS
Density distributions of HCV in untreated serum from patient S6 in different gradient materials.
The density distributions of HCV RNA from patient S6 were compared by isopycnic density gradient centrifugation using iodixanol or sucrose gradients and by sequential ultracentrifugation in NaBr solutions. The HCV RNA contents of fractions collected from the gradients were semiquantified by densitometry. The analysis by RT-PCR of untreated S6 serum in an isotonic iodixanol gradient showed that all HCV RNA had a density below 1.13 g/ml. Indeed, the majority of viral RNA was found toward the top of the gradient with a density below 1.08 g/ml (Fig. 1A). In contrast, when serum from patient S6 was separated on a sucrose gradient, one peak of HCV RNA with low density (ρ ≤ 1.06 g/ml) was observed, along with a second peak of HCV RNA at a higher density, between 1.13 and 1.16 g/ml (Fig. 1B). This bimodal distribution of HCV in sucrose gradients was confirmed by quantitative RT-PCR. Our observation is very similar to data published by Trestard et al. (55), who detected a major peak of HCR RNA at a density of 1.13 g/ml and a minor peak at a density of 1.05 to 1.07 g/ml in a sucrose density gradient from serum of an agammaglobulinemic patient.
FIG. 1.
Density distribution of HCV RNA in serum from immunodeficient patient S6 on iodixanol and sucrose gradients. Two identical serum samples with 6 × 106 IU of HCV RNA were applied to either an iodixanol (A) or a sucrose (B) density gradient. The two gradients were centrifuged and harvested under identical conditions. Fractions are numbered from the bottom, dense end of the gradient. HCV RNA in each fraction was detected by RT-PCR and analyzed by agarose gel electrophoresis. The PCR product from each fraction was semiquantified by densitometry, and values are shown as bar graphs. The linearity of each density gradient is shown as a line graph. The peak fraction of HCV RNA in the iodixanol gradient had a density below 1.04 g/ml. Two peak fractions of HCV RNA in the sucrose gradient were in the range below 1.08 g/ml and 1.12 to 16 g/ml.
After sequential ultracentrifugation of the untreated S6 serum in NaBr, quantitative RT-PCR revealed the distribution of viral RNA. The low-density fraction (ρ ≤ 1.063 g/ml) contained 1 × 107 IU (33%), the intermediate-density fraction (1.063 g/ml < ρ < 1.21 g/ml) contained 1.7 × 107 IU (57%), and the high-density fraction (ρ ≥ 1.21 g/ml) contained 3 × 106 IU (10%). The recovery from HCV RNA applied to the gradient was 50%. As the intermediate-density fraction contained the highest percentage of HCV RNA, the prevalence of low-density HCV in serum S6 observed in the iodixanol gradient is not apparent in the NaBr gradient.
Density distribution of HCV from untreated plasma samples from patients S7 to S9.
The comparison between isotonic iodixanol gradients and hypertonic NaBr gradients was extended to include plasma from three immunocompetent patients infected with HCV of different genotypes. The density distribution of HCV RNA from patient S7 was biased toward the top of the iodixanol gradient, with a peak fraction occurring at a density below 1.06 g/ml (Fig. 2A). Patient S8 showed a peak density around 1.07 g/ml (Fig. 2B), and HCV from patient S9 was biased toward the intermediate-density range, with a peak occurring at 1.14 g/ml (Fig. 2C).
FIG. 2.
Density distribution of HCV RNA in plasma from three immunocompetent patients using iodixanol gradients. All three patients, S7, S8, and S9, had been diagnosed with chronic HCV infection, and antiviral treatment had not been initiated. (A) Patient S7 had moderate HCV infection, and 100 μl plasma with 2.6 × 105 IU of HCV RNA was applied to the gradient. (B) Patient S8 had mild HCV infection, and 100 μl plasma with 1.6 × 104 IU of HCV RNA was applied to the gradient. (C) Patient S9 had been diagnosed with cirrhosis due to HCV infection, and 100 μl plasma with 1.3 × 104 IU of HCV RNA was applied to the gradient. RNA in each fraction from the gradients was detected by RT-PCR and analyzed by agarose gel electrophoresis. The PCR product was semiquantified by densitometry, and values are shown as bar graphs. The linearity of each density gradient is shown as a line graph.
After ultracentrifugation in NaBr gradients, HCV RNA from patients S7 to S9 showed the highest titer of HCV RNA in the intermediate-density fraction (1.063 g/ml < ρ < 1.21 g/ml): S7, 1.2 × 106 IU (52%); S8, 1.3 × 105 IU (65%); and S9, 2.2 × 105 IU (50%). Patient S7 also had a high titer of HCV RNA in the low-density fraction (ρ < 1.063 g/ml; 8.6 × 105 IU [37%]). The low-density titers in patients S8 and S9 were 4 × 104 IU (20%) and 2 × 104 IU (4%), respectively. Thus, a good correspondence between NaBr gradients and iodixanol gradients was observed, but the excess of low-density HCV RNA in the iodixanol gradient from patient S7 was not evident in the NaBr gradient, and fractions of HCV RNA with high density (ρ ≥ 1.21 g/ml) found with NaBr in all three sera were not found with iodixanol. This suggests that the osmolarity of the gradient matrix may contribute to the density heterogeneity of HCV RNA.
Effect of pH on the density distribution of HCV.
Previous studies on the effect of low pH on Pestiviridae (14), which belong to the same family of virus as HCV, led us to test low pH as a method to change the density distribution of HCV from patient S6 in the iodixanol gradient. HCV from patient S6 serum was treated with PIPPS buffer at pH 4.0 and then applied to an iodixanol gradient. The density gradient profile after this treatment showed a peak between 1.10 and 1.12 g/ml (Fig. 3). Thus, the pH treatment had caused a shift in the density of HCV RNA from low density to intermediate density. To test the stability of the virus sample after low-pH treatment, HCV from the peak fractions 8 and 9 were reapplied to an iodixanol gradient prepared with pH 7.4. However, after centrifugation and analysis by RT PCR, small amounts of HCV RNA were found throughout the gradient and the titer had decreased by more than 95% (data not shown). Thus, exposure to low pH changed both the density and stability of HCV RNA, and further tests were not pursued. The effect of high pH (pH = 9.2) on the density distribution of HCV was also tested but was found to be unaltered from that observed at pH 7.4.
FIG. 3.
Effect of low-pH treatment on the density distribution of HCV in an iodixanol density gradient. A serum sample from patient S6 with 6 × 106 IU of HCV RNA was incubated with PIPPS buffer, pH 4.0, and applied to an iodixanol density gradient prepared with the same buffer. The distribution of HCV RNA in the gradient was determined by RT-PCR and semiquantitated by densitometry from an agarose gel. A peak of HCV RNA was observed with a density between 1.10 and 1.12 g/ml, and HCV RNA was also observed toward the top of the gradient with a density below 1.08 g/ml.
Effects of detergents on the density distribution of HCV.
Deoxycholic acid was tested for its effect on the density distribution of HCV RNA in iodixanol gradients, and the titer of HCV RNA in each fraction from the gradient was determined by quantitative real-time PCR. Prince et al. (41) have reported that deoxycholic acid at a concentration of 0.1% in the presence of KBr shifted the density of HCV RNA from <1.06 g/ml to 1.10 g/ml when analyzed by sucrose gradient centrifugation. Agnello et al. (1) reported that deoxycholate at a concentration of 0.1% was unsuccessful in dissociating HCV lipoprotein complexes. Thus, conflicting data on the effect of deoxycholic acid or deoxycholate on HCV have been reported. In our hands, 0.1% deoxycholic acid alone was not sufficient to shift the density of HCV, but inclusion of 400 mM KBr did cause some HCV with low density to shift to a density around 1.12 g/ml. We then tested different concentrations of deoxycholic acid and deoxycholate between 0.1% and 0.5% in the presence or absence of 150 mM NaCl or 400 mM KBr. The optimal procedure to prepare HCV RNA with a density between 1.11 and 1.13 g/ml was found to be treatment of serum with 0.5% deoxycholic acid in the absence of NaCl or KBr. Serum S6 was treated with deoxycholic acid at a concentration of 0.1% in the presence of 400 mM KBr (Fig. 4B) or 0.5% deoxycholic acid in the absence of NaCl and KBr (Fig. 4C). Deoxycholic acid at 0.1% with 400 mM KBr caused a shift in the density distribution of HCV RNA from a pattern where 67% of HCV RNA was below 1.10 g/ml (Fig. 4A) to a distribution where 58% of HCV RNA was between 1.11 and 1.13 g/ml (Fig. 4B). After treatment, the peak fraction 9 contained 33% of all HCV RNA within the gradient and only 15% of HCV RNA was found toward the top of the gradient with a density below 1.06 g/ml. From 15 × 106 IU of HCV RNA treated with deoxycholic acid, 2.9 × 106 IU (19%) was recovered in the fractions. When treated with 0.5% deoxycholic acid alone, most of the HCV RNA shifted position in the gradient from low density to an intermediate density between 1.11 and 1.13 g/ml (Fig. 4C). HCV RNA from peak fractions 7 and 8 represented 55% of all HCV RNA in the gradient. The percentage of HCV RNA remaining in the low-density fractions at the top of the iodixanol gradient was 1%, and the overall recovery of HCV RNA was calculated as 18%. Samples 7 and 8 were harvested and reapplied to a second, identical iodixanol gradient. HCV RNA was again found at a density of 1.12 g/ml, which suggests HCV treated with 0.5% deoxycholic acid is stable and uniform in iodixanol density gradients.
FIG. 4.
Effects of detergents on the density distribution of HCV RNA in iodixanol gradients. A serum sample from patient S6 with 15 × 106 IU of HCV RNA was applied to an iodixanol density gradient either before treatment (A) or after treatment with 0.1% deoxycholic acid with 400 mM KBr (B), 0.5% deoxycholic acid without NaCl or KBr (C), or 0.18% NP-40 (D). Changes in the density profile of HCV were assessed by the amount of HCV RNA in fractions from the gradient as determined by quantitative real-time PCR. The linearity of the density gradient is shown as a line graph, and the bar graphs show the amount of HCV RNA in each density gradient fraction. Each bar is the mean from two determinations.
Miyamoto et al. (36) have used 0.16% nonionic detergent NP-40 at 0°C for 5 min, followed by sucrose density gradient centrifugation, to remove the lipid envelope from HCV. Serum from patient S6 was treated with 0.18% NP-40 at 0°C for 12 min and then analyzed on an iodixanol gradient. A shift in the density of HCV was observed, with a peak in fraction 5 at a density of 1.18 g/ml (Fig. 4D), which is lower than the density of HCV RNA-containing particles produced by Miyamoto et al. (1.25 g/ml) (36). Fraction 5 contained 80% of the HCV RNA within the gradient, and less than 6% of the HCV RNA was found toward the top of the gradient, with a density below 1.06 g/ml. The recovery of HCV RNA from 15 × 106 IU applied to the gradient was 96%.
Both deoxycholic acid and NP-40 increased the density of HCV-containing particles to 1.15 to 1.17 g/ml, above which no HCV RNA could be found. This suggests that both detergents remove lipid from the lipoviroparticles, with NP-40 being the most efficient. However, it is possible that the viral envelope remains intact, as the cell culture-derived HCV particles described by Wakita et al. (58) had a density of 1.17 g/ml.
Sedimentation coefficients for HCV isolated by detergent treatment.
Sedimentation analysis was performed on HCV RNA-containing particles in order to estimate their sedimentation coefficients and diameters. HCV RNA from patient S6 serum, which had been treated with deoxycholic acid and purified on two successive iodixanol gradients, was used in the sedimentation analysis. The peak concentration of viral RNA in this preparation was found at a density of 1.12 g/ml. This HCV sample was analyzed by rate-zonal centrifugation using an iodixanol gradient, and the position of HCV RNA was determined by RT-PCR after 3 and 6 h of centrifugation. The distance traveled by these virus particles was measured at 3.8 cm after 3 h and 7.1 cm after 6 h. This result showed that the virus particles sedimented at an almost constant velocity through the gradient. The data confirm that the virus sample was homogeneous and show that the iodixanol gradient was isokinetic (21). The sedimentation coefficient, s20,w, was calculated as 215S ± 10S, and the diameter was calculated using Stokes' law as 54 nm. The sedimentation coefficient is comparable with values of 170 to 210S determined for intact virus particles within the family Flaviviridae (43). The diameter is comparable with morphological findings from electron microscopy of HCV particles and with values from other members of the family Flaviviridae, which are ∼50 nm in diameter (33). Sedimentation analysis was also performed with HCV RNA from serum that had been treated with NP-40. The detergent-treated serum was applied directly to an iodixanol gradient and analyzed by rate-zonal centrifugation. The distances traveled by these particles after 2 h and 4 h were 3 cm and 6 cm, respectively. The data show that the particles are homogeneous; the sedimentation coefficient (s20w) was calculated as 180S ± 10S, and the diameter was calculated as 42 nm. This value is higher than the sedimentation coefficients for nucleocapsids within the family Flaviviridae (43), which range between 120 and 140S. Therefore, it is not clear if these particles are nucleocapsids or quasi-virus particles with an intact viral envelope.
Precipitation of low- and very-low-density lipoproteins from serum.
To investigate whether the low density of HCV RNA in serum from patient S6 was due to association with LDL/VLDL, immunoprecipitation was performed with a rabbit polyclonal antibody against ApoB. ApoB-100 is a large, 540-kDa protein found on the surfaces of VLDL and LDL particles (20). After immunoprecipitation of LDL/VLDL from patient S6 serum with the polyclonal ApoB antibody, all of the ApoB-100 protein was found in the pellet and was absent from the supernatant (Fig. 5, lanes 2 and 3). Two proteins with molecular masses of 410 kDa and 140 kDa were also immunoprecipitated by the ApoB antibody. Proteins of these masses have been observed in preparations of LDL/VLDL from human serum (9) and were identified as cleavage products of ApoB-100. The enzyme responsible for this cleavage is plasma kallikrein (9), and this enzyme is inhibited by EDTA. LDL purified from plasma collected in the presence of 10 mM EDTA did not contain these ApoB cleavage products (Fig. 5, lane 8). Albumin is abundant in human serum and was observed as an intense band at 66 kDa in lanes 1, 2, and 5, with traces in lanes 3 and 6.
FIG. 5.
SDS-PAGE analysis after precipitation of LDL/VLDL from patient S6 serum, using either a polyclonal antibody against human ApoB or heparin/Mn2+. Protein samples were applied to a gradient polyacrylamide gel (5) and stained with Coomassie brilliant blue. Molecular mass markers (lane M). Immunoprecipitation of LDL/VLDL from 20 μl serum with a polyclonal antibody against human ApoB (lanes 1 to 4). Serum before immunoprecipitation (lane 1), supernatant (lane 2), and pellet (lane 3) after immunoprecipitation. Lane 4 received only the ApoB antibody, and protein bands at 50 kDa and 25 kDa correspond to the heavy and light chains of rabbit IgG. Precipitation of LDL/VLDL from 20 μl serum with heparin/Mn2+ (lanes 5 and 6). The supernatant (lane 5) and pellet (lane 6) after the precipitation were applied to the gel. Purified LDL from normal human plasma collected in the presence of EDTA and protease inhibitors (lanes 7 and 8). The density range of the LDL is between 1.019 and 1.063 g/ml, and two samples with either 2 μg (lane 7) or 10 μg (lane 8) protein were applied to the gel. The position of ApoB-100 with a molecular mass of 540 kDa, as well as two 410-kDa and 140-kDa fragmentation products of ApoB, are indicated with arrows.
The distribution of HCV RNA from serum between the pellet and the supernatant after immunoprecipitation with the ApoB antibody was determined by quantitative real-time PCR. The pellet contained 98% of the HCV RNA. The HCV RNA remaining in the supernatant after immunoprecipitation was analyzed on an iodixanol density gradient and was found to have a density between 1.10 and 1.13 g/ml. All HCV with densities below 1.08 g/ml had been precipitated with the ApoB antibody, which suggests that HCV RNA with a density below 1.08 g/ml is associated with LDL/VLDL. A control immunoprecipitation with normal rabbit IgG did not precipitate HCV-RNA, and the distribution of HCV RNA in the supernatant was similar to that in untreated serum.
Heparin in the presence of Mn2+ (8) was also tested to precipitate LDL/VLDL from patient S6 serum. This method has been used by Thomssen et al. (52) to precipitate HCV RNA in association with LDL/VLDL. Following heparin/Mn2+ precipitation, the resuspended pellet and the supernatant were analyzed by SDS-PAGE, and the distribution of HCV RNA between the pellet and the supernatant was measured by PCR. ApoB-100 was present in the pellet, as were the 410-kDa and 140-kDa fragmentation products of ApoB-100 (Fig. 3, lane 6). ApoB-100 was not detected in the supernatant (Fig. 3, lane 5), which confirmed that the precipitation of LDL/VLDL was complete. Analysis of the distribution of HCV RNA between the pellet and the supernatant by quantitative real-time PCR showed that all HCV RNA was precipitated by heparin/Mn2+.
Immunoprecipitation of HCV structural proteins.
The level of HCV structural proteins in the serum from patient S6 was insufficient for detection of viral proteins by Western blotting. However, the transplanted liver from the same patient had a very high titer of HCV, and glycoproteins E1 and E2, as well as the core protein, were detected in a liver macerate by Western blotting (39). HCV lipoviroparticles with densities below 1.06 g/ml were obtained from this liver macerate by iodixanol density gradient centrifugation (39). The LVPs were dissociated by suspension in buffer containing 1% NP-40 and were used for immunoprecipitations. The quantity of HCV viral protein immunoprecipitated by each antibody was determined by densitometry and compared with standards containing known amounts of viral protein (Fig. 6). This procedure allowed the efficiency of each antibody to be evaluated and expressed as a percentage (Table 2). Five antibodies against HCV E2 protein were found to immunoprecipitate E2 from the liver macerate with efficiencies between 16% (CBH-7) and 67% (CBH-2). The E2 protein immunoprecipitated by these antibodies had a molecular mass of 62 kDa, which is identical to that of E2 from HCV-infected human liver determined previously (39). Two broad bands with molecular masses of 300 kDa and >550 kDa were present in the immunoprecipitate with E2 antibody B65581G. As these bands were also present in the control reaction with normal goat IgG (Fig. 6, lanes 4 and 5), they are nonspecific and may be aggregates of goat antibodies released from protein G-Sepharose beads. Two MAbs against HCV E1, clone 11B7D8 and 15G6B2, did not immunoprecipitate the HCV glycoprotein, and two antibodies against HCV core protein, MAb Φ126 and goat polyclonal HG-22, were tested for immunoprecipitation using the liver macerate S6b but were found not to precipitate core protein (data not shown).
FIG. 6.
Western blot analysis of HCV glycoprotein E2 from transplant liver S6b. Samples from the low-density fraction (ρ ≤ 1.06 g/ml) of liver macerate were analyzed either undiluted (lane 1), diluted 1:2 (lane 2), or diluted 1:4 (lane 3). The Western blot was immunostained using mouse MAb AP33 against HCV E2. The positions of E2 and the IgG light chain (L) are indicated with arrows. Immunoprecipitations of E2 from undiluted samples of liver macerate were performed using monoclonal and polyclonal antibodies. Normal goat IgG (lane 4), B65581G goat anti-E2 (lane 5), MAb 2G12-2 (negative control; lane 6), MAb AP33 anti-E2 (lane 7), human MAb CBH-2 anti-E2 (lane 8), and human MAb CBH-5 anti-E2 (lane 9). Biotinylated molecular mass markers (lane M).
TABLE 2.
Evaluation of antibodies for immunoprecipitation of LVPs
| Antibody | Host species | Antigen | % Antigen precipitateda | % HCV RNA precipitatedb |
|---|---|---|---|---|
| Normal IgG | Rabbit | Negative control | 0 | 11.4 |
| Anti-ApoB | Rabbit | Human ApoB | 100 | 91.8 |
| Anti-ApoE | Rabbit | Human ApoE | NTc | 95.0 |
| Normal IgG | Goat | Negative control | 0 | 11.0 |
| B65581G | Goat | HCV E2 | 36 | 24.9 |
| 2G12-2 | Mouse | Negative control | 0 | 11.6 |
| AP33 | Mouse | HCV E2 | 27 | 8.5 |
| 3C7-C3 | Mouse | HCV E2 HVR-1 | NTc | 25.0 |
| Normal IgG | Human | Negative control | 0 | 7.7 |
| CBH-5 | Human | HCV E2 | 52 | 23.9 |
| CBH-2 | Human | HCV E2 | 67 | 19.1 |
| CBH-7 | Human | HCV E2 | 10 | 16.0 |
| CBH-4D | Human | HCV E2 | 0 | 7.7 |
Calculated as the amount of antigen in the immunoprecipitate (percentage of antigen available in the sample prior to immunoprecipitation) (Fig. 5 and 6).
Calculated as the titer of HCV RNA in the immunoprecipitate (percentage of total HCV RNA in the immunoprecipitate plus supernatant).
NT, not tested.
Immunoprecipitation of HCV RNA with low, intermediate, and high density.
HCV RNA from patient S6 serum was separated on an iodixanol gradient to obtain a density distribution of virus similar to that in Fig. 4A. Fractions 13 and 14 at the top of the gradient, which contained HCV RNA with a density below 1.06 g/ml, were combined and used for further analysis by immunoprecipitation. The polyclonal antibody against ApoB precipitated 91.8% of the HCV RNA, and the polyclonal antibody against ApoE precipitated 95.0% of the HCV RNA (Table 2). In the control reaction, normal rabbit IgG precipitated 11.4% of the HCV RNA. This level of nonspecific precipitation was consistently observed with normal IgG from goat, rabbit, mouse, or human (Table 2). These results show that HCV LVP with a density of ≤1.06 g/ml contains ApoB and ApoE, which indicates that the particle resembles VLDL. The polyclonal HCV E2 antibody B65581G precipitated 24.9% of the HCV RNA, which was higher than the nonspecific precipitation observed with goat IgG (Table 2). Monoclonal antibody 3C7-C3 against the hypervariable region of E2 (10) precipitated a similar proportion of HCV RNA (25.0%). Three human monoclonal antibodies against HCV E2 (30) also precipitated HCV RNA above the level observed with normal human IgG. The highest percentage was obtained with clone CBH-5, which precipitated 23.9% of the HCV RNA. These results show that E2 is present and accessible for antibody binding in a proportion of lipoviroparticles. Recovery of HCV RNA in each immunoprecipitation reaction was calculated from real-time PCR data for both the pellet and the supernatant and compared with a control reaction which did not receive antibody. The average recovery of HCV RNA was 88.9%.
HCV particles with a density of 1.12 g/ml were obtained from an iodixanol gradient of serum treated with 0.5% deoxycholic acid. The distribution of HCV RNA was similar to that shown in Fig. 4C, and fraction 8 was chosen for immunoprecipitation experiments. The polyclonal antibody against ApoB precipitated 68.4% of the HCV RNA compared to nonspecific precipitation with normal rabbit IgG of 10.0%. The polyclonal antibody against ApoE precipitated 12.6% of the HCV RNA, which was similar to the nonspecific precipitation with normal rabbit IgG.
HCV particles with a density of 1.18 g/ml were obtained from an iodixanol gradient of serum treated with NP-40 at 0°C. The distribution of HCV was similar to that shown in Fig. 4D, and fraction 5 was chosen for immunoprecipitation experiments. The polyclonal antibody against ApoB precipitated 78.0% of the HCV RNA, which was higher than the nonspecific precipitation with normal rabbit IgG (6.8%). The polyclonal antibody against ApoE precipitated 9.1% of the HCV RNA, which was similar to the precipitation observed with normal rabbit IgG. These results show that ApoB remains associated with HCV after treatment of the virus with either deoxycholic acid or NP-40, whereas ApoE is displaced by the two detergents. Thus, firm binding exists between HCV and ApoB, although further experiments are required to determine how ApoB is linked to the HCV particle.
DISCUSSION
Iodixanol is a density gradient medium that can form solutions of a density up to 1.31 g/ml while retaining osmolarity at or below 240 mosmol/kg; it is isotonic and isosmotic with blood (16). Solutions of sucrose are only isosmotic at densities below 1.12 g/ml; above this density, the osmolarity increases exponentially (data not shown). Sequential ultracentrifugation of HCV in NaBr take place in solutions of high ionic strength that are hypertonic. Separation of HCV RNA in one serum from a patient with common variable immunodeficiency by an iodixanol, sucrose, or NaBr gradient and of HCV RNA in three sera from immunocompetent patients by NaBr and iodixanol gradients produced markedly different results. This suggests that the density heterogeneity of HCV is partially due to the matrix of the gradients and that iodixanol provides better preservation of the lower-density HCV-host lipoprotein complexes.
Even when separated on iodixanol, however, sera from immunocompetent patients yield HCV RNA-containing particles that are markedly heterogeneous in density, with patterns varying widely from patient to patient. This heterogeneity has been attributed to a varying ratio of virions and virus cores, which may be variably associated with immunoglobulins and/or with host lipoproteins, which themselves form a complex population of particles varying in composition and density (4, 12, 52, 53, 59). That much of this complexity may be due to the immune response against infection is demonstrated by the relatively simple density profile of viral RNA in the serum of the immunodeficient and immunosuppressed, antiviral-antibody-negative patient S6, who was suffering an acute infection of the liver graft. A similarly narrow density range has been observed in acute infection prior to the onset of immune responses (25).
In patient S6, essentially all of the circulating HCV RNA was found complexed to ApoB, as demonstrated by immunoprecipitation and precipitation with manganese and heparin. Such low-density viral RNA complexes with lipoprotein have been characterized as LVPs by André et al. (3, 4). Our immunoprecipitation analysis showed that the LVPs contain both ApoB and ApoE. ApoB is found on all low-density lipoproteins, but the presence of ApoE is characteristic of VLDL particles (20), and its presence indicates that LVPs resemble VLDL. Such an observation was made earlier by Prince et al. (41). The diameter of VLDL is heterogeneous, between 35 nm and 80 nm, and the particles are processed in the circulation via the VLDL/LDL lipolytic cascade, losing lipid and ApoE to be eventually taken up by hepatocytes as an LDL particle. It is not clear if LVPs participate in this lipolytic cascade. ApoB is the only lipoprotein on LDL, and ApoB also remains bound to LVPs after detergent treatment, which shows that an LVP particle resembling LDL can be produced.
We have found that HCV core, as well as glycoproteins E1 and E2, is present in fractions with densities below 1.08 g/ml from liver S6b (42) (Fig. 6), and this has allowed us to seek antibodies capable of immunoprecipitating these viral proteins. To date, we have been unable to locate antibodies capable of satisfactory immunoprecipitation of either core or E1 glycoprotein in this system. However, five polyclonal and monoclonal antibodies have been shown to immunoprecipitate glycoprotein E2. These antibodies provided evidence for the presence of E2 in LVPs with densities below 1.08 g/ml from serum. This shows that E2 protein is present and accessible to antibodies on a proportion of serum-derived LVPs. The efficiency of immunoprecipitation of liver-derived antigen released by NP-40 can be fruitfully compared to the immunoprecipitation of LVP particles from serum in the absence of detergents. We found that the antibodies which immunoprecipitated E2 from S6b liver macerate were also those which performed best in immunoprecipitation of LVPs; however, the efficiency of the latter was always reduced. Two reasons can be suggested for this incomplete precipitation, either that only a proportion of LVPs in serum S6 contain glycoprotein E2 or that regions of the E2 molecules in LVPs are hidden or masked.
Several of the antibodies used in the present study have been used successfully by others to precipitate virus-like particles and pseudo-viral particles. Clayton et al. (13) have shown that the HCV E2 antibody AP33 can recognize HCV virus-like particles derived from a recombinant baculovirus. This antibody, which recognized E2 glycoprotein from S6b liver well in Western blotting, immunoprecipitated NP-40-released E2 from liver relatively poorly and did not immunoprecipitate LVPs from serum. Two of the human MAbs from Keck et al. (30), CBH-5 and CBH-2, also immunoprecipitated HCV pseudo-viral particles. These antibodies recognize a conformational epitope of HCV E2 and have strong reactivity against cell-associated E2 protein. This observation was confirmed here, as both antibodies precipitated E2 from liver S6b with high efficiency. Antibodies to the E2 HVR1 region have been shown to immunoprecipitate HCV particles from human serum by Cerino et al. (10) using MAb 3C7-C3 and by Esumi et al. (16) using a mouse polyclonal antibody to HVR-1. Here, MAb 3C7-C3 gave the most efficient immunoprecipitation of LVP from S6 serum observed.
For physicochemical characterization of the HCV virion derived from serum, it will be necessary to dissociate the virus from complexes with VLDL/LDL. Dissolution of host lipoprotein particles in serum S6 by treatment at low pH was attempted in the hope that this might release HCV virions. Although low-pH treatment did release particles of increased density, they proved unstable, and the recovery of HCV RNA was low. Enzymatic digestion of lipoviroparticles with lipoprotein lipase has been used by Thomssen et al. (51), but this method was also found to produce viral particles which were unstable and led to low recovery of HCV RNA. Prince et al. (41) used deoxycholic acid between 0.05% and 1.0% in the presence of KBr to release HCV particles from association with lipoprotein. They found that HCV RNA-containing particles with a density of 1.10 g/ml were released with 0.05% and 0.1% deoxycholic acid. When the concentration of deoxycholic acid was increased above 0.2% and KBr was present, increasing amounts of HCV RNA with a density between 1.18 and 1.20 was observed (41). In our experiments, using deoxycholic acid between 0.1% and 0.5%, we found that 0.5% deoxycholic acid in the absence of NaCl and KBr gave the best yield and density shift, releasing HCV RNA-containing particles with a density between 1.11 and 1.13 g/ml in iodixanol, which is similar to the density of the novel peak of viral RNA generated in sucrose gradient centrifugation of the same serum. The density shift indicates that lipid had been removed from the LVP, and the observed density is in agreement with previous estimates of 1.08 to 1.11 g/ml for the density of free virions recovered from plasma or serum by Hijikata et al. (25) and Kanto et al. (28) using sucrose gradients. Fujita et al. (18) prepared HCV particles from serum treated with paraformaldehyde and estimated the density as 1.13 g/ml, and Li et al. (32) detected virus-like particles with a density of 1.11 g/ml in serum from a liver transplant patient. Particles released from LVP by deoxycholic acid are not free virions, however, as immunoprecipitation experiments showed that they remained associated with ApoB, although they had lost surface ApoE. Deoxycholic acid-treated particles prepared by Prince et al. (41) also appeared in electron microscopy to have residual fragments of LDL or VLDL on the surface. Thus, treatment with deoxycholic acid appears to dissociate host lipid from the lipoviroparticle to produce LVPs containing ApoB and possibly some residual lipoprotein lipid. However, treatment with deoxycholic acid destroys over 80% of the particles, and it remains unclear whether those that remain are typical of the whole. The sedimentation coefficient of deoxycholic acid-treated particles is within the range of sedimentation coefficients (200 to 280S) determined for non-A, non-B hepatitis virus by Bradley (7) but higher than the sedimentation coefficient for another Flavivirus, Rio-Bravo virus (201.5S) (24). The calculated particle diameter compares well with the 55- to 65-nm flavivirus-like particles observed by immunoelectron microscopy in a sucrose density gradient fraction of infected serum with a density of 1.14 to 1.16 g/ml (26).
NP-40 caused HCV RNA to shift to a higher density, 1.18 g/ml, than the anionic detergent deoxycholic acid. Such an observation has also been made with the flavivirus Saint Louis encephalitis virus (54). The nonionic detergent Brij-58 produced nucleocapsids that contained capsid protein and RNA, whereas deoxycholate produced virus particles of lower density that contained envelope glycoprotein, as well as capsid protein and RNA. The density of particles generated by NP-40 treatment here, 1.18 g/ml, was lower than the 1.25-g/ml particles reported by Miyamoto et al. (36) and Kanto et al. (28) in sucrose gradients and the 1.35-g/ml particles produced by Shindo et al. (47) and Maillard et al. (35) in CsCl density gradients. The diameter of our NP-40-treated HCV RNA-containing particles is higher than the diameter of putative HCV nucleocapsids released from serum with 0.05% Tween 80 (33 nm) (49) but similar to those of the 38- to 43-nm nucleocapsid particles observed in serum by Maillard et al. (35). Thus, differences are apparent in both the reported size and density of the putative HCV nucleocapsid prepared from in vivo sources, and this may, in part, be due to residual lipoprotein and/or viral envelope on the particle. The physicochemical properties of the particle produced may also depend on the conditions used for the detergent treatment. Kanto et al. (28) used 3% NP-40 for 45 min at 0°C, Shindo et al. (47) used 0.1% NP-40 for 3 h at room temperature, and Stollar (48) used 0.1% NP-40 for 20 min at 0°C. It is intriguing that the density of our NP-40-treated HCV RNA-containing particles is similar to the density of cell culture-derived HCV particles (58). It is possible that ice-cold NP-40 treatment of HCV LVPs, followed by separation by iodixanol gradient centrifugation, produces quasi-virus particles minimally loaded with host lipoprotein.
Our immunoprecipitation studies demonstrate that HCV particles treated with either deoxycholic acid or NP-40 remain associated with ApoB. This observation can be correlated with earlier data which showed that deoxycholic acid and NP-40 removed lipids from ApoB in the LDL particle when the ratio between detergent and ApoB was above 12:1 (23). In our experiments, the ratio between deoxycholic acid and ApoB was 40:1 and the ratio between NP-40 and ApoB was 16:1. Therefore, the interaction between ApoB and the HCV virion appears to be resistant to the loss of lipoprotein lipid after detergent treatment of LVPs. It is unclear why yields following treatment of LVPs with deoxycholic acid were much lower than those following NP-40 treatment.
In conclusion, we have determined sedimentation coefficients and diameters for HCV RNA-containing particles with a density of either 1.12 g/ml or 1.18 g/ml. We have also confirmed the observation that HCV of low density is associated with lipoprotein in a hybrid lipoviroparticle resembling VLDL (4) and presented evidence that at least 25% of these LVPs contain HCV glycoprotein E2. The association with host lipoproteins may assist virus secretion from infected cells and protect virus from antibody-mediated clearance, as well as facilitate virus uptake by lipoprotein receptors.
Acknowledgments
This work was supported by the Wellcome Trust, London (reference no. 059376).
We acknowledge the kind gift of antibodies from A. Patel, Glasgow, United Kingdom; E. Depla, Ghent, Belgium; S. Foung, Palo Alto, California; and M. Mondelli, Pavia, Italy. The research was initiated by D. Bevitt and J. Watson and performed with technical assistance from F. Fenwick.
REFERENCES
- 1.Agnello, V., G. Ábel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnello, V., R. T. Chung, and L. M. Kaplan. 1992. A role for hepatitis C virus infection in type II cryoglobulinemia. N. Engl. J. Med. 327:1490-1495. [DOI] [PubMed] [Google Scholar]
- 3.André, P., G. Perlemuter, A. Budkowska, C. Bréchot, and V. Lotteau. 2005. Hepatitis C virus particles and lipoprotein metabolism. Semin. Liver Dis. 25:93-104. [DOI] [PubMed] [Google Scholar]
- 4.André, P., F. K. Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Bréchot, G. P. Baccalà, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews, A. T. 1989. Electrophoresis. Theory, techniques, and biochemical and clinical applications, 2nd ed., p. 93-116. Oxford University Press, Oxford, England.
- 6.Bradley, D., K. McCaustland, K. Krawczynski, J. Spelbring, C. Humphrey, and E. H. Cook. 1991. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J. Med. Virol. 34:206-208. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, D. W. 1985. The agents of non-A, non-B hepatitis. J. Virol. Methods 10:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burstein, M., H. R. Scholnick, and R. Morfin. 1970. Rapid method for the isolation of lipoproteins from human serum by precipitation with polyanions. J. Lipid Res. 11:583-595. [PubMed] [Google Scholar]
- 9.Cardin, A. D., K. R. Witt, J. Chao, H. S. Margolius, V. H. Donaldson, and R. L. Jackson. 1984. Degradation of apolipoprotein B-100 of human plasma low density lipoproteins by tissue and plasma kallikreins. J. Biol. Chem. 259:8522-8528. [PubMed] [Google Scholar]
- 10.Cerino, A., A. Meola, L. Segagni, M. Furione, S. Marciano, M. Triyatni, T. J. Liang, A. Nicosia, and M. U. Mondelli. 2001. Monoclonal antibodies with broad specificity for hepatitis C virus hypervariable region 1 variants can recognize viral particles. J. Immunol. 167:3878-3886. [DOI] [PubMed] [Google Scholar]
- 11.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Choo, S.-H., H.-S. So, J. M. Cho, and W.-S. Ryu. 1995. Association of hepatitis C virus particles with immunoglobulin: a mechanism for persistent infection. J. Gen. Virol. 76:2337-2341. [DOI] [PubMed] [Google Scholar]
- 13.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depner, K., T. Bauer, and B. Liess. 1992. Thermal and pH stability of pestiviruses. Rev. Sci. Tech. Off. Int. Epizoot. 11:885-893. [DOI] [PubMed] [Google Scholar]
- 15.Eivindvik, K., and C. E. Sjøgren. 1995. Physicochemical properties of iodixanol. Acta Radiol. Suppl. 399:32-38. [DOI] [PubMed] [Google Scholar]
- 16.Esumi, M., M. Ahmed, Y. Zhou, H. Takahashi, and T. Shikata. 1998. Murine antibodies against E2 and hypervariable region 1 cross-reactively capture hepatitis C virus. Virology 251:158-164. [DOI] [PubMed] [Google Scholar]
- 17.Fenwick, F. I., M. F. Bassendine, K. Agarwal, D. Bevitt, W. Pumeechockchai, A. D. Burt, and G. L. Toms. Immunohistochemical assessment of hepatitis C virus antigen in cholestatic hepatitis after liver transplantation. J. Clin. Pathol., in press. [DOI] [PMC free article] [PubMed]
- 18.Fujita, N., M. Kaito, S. Ishida, N. Nakagawa, J. Ikoma, Y. Adachi, and S. Watanabe. 2001. Paraformaldehyde protects hepatitis C virus particles during ultracentrifugation. J. Med. Virol. 63:108-116. [PubMed] [Google Scholar]
- 19.Germi, R., J.-M. Crance, D. Garin, J. Guimet, H. L. Jacob, R. W. H. Ruigrok, J.-P. Zarski, and E. Drouet. 2002. Cellular glycosaminoglycans and low density lipoprotein receptor are involved in hepatitis C virus adsorption. J. Med. Virol. 68:206-215. [DOI] [PubMed] [Google Scholar]
- 20.Ginsberg, H. N., J. L. Dixon, and I. J. Goldberg. 1999. VLDL/LDL cascade system: assembly, secretion and intravascular metabolism of apoprotein B-containing lipoproteins, p. 55-70. In D. J. Betteridge, D. R. Illingworth, and J. Shepherd (ed.), Lipoproteins in health and disease. J. Arnold, London, United Kingdom.
- 21.Hames, B. D. 1984. Choice of conditions for density gradient centrifugation, p. 45-93. In. D. Rickwood (ed.), Centrifugation: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 22.Harlow, E., and D. Lane. 1988. Antibodies. A laboratory manual, p. 421-470. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Helenius, A., and K. Simons. 1971. Removal of lipids from human plasma low-density lipoprotein by detergents. Biochemistry 10:2542-2547. [DOI] [PubMed] [Google Scholar]
- 24.Hendricks, D. A., A. K. Patick, L. M. Petti, and A. J. Hall. 1988. Biochemical and biophysical characteristics of Rio Bravo virus (Flaviviridae). J. Gen. Virol. 69:337-347. [DOI] [PubMed] [Google Scholar]
- 25.Hijikata, M., Y. K. Shimizu, H. Kato, A. Iwamoto, J. W. Shih, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1993. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J. Virol. 67:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaito, M., S. Watanabe, K. T. Kohara, K. Yamaguchi, Y. Kobayashi, M. Konishi, M. Yokoi, S. Ishida, S. Suzuki, and M. Kohara. 1994. Hepatitis C virus particle detected by immunoelectron microscopic study. J. Gen. Virol. 75:1755-1760. [DOI] [PubMed] [Google Scholar]
- 27.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Density analysis of hepatitis C virus particle population in the circulation of infected hosts: implications for virus neutralization or persistence. J. Hepatol. 22:440-448. [DOI] [PubMed] [Google Scholar]
- 28.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1994. Buoyant density of hepatitis C virus recovered from infected hosts: two different features in sucrose equilibrium density gradient centrifugation related to degree of liver inflammation. Hepatology 19:296-302. [PubMed] [Google Scholar]
- 29.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Oshita, K. Katayama, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Serial density analysis of hepatitis C virus particle populations in chronic hepatitis C patients treated with interferon-α. J. Med. Virol. 46:230-237. [DOI] [PubMed] [Google Scholar]
- 30.Keck, Z. Y., A. Op De Beeck, K. G. Hadlock, J. Xia, T. K. Li, J. Dubuisson, and S. K. H. Foung. 2004. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J. Virol. 78:9224-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laude, H. 1979. Nonarbo-togaviridae: comparative hydrodynamic properties of the pestivirus genus. Arch. Virol. 62:347-352. [DOI] [PubMed] [Google Scholar]
- 32.Li, X., L. J. Jeffers, L. Shao, K. R. Reddy, M. Medina, J. Scheffel, B. Moore, and E. R. Schiff. 1995. Identification of hepatitis C virus by immunoelectron microscopy. J. Viral. Hepat. 2:227-234. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbach, B. D., and C. M. Rice. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23-61. [DOI] [PubMed] [Google Scholar]
- 34.Mackness, M. I., and P. N. Durrington. 1992. Lipoprotein separation and analysis for clinical studies, p. 1-42. In C. A. Converse, and E. R. Skinner (ed.), Lipoprotein analysis: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 35.Maillard, P., K. Krawczynski, J. Nitkiewicz, C. Bronnert, M. Sidorkiewicz, P. Gounon, J. Dubuisson, G. Faure, R. Crainic, and A. Budkowska. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 75:8240-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamoto, H., H. Okamoto, K. Sato, T. Tanaka, and S. Mishiro. 1992. Extraordinarily low density of hepatitis C virus estimated by sucrose density gradient centrifugation and the polymerase chain reaction. J. Gen. Virol. 73:715-718. [DOI] [PubMed] [Google Scholar]
- 37.Monazahian, M., I. Böhme, S. Bonk, A. Koch, C. Scholz, S. Grethe, and R. Thomssen. 1999. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 57:223-229. [DOI] [PubMed] [Google Scholar]
- 38.Monazahian, M., S. Kippenberger, A. Müller, H. Seitz, I. Böhme, S. Grethe, and R. Thomssen. 2000. Binding of human lipoproteins (low, very low, high density lipoproteins) to recombinant envelope proteins of hepatitis C virus. Med. Microbiol. Immunol. 188:177-184. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, S. U., M. F. Bassendine, A. D. Burt, D. J. Bevitt, and G. L. Toms. 2004. Characterization of the genome and structural proteins of hepatitis C virus resolved from infected human liver. J. Gen. Virol. 85:1497-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petit, M. A., M. Lièvre, S. Peyrol, S. D. Sequeira, P. Berthillon, R. W. H. Ruigrok, and C. Trépo. 2005. Enveloped particles in the serum of chronic hepatitis C patients. Virology 336:144-153. [DOI] [PubMed] [Google Scholar]
- 41.Prince, A. M., T. H. Byron, T. S. Parker, and D. M. Levine. 1996. Visualization of hepatitis C virions and putative defective interfering particles isolated from low density lipoproteins. J. Viral Hepat. 3:11-17. [DOI] [PubMed] [Google Scholar]
- 42.Pumeechockchai, W., D. Bevitt, K. Agarwal, T. Petropoulou, B. C. A. Langer, B. Belohradsky, M. F. Bassendine, and G. L. Toms. 2002. Hepatitis C virus particles of different density in the blood of chronically infected immunocompetent and immunodeficient patients: implications for virus clearance by antibody. J. Med. Virol. 68:335-342. [DOI] [PubMed] [Google Scholar]
- 43.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-959. In B. N. Fields, D. M. Knipe, and P. M Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 44.Routledge, E. G., J. McQuillin, A. C. R. Samson, and G. L. Toms. 1985. The development of monoclonal antibodies to respiratory syncytial virus and their use in diagnosis by indirect immunofluorescence. J. Med. Virol. 15:305-320. [DOI] [PubMed] [Google Scholar]
- 45.Sansonno, D., R. Iacobelli, V. Cornacchiulo, G. Lauletta, M. A. Distasi, P. Gatti, and F. Dammacco. 1996. Immunochemical and biomolecular studies of circulating immune complexes isolated from patients with acute and chronic hepatitis C virus infection. Eur. J. Clin. Investig. 26:465-475. [DOI] [PubMed] [Google Scholar]
- 46.Schneider, C., R. A. Newman, D. R. Sutherland, U. Asser, and M. F. Greaves. 1982. A one step purification of membrane proteins using a high efficiency immunomatrix. J. Biol. Chem. 257:10766-10769. [PubMed] [Google Scholar]
- 47.Shindo, M., A. M. Di Bisceglie, T. Akatsuka, T. L. Fong, L. Scaglione, M. Donets, J. H. Hoofnagle, and S. M. Feinstone. 1994. The physical state of the negative strand of hepatitis C virus RNA in serum of patients with chronic hepatitis C. Proc. Natl. Acad. Sci. USA 91:8719-8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stollar, V. 1969. Studies on the nature of dengue viruses. Virology 39:426-438. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi, K., S. Kishimoto, H. Yoshizawa, H. Okamoto, A. Yoshikawa, and S. Mishiro. 1992. p26 protein and 33 nm particle associated with nucleocapsid of hepatitis C virus recovered from the circulation of infected hosts. Virology 191:431-434. [DOI] [PubMed] [Google Scholar]
- 50.Thomson, M., and T. J. Liang. 2000. Molecular biology of hepatitis C virus, p. 1-23. In T. J. Liang, and J. H. Hoofnagle (ed.), Hepatitis C. Biomedical Research Reports. Academic Press, San Diego, Calif.
- 51.Thomssen, R., and S. Bonk. 2002. Virolytic action of lipoprotein lipase on hepatitis C virus in human sera. Med. Microbiol. Immunol. 191:17-24. [DOI] [PubMed] [Google Scholar]
- 52.Thomssen, R., S. Bonk, C. Propfe, K.-H. Heermann, H. G. Köchel, and A. Uy. 1992. Association of hepatitis C virus in human sera with β-lipoprotein. Med. Microbiol. Immunol. 181:293-300. [DOI] [PubMed] [Google Scholar]
- 53.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of β-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 182:329-334. [DOI] [PubMed] [Google Scholar]
- 54.Trent, D. W., and A. A. Qureshi. 1971. Structural and nonstructural proteins of Saint Louis encephalitis virus. J. Virol. 7:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trestard, A., Y. Bacq, L. Buzelay, F. Dubois, F. Barin, A. Goudeau, and P. Roingeard. 1998. Ultrastructural and physicochemical characterization of the hepatitis C virus recovered from the serum of an agammaglobulinemic patient. Arch. Virol. 143:2241-2245. [DOI] [PubMed] [Google Scholar]
- 56.Triyatni, M., B. Saunier, P. Maruvada, A. R. Davis, L. Ulianich, T. Heller, A. Patel, L. D. Kohn, and T. J. Liang. 2002. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J. Virol. 76:9335-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. H. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 58.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Kräusslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 7:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wünschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young, B. D. 1987. Measurement of sedimentation coefficients and computer simulation of rate-zonal separations, p. 127-159. In D. Rickwood (ed.), Centrifugation: a practical approach. Oxford University Press, Oxford, United Kingdom.








