Abstract
The restriction factors Fv1 and TRIM5α provide dominant blocks to retroviral infection, targeting incoming capsids at a postentry, preintegration step. They both restrict N-tropic murine leukemia virus with similar specificity yet act at different points in the viral life cycle. TRIM5α-restricted virus is usually unable to reverse transcribe, whereas Fv1-restricted virus reverse transcribes normally. Here we investigate the relationship between these two restriction factors by expressing Fv1 alleles in human cells. We demonstrate that Fv1 is able to compete with TRIM5α for virus before reverse transcription. In human cells expressing Fv1b, N-tropic restricted virus becomes less infectious but reverse transcribes more efficiently, indicating competition between the two antiviral molecules and protection of the virus from TRIM5α by Fv1. Our findings suggest that, like TRIM5α, Fv1 interacts with virus before reverse transcription, but the consequences of this interaction are not realized until a later stage of the life cycle. We also demonstrate that Fv1 is functionally independent of TRIM5α when expressed in human cells.
Selective pressure imposed by retroviral infection has driven the evolution of antiviral cellular factors that contribute to defense mechanisms against retroviruses. Products of the tripartite motif 5 (TRIM5) gene in primates and the Friend virus susceptibility factor-1 (Fv1) gene in mice constitute a class of restriction factors that inhibit retroviral infection, targeting incoming viral capsids and preventing the establishment of a provirus (6, 12, 14, 17, 20, 23, 26, 31). Restriction of viral infectivity by such factors determines retroviral tropism at the species level, and zoonotic viral transfer between species is likely to require insensitivity to these antiviral mechanisms.
Fv1 was first described as one of a series of loci controlling mouse susceptibility to leukemia induced by the Friend strain of murine leukemia virus (MLV) (17). Two main alleles of Fv1 have been described, Fv1n from NIH mice and Fv1b from BALB/c mice. Fv1 enabled division of MLVs into subgroups. N-tropic MLV (MLV-N) strains are able to infect Fv1n/n cells (or NIH mice) but not Fv1b/b cells (or BALB/c mice), whereas B-tropic MLV strains (MLV-B) display the opposite phenotype, infecting Fv1b/b cells (or BALB/c mice) but not Fv1n/n cells (or NIH mice) (24). NB-tropic strains (MLV-NB), which include Moloney MLV, constitute a third Fv1 sensitivity phenotype able to infect cells of any Fv1 genotype.
Fv1 blocks MLV infection in a saturable way after the virus has entered target cells and after viral reverse transcription but before the establishment of a provirus (13). The target of Fv1 restriction is the viral capsid protein, as sensitivity to Fv1 depends specifically on residues within the capsid (CA). The mechanism by which Fv1 blocks retroviral infection and the point in the viral life cycle at which Fv1 recognizes capsid remain obscure. Humans lack an Fv1 orthologue yet display an Fv1-like activity against MLV-N (28). The human gene encoding this activity was originally called Ref1 and is now known as TRIM5α (12, 14, 20, 31). TRIM5α is a member of the tripartite motif (TRIM) protein family that is defined by a cluster of three motifs with a characteristic ordering and spacing: an N-terminal RING domain, one or two B-Box type 1 or type 2 domains, and a coiled-coil region (21). Notably, sensitivity to both Fv1b and human TRIM5α (huTRIM5α) is determined by CA residue 110, where arginine denotes N tropism and glutamate B tropism (15, 28). Thus, CA mutation R110Q in N-tropic AKV WN41 leads to complete insensitivity to both Fv1b and huTRIM5α. Residues around this site, however, are also important in determining sensitivity to restriction, particularly for the attainment of an Fv1-insensitive NB phenotype (16, 25). Moreover, certain mutations are able to distinguish between Fv1 and huTRIM5α sensitivity, for example, CA D92E mutation in MLV Friend strain renders it sensitive to Fv1b but not to human TRIM5α (16).
Mutational analysis of TRIM5α has contributed to a model for restriction. Swapping SPRY domains between TRIM5α alleles or SPRY domain mutagenesis demonstrates that it encodes the antiviral specificity determinant (18, 19, 22, 27, 32). The importance of the SPRY domain is also illustrated by the owl monkey example, where the TRIM5 SPRY domain has been replaced by an in-frame cyclophilin A pseudogene (23). The CypA domain recruits the TRIM5 RBCC domain to incoming human immunodeficiency virus type 1 (HIV-1) capsids, leading to restriction of HIV-1 infectivity. The molecular details of the TRIM5α antiviral mechanism remain to be solved, but the simplest model is SPRY domain-dependent recognition of incoming capsids and interference with subsequent core maturation and uncoating.
It is intriguing that Fv1 and TRIM5α, two proteins with no apparent homology, have antiviral activities against MLV-N, both with specificity for the MLV CA at position 110. It is also striking that despite similar antiviral specificity, Fv1-restricted MLV is able to synthesize DNA, whereas TRIM5α-restricted MLV does not. Here we demonstrate that Fv1 and TRIM5α operate independently and that they can compete for retroviral particles at a pre-reverse transcription step.
MATERIALS AND METHODS
Cell lines and viral vectors.
Cells were maintained in Dulbecco's modified Eagle medium with 10% fetal calf serum and 1 U/ml penicillin and streptomycin. SC1 is a feral mouse embryo cell line that restricts neither MLV-N, MLV-B, nor MLV-NB. TEN cells have been described previously (5). The TEB stable line was generated by transducing TE671 cells at high multiplicities of infection (MOI) with a Moloney MLV-based vector encoding red fluorescent protein (RFP) and Fv1b as described previously (5). Transduced cells were cloned by limiting dilution and were tested for RFP fluorescence and the ability to restrict MLV-N. The TEBT5KD stable line was generated by transducing TEB cells with an HIV-1-based vector carrying a short hairpin RNA (shRNA) against TRIM5 as described previously (33). Vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped MLV, equine infectious anemia virus (EIAV), and HIV-1 vectors were prepared by transient triple transfection of 293T cells with FuGENE 6 (Roche) as described previously (4). MLV stocks were quantified by titration on permissive SC1 cells, and, where relevant, titers are described as SC1 infectious units. HIV-1 and EIAV viral reverse transcriptase activity was measured by using an enzyme-linked immunosorbent assay (CavidiTech, Uppsala, Sweden).
Infection assays and flow cytometry.
Infection assays were performed as described previously (14, 33). Arsenic trioxide (As2O3) (Sigma) was prepared and used at 2 μM as described previously (3). The human TRIM5δ splice variant was PCR cloned from a plasmid kindly provided by P. Pelicci (Istituto Europeo di Oncologia, Milan, Italy) using forward primer 5′-CAGACGAATTCCACCATGGCTTCTGGAATCCTGGTTAATG-3′ and reverse primer 5′-TCGATTTCGAACTACTTGGGAGGCTGAGGCAGGAG-3′. The cDNA was cloned into the MLV expression vector CXCR, which also encodes red fluorescent protein (RFP), using EcoRI and Csp45I (underlined sequences). This plasmid was called CTdCR. DNA sequence was analyzed using DNA Cowboy (DNA sequence analysis software). TRIM5δ was then expressed by infecting target cells at high multiplicities of infection (MOI) with a Moloney MLV virus packaging CTdCR as previously described (14). Forty-eight hours later cells were replated, and 24 h after that they were challenged with the relevant green fluorescent protein (GFP)-encoding vectors as described previously (14).
Restriction factor saturation experiments.
Restriction factors such as TRIM5α and Fv1 can be saturated by treatment with an excess of virus-like particles (VLPs). VLPs were serially diluted and added to target cell cultures with a fixed dose of VSV-G-pseudotyped GFP-encoding reporter virus. The amount of GFP reporter virus used varied according to each cell line and from experiment to experiment but was selected so that infection with a restricted virus gave low, but accurately measurable, levels of infection (0.2 to 2% GFP-positive cells), so that an increase in permissivity could easily be detected. After 6 h, virus was removed and replaced with fresh medium. Infected cells were analyzed by fluorescence-activated cell sorting (FACS) as described above.
RNA interference.
Cells were infected at high MOIs with an HIV-1-based vector carrying an shRNA against TRIM5 (26, 33) or a control shRNA against red fluorescent protein (33). This vector was packaged into VSV-G-pseudotyped HIV-1 virus by transfection of 293T cells as described previously (4). Forty-eight hours after infection with shRNA-carrying virus, cells were replated and left overnight and then infected with GFP-encoding reporter vectors. The vector dose was selected, based on previous titrations, to give a low but measurable number of GFP-positive cells (0.2 to 2%) such that any enhancement could be readily measured.
Quantitative PCR.
TaqMan PCR to measure viral DNA synthesis was performed using primers and probe sequences specific to GFP as described previously (5). Cells (105) were infected in 6-well plates in triplicate with equivalent doses of DNase-treated MLV-N and MLV-B as measured on SC1 cells. Six hours after infection, total DNA was extracted from two samples using a DNeasy kit (QIAGEN, Chatsworth, CA). The third sample was subjected to FACS analysis 48 h after infection to enumerate infected cells. DNA (100 ng) was subjected to TaqMan quantitative PCR as described previously (5, 30).
RESULTS
Expression of Fv1b in human cells leads to additive restriction of MLV-N.
In order to investigate the relationship between Fv1 and TRIM5α, we generated a human TE671 cell line exogenously expressing Fv1b via a retroviral expression vector. We challenged TEB cells with serial dilutions of MLV-N, MLV-B, and MLV-NB GFP (Fig. 1A). Parallel titrations on unmodified TE671 cells are shown as a control (Fig. 1B). TEB cells were able to restrict significantly higher doses of MLV-N than unmodified TE671 cells, indicating that Fv1b and TRIM5α act additively. We also observed a slight decrease in the titer of MLV-NB in TEB cells, confirming that Fv1b can restrict MLV-NB infectivity when overexpressed (7).
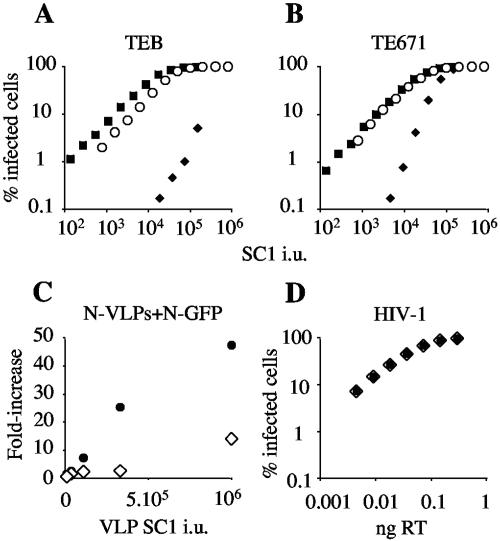
FIG. 1.
Expression of Fv1b in human cells leads to additive restriction of MLV-N. (A and B) Serial dilutions of GFP-encoding MLV-N (⧫), MLV-B (▪), and MLV-NB (○) were titrated onto human TE671 cells expressing Fv1b (TEB) (A) and unmodified TE671 cells (B). (C) Saturation experiment. TE671 (•) and TEB (⋄) cells were coinfected with a fixed dose of MLV-N encoding GFP and serial dilutions of MLV-N virus-like particles (N-VLPs). VLP dose in SC1 infectious units (i.u.) (x axis) is plotted against the fold increase in MLV-N GFP infectivity (y axis). (D) HIV-1 encoding GFP was titrated onto TE671 (•) and TEB (⋄) cells. Infected cells were enumerated 48 h later by FACS. Results are representative of at least two independent experiments performed with two independent preparations of virus. RT, reverse transcriptase.
Dominant restriction factors such as Fv1 and TRIM5α are generally saturable (8, 9, 29). Thus, treatment of target cells with high levels of restricted virus-like particles (VLPs) can saturate the factor and facilitate infection by a second restricted virus. In order to test whether Fv1b expression in human cells leads to an increase in the pool of active restriction factor molecules and increases the amount of VLPs required to saturate MLV-N restriction, we performed a saturation experiment. We titrated MLV-N VLPs onto TE671 and TEB cells in the presence of a fixed dose of MLV-N encoding GFP. Forty-eight hours later, GFP-expressing cells were counted by FACS and the fold increase in MLV-N titer was plotted against VLP dose (Fig. 1C). In concordance with Fig. 1A, TEB cells which express Fv1b in addition to endogenous TRIM5α require more MLV-N VLPs to saturate restriction. At the highest dose of VLPs, restriction of MLV-N is almost completely saturated in TE671 cells, whereas in TEB cells restriction is only partially saturated. Expression of Fv1b had no effect on the infectivity of MLV-B (Fig. 1A and B) or HIV-1 GFP (Fig. 1D), showing that the increased restriction in TEB cells is strictly virus specific. These findings show that Fv1b is functional when expressed in human cells and acts additively with TRIM5α. They also show that Fv1b cannot restrict HIV-1, confirming previous reports (2, 11) (Fig. 1D).
MLV-N is less infectious but reverse transcribes more efficiently in human cells expressing Fv1b.
TRIM5α and Fv1b both restrict MLV-N after viral entry and before integration and work additively when coexpressed (Fig. 1). They appear to have a critical mechanistic difference in that Fv1b-restricted MLV-N is able to reverse transcribe, whereas TRIM5α-restricted MLV-N is not. It is known that DNA synthesis is not strictly required for viral cores to interact with Fv1, because polymerase-deleted VLPs are able to saturate Fv1-mediated restriction (1). However, this result does not reveal whether Fv1-restricted virus is able to reverse transcribe because it interacts with Fv1 after reverse transcription has been completed or whether the consequence of an early interaction with Fv1 is only realized after reverse transcription. To address this question, we compared levels of MLV-N DNA synthesis after infection of TE671, TEB cells, and a TEB cell line stably expressing small interfering RNA (siRNA) to TRIM5 named TEBT5KD (Fig. 2A). FACS analysis of cells infected in parallel to those used for PCR are shown to indicate the viral infectivity. We used equal doses of MLV-N and MLV-B, determined on permissive SC1 cells. Six hours after exposure to virus, we measured the amount of viral cDNA by TaqMan quantitative PCR (30). Remarkably, MLV-N reverse transcription was considerably rescued in TEB cells compared to TE671 cells (Fig. 2A), despite the fact that TEB cells are around 10 times less permissive to MLV-N infection (Fig. 2B). As expected, MLV-N DNA synthesis was completely rescued in TEB cells expressing reduced levels of TRIM5 (TEBT5KD), but the cells remained nonpermissive to MLV-N due to Fv1b activity. Infectivity and DNA synthesis by unrestricted MLV-B was comparable in all three cell lines.
FIG. 2.
MLV-N is less infectious but reverse transcribes more efficiently in human cells expressing Fv1b. (A) Measurement of viral cDNA synthesis after restricted and unrestricted infection by TaqMan quantitative PCR. TE671, TE671 expressing Fv1b (TEB), and TEB expressing reduced levels of TRIM5 (TEBT5KD) were infected in triplicate with equal doses of GFP-encoding MLV-N (black bars) or MLV-B (white bars). Six hours after infection, DNA was extracted from two samples and subjected to quantitative PCR in duplicate using primers and probe specific for GFP. (B) The third sample was analyzed by FACS 48 h after infection to measure permissivity. Errors are standard errors of the means. Results are representative of two independent experiments performed on two different TEB clones. SSC, side scatter.
These data show that while TEB cells more strongly restrict infection by MLV-N, viral reverse transcription is substantially more efficient. This observation suggests that expression of Fv1b in human cells protects the virus from TRIM5α and allows reverse transcription to proceed. It implies that Fv1b interacts with viral cores early after they enter target cells and can compete with TRIM5α for interaction with cores before significant reverse transcription has occurred.
Fv1b is functionally independent from TRIM5α in human cells.
We next examined the relationship between Fv1 and TRIM5α. We performed a series of experiment where we manipulated restriction by TRIM5α and examined the effect on restriction by Fv1b in the same cells. We first reduced TRIM5 expression in TEB and TE671 cells by expressing TRIM5-specific siRNA using a lentiviral vector delivery system (33). TEB or TE671 cells were then challenged with GFP-encoding MLV-N, MLV-B, or EIAV vectors (Fig. 3A). As expected, TE671 cells restricted MLV-N and EIAV, and expressing TRIM5-specific siRNA rescued infectivity of both viruses. The titer of unrestricted MLV-B remained unaffected. In concordance with Fig. 1, the titer of MLV-N was lower on TEB cells than on TE671 cells. Reducing TRIM5α expression in TEB cells only slightly rescued infectivity of MLV-N, which is still strongly restricted by Fv1b. No effect was seen with a control siRNA against RFP, indicating that this result is TRIM5 specific. Effective reduction of TRIM5 expression in both TE671 and TEB cells is indicated by an equal rescue of EIAV infectivity in each cell line. These data show that reduction of TRIM5 expression has no effect on restriction by Fv1b in the same cells. They also show that, despite having a similar specificity to human TRIM5α in terms of MLV-N, Fv1b does not restrict the TRIM5α-sensitive horse lentivirus EIAV.
FIG. 3.
Fv1b is functionally independent of TRIM5α in human cells. (A) TE671 and TE671 cells expressing Fv1b (TEB) were infected with a lentiviral vector carrying an shRNA against TRIM5 (T5) or a control shRNA against RFP. Unmodified cells were also infected as a control (Ct). Seventy-two hours later, cells were challenged with serial dilutions of MLV-N (black bars), MLV-B (white bars), or EIAV (dotted bars), and infectious titers per milliter are plotted. Errors are standard errors of the means. (B) TE671 (circles) and TEB cells (triangles) were infected with serial dilutions of MLV-N GFP in the absence (hollow symbols) or presence (filled symbols) of 2 μM arsenic trioxide (As2O3). Permissivity was measured 48 h after by FACS. (C) Saturation experiment. TE671 (circles) and TEB cells (triangles) were coinfected with a fixed dose of MLV-N encoding GFP and serial dilutions of EIAV VLPs. VLP dose (in nanograms of reverse transcriptase [RT]) (x axis) is plotted against the fold increase in MLV-N GFP infectivity (y axis). (D and E) The TRIM5δ splice variant was expressed in TE671 (D) and TEB cells (E) using a retroviral vector delivery system. Seventy-two hours later, cells were challenged with MLV-N or MLV-B encoding GFP. MLV-N and MLV-B infectivity on unmodified cells is also shown as a control. Infected cells were enumerated 48 h later by FACS. Results are representative of at least two independent experiments performed with two independent preparations of virus. i.u., infectious units. SSC, side scatter.
Arsenic trioxide (As2O3) is known to interfere with TRIM5α activity by an unknown mechanism (3, 14). In order to examine the possibility of shared regulation between Fv1 and TRIM5α, we tested whether As2O3 treatment can affect Fv1 activity when it is expressed in human cells. We titrated GFP-encoding MLV-N onto TE671 and TEB cells in the presence or absence of 2 μM As2O3 (Fig. 3B). As2O3 rescued MLV-N infectivity in TE671 cells, as previously described (3), but it did not rescue MLV-N infectivity in TEB cells. This demonstrates that expression of Fv1b in human cells does not render it sensitive to As2O3 treatment and is in accordance with the insensitivity of endogenous Fv1 to As2O3 in murine cells (3). Notably, MLV-N infectivity is equal on TE671 and TEB cells at very low multiplicities of infection (Fig. 3B), but MLV-N is more restricted in TEB cells at higher multiplicities (Fig. 3A; also compare titration of restricted MLV-N in Fig. 1A and B). This difference is a consequence of dose-dependent infectivity in strongly restrictive cells illustrated by the nonlinear relationship between virus dose and infection seen, for example, in Fig. 1.
We next examined the effect of specifically saturating TRIM5α in TEB cells with TRIM5α-sensitive EIAV VLPs. We coinfected TE671 or TEB cells with a fixed dose of MLV-N GFP and a serial dilution of EIAV VLPs. As expected, EIAV VLPs rescued MLV-N GFP infectivity in a dose-dependent manner in TE671 cells (Fig. 3C). However, TEB cells still restricted MLV-N in the presence of EIAV VLP, suggesting that EIAV VLPs are not able to abrogate Fv1b restriction, probably because they cannot bind to it and are not restricted by it.
Stremlau and colleagues showed that expression of the γ splice variant of TRIM5 acts in a dominant-negative fashion to TRIM5α antiviral function (26). The TRIM5δ splice variant is slightly shorter than the γ variant and has a different carboxy terminus (21). First, we tested whether TRIM5δ can act dominant negatively against the TRIM5α anti-MLV-N activity. We infected TE671 cells at high multiplicities of infection with a Moloney retroviral vector encoding RFP and TRIM5δ. Seventy-two hours later we challenged cells with MLV-N or MLV-B encoding GFP. Expression of TRIM5δ markedly increased the permissivity of TE671 cells to MLV-N infection, by two orders of magnitude, demonstrating that TRIM5δ is dominant negative to TRIM5α function (Fig. 3D). We also performed the same experiment in TEB cells (Fig. 3E). MLV-N infectivity was not rescued in TEB cells expressing dominant-negative TRIM5δ, further confirming functional independence between Fv1b and TRIM5α. The titer of unrestricted MLV-B was unaffected in either TE671 or TEB cells by TRIM5δ expression at either high MLV-B MOI (Fig. 3D and E) or low MOI (data not shown). These findings provide further evidence that Fv1b can restrict MLV-N in human cells when TRIM5α activity has been functionally abrogated, consistent with the notion that it acts independently from TRIM5α when expressed in human cells.
Fv1n is also functionally independent from TRIM5α.
Fv1n has also been shown to be active when expressed in human TE671 cells, blocking MLV-B after reverse transcription as it does in NIH 3T3 cells (5). Previous work has shown that reduction of TRIM5α expression in TEN cells by transiently transfecting TRIM5-specific siRNA oligonucleotides partially rescued the Fv1n-restricted infectivity of MLV-B (14). This result is reproducible, but we have observed that transfection of shRNA can increase the permissivity of target cells to viral infection in a nonspecific manner (data not shown). We therefore reexamined the relationship between Fv1n and TRIM5α in TEN cells by using a lentiviral vector-based siRNA delivery system (33). We infected TEN cells with an HIV-1 vector carrying a TRIM5-specific shRNA and compared the titers of MLV-N, MLV-B, and MLV-NB encoding GFP on these cells and on untreated controls (Fig. 4A). Down-regulation of TRIM5 expression completely rescued infectivity of TRIM5α-sensitive MLV-N but not of Fv1n-restricted MLV-B, indicating that TEN cells still restrict MLV-B through Fv1n when TRIM5α levels are reduced. The titer of unrestricted MLV-NB was unaffected.
FIG. 4.
Fv1n is also functionally independent from TRIM5α. (A) TE671 cells expressing Fv1n (TEN) were infected with a lentiviral vector carrying an shRNA against TRIM5 (white bars) or left untreated (black bars). Seventy-two hours later cells were challenged with serial dilutions of MLV-N, MLV-B, or MLV-NB encoding GFP. Infection was analyzed by FACS, and data are presented as viral titers (infectious units [i.u.]/ml). Errors are standard errors of the means. (B) TEN cells were infected with serial dilutions of GFP-encoding MLV-N (circles) or MLV-B (triangles) in the absence (hollow symbols) or presence (filled symbols) of 2 μM arsenic trioxide (As2O3). Infected cells were measured 48 h after infection by FACS. (C and D) Saturation experiments. TEN cells were coinfected with a fixed dose of GFP-encoding MLV-N (circles) or MLV-B (triangles) and serial dilutions of either MLV-N (C) or EIAV (D) VLPs. VLP dose in SC1 i.u. or nanograms of reverse transcriptase (RT) (x axis) is plotted against the fold increase in MLV GFP infectivity (y axis). (E) The TRIM5δ splice variant was expressed in TEN cells using a retroviral vector delivery system. Seventy-two hours later cells were challenged with MLV-N or MLV-B encoding GFP. MLV-N and MLV-B infectivity on unmodified TEN cells is shown as a control. Infected cells were enumerated 48 h after infection by FACS. Results are representative of at least two independent experiments performed with two independent preparations of virus. SSC, side scatter.
To further examine the relationship between Fv1n and TRIM5α in TEN cells, we tested the effect of As2O3 on Fv1n-mediated restriction of MLV-B. We titrated GFP-expressing MLV-N and MLV-B on TEN cells in the presence or absence of 2 μM As2O3 (Fig. 4B). In concordance with data in Fig. 3, As2O3 treatment rescued infectivity of TRIM5α-sensitive MLV-N in TEN cells but not of Fv1n-restricted MLV-B, indicating that this drug does not regulate Fv1n activity.
Next we tested whether saturation of TRIM5α with either MLV-N or EIAV VLPs could affect Fv1n-mediated restriction of MLV-B in TEN cells. We coinfected TEN cells with a fixed dose of GFP-encoding MLV-N or MLV-B in the presence of a serial dilution of MLV-N VLPs (Fig. 4C) or EIAV VLPs (Fig. 4D). Both MLV-N and EIAV VLPs were able to efficiently saturate TRIM5α and rescue infectivity of MLV-N, as expected. However, Fv1n-restricted MLV-B remained restricted in the presence of a saturating dose of either MLV-N or EIAV VLP.
We then tested the effect of TRIM5δ expression on MLV-N and MLV-B restriction in TEN cells (Fig. 4E). The titer of TRIM5α-sensitive MLV-N was substantially increased by expression of TRIM5δ. However, TRIM5δ expression did not affect restriction of MLV-B by Fv1n. We conclude that TRIM5α is not required for Fv1n function in human cells and that Fv1n expressed in human cells is not sensitive to As2O3 treatment.
DISCUSSION
Innate immune strategies that defend mammals against retroviruses are of considerable medical and evolutionary importance. Here we have examined the relationship between two unrelated antiretroviral restriction factors, Fv1 and TRIM5α, and demonstrated that they operate independently and can compete for viral cores when coexpressed in human cells. TRIM5α-restricted virus does not reverse transcribe, so TRIM5α must recognize the viral capsid before reverse transcription has started or very early afterwards. On the other hand, Fv1-restricted virus reverse transcribes normally. We have shown that expression of Fv1b in human cells renders MLV-N significantly less infectious but able to reverse transcribe more efficiently (Fig. 2). This implies that Fv1 is able to compete with TRIM5α for retroviral particles and that the particles that are recognized by Fv1 are protected from TRIM5α and can undergo reverse transcription. However, despite being able to reverse transcribe, they are subject to the consequences of interacting with Fv1 and are eventually noninfectious. It remains unclear whether this is because Fv1 affects the maturation of the restricted reverse transcription complex (RTC) that occurs during reverse transcription (10) or whether RTC trafficking is influenced by Fv1 interaction, leading to reverse transcription occurring in the wrong, nonpermissive cellular compartment. Although Fv1 expression levels may be higher than those of TRIM5α in TEB cells, the fact that Fv1 blocks infection after reverse transcription, even when exogenously expressed in human cells, indicates that expression levels are unlikely to influence the timing of the interaction with virus.
Whether Fv1 and TRIM5α act alone or require specific cofactors remains to be determined. It remains unexplained why murine cells do not restrict both MLV-N and MLV-B when Fv1b is overexpressed in NIH 3T3 cells (7). One explanation for this result is that the high levels of exogenously expressed Fv1b compete with the low levels of endogenously expressed Fv1n for a limiting cellular cofactor. A low level of both proteins presumably explains the codominance of Fv1 restriction in progeny mice from NIH-BALB crosses. A cofactor for Fv1 therefore remains a possibility, but TRIM5α is no longer a candidate.
In conclusion, we have defined the time point at which Fv1 interacts with virus and shown that it can work additively and independently of TRIM5α. We conclude that Fv1 and TRIM5α can both recognize retroviral particles early after cell entry and before reverse transcription. Further characterization of these antiviral factors promises to reveal new viral biology and to form the basis for new therapeutic strategies against pathogenic retroviruses.
Acknowledgments
We thank Ben Webb, Sam Wilson, Laura Ylinen, Neruban Kumaran, Matteo Rizzi Brignoli, and Massimo Slavich for reagents and helpful discussion and P. G. Pelicci for the TRIM5δ cDNA.
This work was funded by a Wellcome Trust RCDF, number 064257, to G.J.T., and a UCL Graduate School Scholarship to Z.K.
REFERENCES
- 1.Bassin, R. H., B. I. Gerwin, J. G. Levin, G. Duran-Troise, B. M. Benjers, and A. Rein. 1980. Macromolecular requirements for abrogation of Fv-1 restriction by murine leukemia viruses. J. Virol. 35:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, J. G., D. Unutmaz, M. D. Miller, S. K. Breun, S. M. Grill, J. Mirro, D. R. Littman, A. Rein, and V. N. KewalRamani. 2004. Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 78:12537-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthoux, L., G. Towers, C. Gurer, P. Salomoni, P. P. Pandolfi, and J. Luban. 2003. As2O3 enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J. Virol. 77:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besnier, C., L. Ylinen, B. Strange, A. Lister, Y. Takeuchi, S. P. Goff, and G. Towers. 2003. Characterization of murine leukemia virus restriction in mammals. J. Virol. 77:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 7.Bock, M., K. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boone, L. R., P. L. Glover, C. L. Innes, L. N. Niver, M. C. Bondurant, and W. K. Yang. 1988. Fv-1 N and B tropism-specific sequences in murine leukemia virus and related endogenous proviral genomes. J. Virol. 62:2644-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duran-Troise, G., R. H. Bassin, A. Rein, and B. I. Gerwin. 1977. Loss of Fv-1 restriction in Balb/3T3 cells following infection with a single N tropic murine leukemia virus particle. Cell 10:479-488. [DOI] [PubMed] [Google Scholar]
- 10.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatziioannou, T., S. Cowan, and P. D. Bieniasz. 2004. Capsid-dependent and independent postentry restriction of primate lentivirus tropism in rodent cells. J. Virol. 78:1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolicoeur, P., and E. Rassart. 1980. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J. Virol. 33:183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300-305. [DOI] [PubMed] [Google Scholar]
- 16.Lassaux, A., M. Sitbon, and J. L. Battini. 2005. Residues in the murine leukemia virus capsid that differentially govern resistance to mouse Fv1 and human Ref1 restrictions. J. Virol. 79:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lilly, F. 1970. FV-2: identification and location of a second gene governing the spleen focus response to friend leukemia virus in mice. J. Natl. Cancer Inst. 45:163-169. [PubMed] [Google Scholar]
- 18.Nakayama, E. E., H. Miyoshi, Y. Nagai, and T. Shioda. 2005. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5α determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79:8870-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573.15243629 [Google Scholar]
- 24.Steeves, R., and F. Lilly. 1977. Interactions of host and viral genomes in mouse leukemia. Annu. Rev. Genet. 11:277-296. [DOI] [PubMed] [Google Scholar]
- 25.Stevens, A., M. Bock, S. Ellis, P. LeTissier, K. N. Bishop, M. W. Yap, W. Taylor, and J. P. Stoye. 2004. Retroviral capsid determinants of Fv1 NB and NR tropism. J. Virol. 78:9592-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in old world monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 27.Stremlau, M., M. J. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299.11027299 [Google Scholar]
- 29.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref1 restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Towers, G. J., D. Stockholm, V. Labrousse-Najburg, F. Carlier, O. Danos, and J. C. Pages. 1999. One step screening of retroviral producer clones by real time quantitative PCR. J. Gene Med. 1:352-359. [DOI] [PubMed] [Google Scholar]
- 31.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 33.Ylinen, L., Z. Keckesova, S. J. Wilson, S. Ranasinghe, and G. J. Towers. 2005. Differential restriction of HIV-2 and SIVmac by TRIM5α alleles. J. Virol. 79:11580-11587. [DOI] [PMC free article] [PubMed] [Google Scholar]