Abstract
The gene expression of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) was examined in two types of mammalian cells, human HeLa14 and hamster BHK cells. DNA microarray analysis followed by reverse transcription-PCR identified at least 12 viral genes transcribed in both HeLa14 cells and BHK cells inoculated with AcMNPV. 5′ rapid amplification of cDNA ends was carried out to examine the transcriptional fidelity of these genes in HeLa14 cells. The transcription of ie-1, ie-0 and gp64 was initiated at a baculovirus early gene motif, CAGT, accompanied by a TATA motif. In addition, the same splicing observed for ie-0 mRNA in Sf9 cells occurred in HeLa14 cells. While the transcription initiation sites for pe38 and p6.9 were not located in the CAGT motif, most of them were in a typical eukaryotic RNA polymerase II promoter structure (a conventional TATA motif and/or an initiator). Interestingly, the expression of β-actin was upregulated in the mammalian cells inoculated with AcMNPV. Subsequent experiments using UV-inactivated virus confirmed the upregulation, suggesting that de novo synthesis of viral products is not required for the event. These results indicated that the AcMNPV genome acts as a template for transcription in mammalian cells through the usual infection pathway, though there is no evidence for the functional expression of viral genes at present.
Autographa californica multiple nucleopolyhedrovirus (AcMNPV), a type member of the insect-specific virus family Baculoviridae (genus Nucleopolyhedrovirus [NPV]), infects several important pest insects and has been used as a safer biopesticide in Integrated Pest Management program. The nucleopolyhedroviruses are also used as a gene transfer vector in insect cells, providing one of the most efficient systems for the expression of foreign genes in eukaryotic cells (19). Furthermore, recent studies demonstrated that AcMNPV can enter the nucleus of mammalian cells independently of the cell cycle without multiplication (28, 29) and effectively express foreign genes (15) without expressing its own genes (28), suggesting that the baculovirus can be used as a low risk viral vector in the field of gene therapy (10). Recently, AcMNPV was shown to be capable of stimulating production of interferon in mammalian cell lines and when injected into mice conferred protection from lethal infections of encephalomyocarditis virus (13). It was also reported that the intranasal inoculation of a wild-type AcMNPV induced a strong innate immune response, which protected mice from a lethal challenge of influenza virus, through the Toll-like receptor 9 signaling pathway (1, 3). These reports indicate that baculoviruses affects the physiology of mammalian cells though they are not pathogenic to and have no ability to replicate in mammalian cells.
NPV is a large rod-shaped virus with a double-stranded closed circular DNA genome (130 to ∼160 kbp) containing approximately 150 genes (4). NPV genes are usually categorized into four groups: immediate-early, delayed-early, late, and very late (8). After infection, the immediate-early and delayed-early genes are transcribed by host RNA polymerase II and their promoters are characterized by the transcription initiation site consensus motif CAGT (25). Most of them are thought to code for the transcriptional transactivator of viral genes (33). Late and very-late genes are transcribed by viral RNA polymerase from the late promoter motif TAAG (22, 24), whose expression is regulated by the early genes. Thus, the senseful expression of early genes is key to NPV infection. The mechanism of transcription by RNA polymerase II is highly conserved among eukaryotes (14). Indeed, the promoter for immediate-early gene 1 (ie-1) of Bombyx mori NPV (BmNPV) showed the ability to promote transcription in mammalian cells in transfection experiments using reporter plasmids (21). Furthermore, the viral essential regulatory protein encoded by ie-1 (IE-1) is functional in mammalian cells (21, 23). More recently, the acidic activation domains of AcMNPV IE-1 were demonstrated to be functional for transcriptional activation in mammalian cells (12). These observations suggest that NPV could express at least part of the viral genomic information in mammalian cells and may affect the physiology of the host cells. Progress in the application of baculoviruses requires more information about the functions of AcMNPV in mammalian cells. In this study, we reexamined the gene expression of AcMNPV in mammalian cells using a DNA microarray, which suggested transcription from several viral genes and increased transcription from cellular β-actin.
MATERIALS AND METHODS
Cells and viral inoculation.
Spodoptera frugiperda (Sf9 cells) were grown in TC-100 medium (Sigma) containing 10% fetal bovine serum (FBS) at 26°C. Mammalian cells (HeLa14 and BHK) were maintained in modified eagle medium (Nissui) containing 10% FBS in a CO2 incubator. Treatment (inoculation) of the mammalian cells with AcMNPV was carried out by incubating the cells in Dulbecco’s modified eagle medium (DMEM) (FBS-free) containing AcMNPV for 2 h at 37°C followed by culture in DMEM containing 10% FBS at 37°C.
Viruses.
AcMNPV was grown in Sf9 cells in TC-100 medium containing 10% FBS at 26°C. We constructed the recombinant AcMNPV, Ac-dhspp-EGFP, using the Bac-to-Bac system (Invitrogen). This virus has an EGFP-coding sequence under the control of the Drosophila heat shock protein 70 promoter (nucleotides 1 to 1927; GenBank accession number AY032740). Viral titers were determined in Sf9 cells by plaque assay according to Maeda's method (19).
Inactivation of viruses was carried out by exposing the viral suspension (3 × 107 PFU/ml DMEM) in a sterile tissue culture dish to shortwave UV (UV-band, light 254 nm) at a distance of 30 cm for 30 min in a laminar flow hood. The inactivation of infectivity was verified by a plaque assay with Sf9 cells. The recombinant virus carrying luciferase under the control of the ie-1 promoter (rBACie1-luc) (2) was also used to examine the inactivation of viruses and demonstrated that UV-irradiation for 30 min under such conditions resulted in the loss of 99.7% of the activity to express luciferase and a lack of plaque formation (data not shown). The approach (irradiation for 30 min) was then used to prepare UV-inactivated virus.
DNA microarray.
A microarray analysis using a baculovirus DNA chip consisted of 192 spots including 140 AcMNPV genes was carried out as described elsewhere (32). In brief, total RNA was isolated using Trizol reagent (Invitrogen) and 400 pg of synthesized RNA, λpolyA+RNA-A (Takara), was added in each series for normalization. The fluorescent-labeled cDNA prepared for hybridization was generated with an RNA Fluorescent Labeling Core Kit (Takara) using 30 μg of total RNA and oligo(dT) according to the manufacturer's instructions. DNA microarrays were hybridized for 10 h under coverslips with Cy5- or Cy3-dCTP (Amersham)-labeled cDNA probes, washed, dried and scanned immediately in an Affymetrix 428 Array Scanner (Affymetrix Instruments). Data were analyzed using ImaGene software (Biodiscovery Inc.).
5′ RACE and Northern blot analysis.
Total RNA was isolated from Sf9 or HeLa14 cells at 48 hours postinfection (hpi) as described above. 5′ rapid amplification of cDNA ends was performed using the GeneRacer core Kit (Invitrogen) according to the manufacturer's instructions. Briefly, mRNAs were dephosphorylated, decapped and ligated with an adaptor oligo RNA supplied in the kit. A first PCR was then carried out with an adaptor primer from the kit and gene-specific first PCR primers (ie-1 and ie-0, 5′-GTCTGTTCAAGGGTTGCACAGC-3′; pe38, 5′-GGCTGGCGCACTGTCGTCAC-3′; gp64, 5′-AGACTGGTGCCGACGCCGCC-3′; p6.9, 5′-GCGTGTTCTGTAACTTCGGCGACC-3′). A second PCR was performed using an adaptor-specific nested primer and gene-specific nested primers (ie-1 and ie-0, 5′-AACTGGCCCACCACACCTTGTG-3; pe38, 5′-CCGTAATGCCACGTTGCGGC-3′; gp64, 5′-CGACCAGCCGCTGGCATCTTTC-3; p6.9, 5′-GGGGTCTACCCGGGCGGCGT-3′). PCR products were cloned using an AT cloning pGEM-T Easy vector system (Invitrogen) and at least four cDNA were sequenced using the ABI PRISM 310 Genetic Analyzer for each gene.
Total RNA (10 μg) isolated at the time presented in the figure was denatured and separated by electrophoresis in a 1% agarose gel, transferred to a nylon membrane (Hybond-N+; Amersham) and hybridized with oligonucleotide probes (human β-actin, 5′-ACGTCACACTTCATGATGGAGTTGAAGGTAGTTTCGTGGATGCCACAGGACTCC ATGCCT-3′; human 18S rRNA, 5′-CTGGACCGGCCCTGCGTACTTAGACATGTATGGCTTAATCTTTGAGACAAGCATATGGTT-3′) labeled with [γ32-P]ATP (Amersham) using MEGALABEL (Takara). Quantitative analysis was performed with an Image Reader BAS1000 (Fuji film).
RESULTS
DNA microarray analysis in mammalian cells.
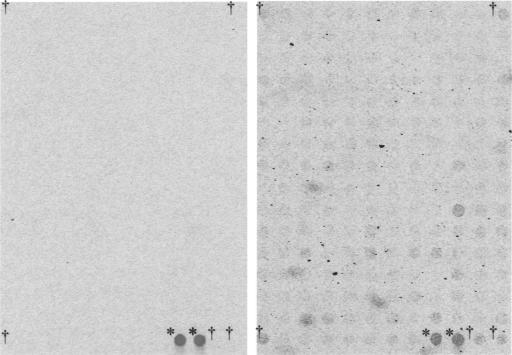
We adopted a DNA microarray to analyze the transcripts of AcMNPV in the mammalian cells. Cy3- or Cy5-dCTP-labeled cDNA probes were simultaneously hybridized to probe sequences on the microarray, and the amount of fluorescence seen with the individual dyes was determined by microarray scanner. The positivity of the signals of the gene spots on the DNA array chips was judged under the criteria “significantly positive: signal mean - (background mean + 2 × background standard deviation) > 0” (Fig. 1A). Forty-three viral genes were detected in the HeLa14 cells and 14 genes in the BHK cells (Table 1). The experiment was duplicated and similar results were obtained. Twelve of these genes overlapped in the two cell types, as confirmed by nested reverse transcription-PCR using gene-specific primers (data not shown).
FIG. 1.
DNA microarray analysis using Cy5-labeled cDNA probes. The cDNA probes were synthesized from total RNA extracted from mock-infected cells (left) or AcMNPV-infected BHK cells (at a MOI of 30) (right). Daggers indicate the positions of the spots for β-actin. Asterisks indicate the internal control, λpolyA.
TABLE 1.
Viral genes judged as positive (transcribed) in DNA microarray analysis
| Cell | Promotersa | Genes | |||||
|---|---|---|---|---|---|---|---|
| HeLa14 | E | orf149 | egt | ie-1 | |||
| E and L | pe38 | ie-2 | ie-0 | gp64 | me53 | ||
| L | orf91 | he65 | p6.9 | orf142 | ptp | odv-e56 | |
| ets | orf19 | vp80 | arif-1 | orf38 | orf120 | ||
| orf4 | orf66 | orf102 | orf92 | orf75 | orf78 | ||
| orf13 | orf73 | orf108 | p35 | ||||
| Others | orf140 | orf152 | odv-e26 | pp34 | dbp | lef-6 | |
| p47 | lef-3 | orf29 | orf41 | orf97 | orf45 | ||
| orf117 | |||||||
| BHK | E | orf149 | |||||
| E and L | pe38 | ie-2 | ie-0 | gp64 | |||
| L | orf91 | he65 | p6.9 | orf142 | hisp | ||
| Others | orf140 | orf152 | odv-e26 | 94k | |||
E or L indicates the presence of the following promoter motifs: E, early promoter motif (TATA box followed by CAGT motif); L, late promoter motif [(A/T/G)TAAG]. Underlining indicates the genes which were detected in both HeLa14 and BHK cells.
Fidelity in transcription.
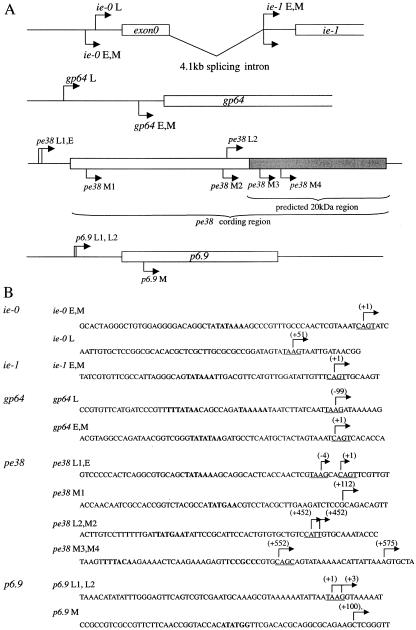
The fidelity of transcription initiation for the AcMNPV genes in HeLa14 cells was then analyzed by 5′ RACE. First of all, transcription initiation sites of ie-0, ie-1, gp64 and pe38 in Sf9 cells were determined at 48 hpi (Fig. 2). The transcription start site of ie-1 in Sf9 cells was located in a CAGT motif (early gene motif) located 77 nts upstream of the translation start codon, consistent with that reported previously (25). In HeLa14 cells, ie-1 was also transcribed from this site (Fig. 2). ie-0 and gp64 of AcMNPV have both a CAGT early motif (16, 9) and a TAAG late motif (4). 5′ RACE using mRNA purified from Sf9 cells at 48 hpi demonstrated that both genes were transcribed from the TAAG motif as previously reported (16, 7, 34) while the transcription start sites of these genes in HeLa14 cells were located in the CAGT early motif (Fig. 2). Furthermore, sequencing of the cDNA synthesized against the 5′ terminal region of ie-0 transcripts, which are known as the only baculoviral transcripts produced by splicing (11), revealed that the intron sequence (nts 122946 to 127149; GenBank accession number L22858) was precisely spliced out in HeLa14 cells. On the other hand, the transcription of pe38 was rather complicated. At 48 hpi, two transcription initiation sites for AcMNPV pe38 were elucidated in Sf9 cells, one in a TAAG motif located 4 nts upstream of the early CAGT motif and another located 452 nts downstream of the early motif (Fig. 2A) (20). This feature of the transcription, i.e., an internal transcription initiation site, corresponded with that observed for the Orgyia pseudotsugata NPV (OpNPV) p34 gene, a homologue of AcMNPV pe38 (31). The internal transcription initiation site generates a small mRNA with the capacity to code for a protein with a calculated molecular mass of 20 kDa both in OpNPV (31) and in AcMNPV. The transcription of pe38 in HeLa14 cells started at several sites which were not located in the CAGT motif (Fig. 2). The transcription start site of the late gene p6.9 in HeLa14 cells was located 99 nts downstream of the late TAAG motif (Fig. 2).
FIG. 2.
Mapping of the 5′ ends of the transcripts for ie-1 and ie-0, gp64, and pe38 in Sf9 and HeLa14 cells at 48 hpi. (A) Schematic representation of the gene structure for ie-1 and ie-0, gp64, and pe38. The open box shows the open reading frame. Arrows on the diagram for each gene indicate the initiation sites in Sf9 cells at the early (E) or late (L) stage and in HeLa14 cells (M), respectively. (B) Sequences around the transcription initiation sites in Sf9 cells and HeLa14 cells. The arrow shows the transcription initiation site and the number in parenthesis indicates the nucleotide position relative to the transcription initiation site in Sf9 cells (+1) at the early stage (for ie-0 [16]), ie-1 (25), gp64 (9), and pe38 (20) or the late stage (for p6.9 [this paper]). The CAGT early motif and the TAAG late motif are underlined. Boldface type indicates TATA-box like and BRE-like sequences.
Upregulation of β-actin expression in mammalian cells inoculated with AcMNPV.
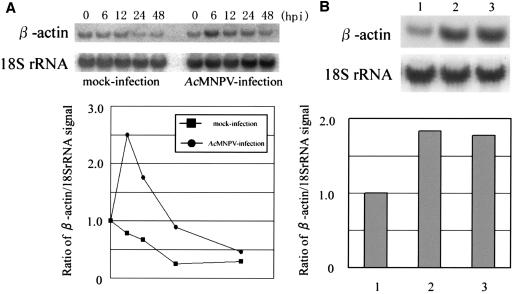
As described elsewhere (32), the NPV DNA microarray contains human β-actin and λ phage sequences as internal controls for normalization between chips. Interestingly, the relative intensity of the signal for β-actin on the DNA array was clearly increased in BHK cells (Fig. 1) and moderately increased in HeLa14 cells (data not shown) inoculated with AcMNPV. We then carried out Northern blot hybridization using total RNA isolated from HeLa14 cells to analyze the kinetics of the transcription of β-actin (Fig. 3). The amount of transcript from β-actin in HeLa14 inoculated with AcMNPV increased until 6 hpi, reached a level about 2.5-fold that of the control and then rapidly decreased, while the amount of transcript in the control cells gradually decreased until 24 hpi. A similar profile for the transcription of β-actin was obtained from BHK cells (data not shown). UV-inactivated AcMNPV also induced upregulation of β-actin expression at a level comparable to that induced by non-irradiated AcMNPV at 6 hpi (Fig. 3B, lanes 1 to 3).
FIG. 3.
Northern hybridization assays using 32P-labeled β-actin and 18S rRNA cDNA probes. (A) Northern hybridization was performed using total RNA purified from AcMNPV-infected HeLa14 cells at the times shown in the panel. Kinetics of the expression of β-actin is presented as a sequential line graph below the autoradiograph and the values are shown as the ratio of β-actin to 18S rRNA on the basis of the signal intensities obtained from the Northern hybridization. Circles and boxes in the graph indicate the kinetics for AcMNPV-infected and mock-infected cells, respectively. (B) Northern hybridization assay using total RNA from mock-infected (1), AcMNPV-infected (MOI of 30) (2) and UV-inactivated virus-infected (3) cells. The bar graph below the autoradiograph shows the ratio of hybridization signal for β-actin to that for 18S rRNA.
DISCUSSION
The host range of baculoviruses is restricted to arthropods including insects and crustaceans. The host specificity of AcMNPV was originally thought to be restricted to cells derived from insects. However, several groups recently reported that AcMNPV can transfer and express foreign genes under the regulation of appropriate eukaryotic promoters in several types of mammalian cells and animal models (18). This observation led us to consider its use in gene therapy given its low cytotoxicity in mammalian cells even at a high multiplicity of infection (MOI), an inherent incapability to replicate in mammalian cells, and the absence of preexisting antibodies against baculoviruses in animals. On the other hand, Beck et al. reported that AcMNPV infection repressed phenobarbital-mediated gene induction and stimulated tumor necrosis factor alpha, interlewkin-1α, interlewkin-1β beta expression in primary cultures of rat hepatocytes (5), indicating that mammalian cells could be physiologically influenced by the experimental attack with AcMNPV. More recently, AcMNPV was shown to induce an innate immunity that confers protection from a lethal challenge of influenza virus in mice through the recognition of the baculovirus genomic DNA by a Toll-like receptor molecule (1, 3). These observations are thus not in contradiction to previous observations that AcMNPV genes were not transcribed in HeLa cells inoculated with AcMNPV at a MOI of 100 PFU per cell (MOI of 100) at 24 hpi or after several passages (28). In contrast, we clearly detected the viral transcripts in mammalian cells inoculated with AcMNPV at a lower MOI (MOI of 30) using a DNA microarray and 5′ RACE technologies. This discrepancy might be due to the difference in the sensitivity of the detection methods used in the two experiments.
5′ RACE analysis revealed that all genes examined in this study, except pe38 and p6.9, were transcribed from an early transcription start site motif (CAGT) preceded by a TATA-motif 30 nts upstream in HeLa14 cells (Fig. 2). The CAGT motif is a part of the eukaryotic transcription initiation site sequence (initiator [INR], YYANT/AYY) (27) and the early promoters for these viral genes with a typical eukaryotic RNA polymerase II promoter structure (a conventional TATA motif and an INR) are functional in HeLa14 cells. Furthermore, the transcripts from ie-0 were observed to be precisely spliced (11) in HeLa14 cells, showing the compatibility of the splicing machinery between mammalian cells and insect cells. These observations suggested that there was no critical disadvantage in the transcription of these genes in mammalian cells. A late gene, p6.9 was also transcribed from 25 nts downstream of a TATA-like motif in the HeLa14 cells. However, the transcription initiation site was located in the coding sequence (Fig. 2). Four transcription initiation sites (nucleotide positions +112, +454, +552 and +575) were identified for pe38 in HeLa14 cells and none of them corresponded with the start sites in Sf9 cells. The most upstream site (+112) and the second site (+454) were located in the INR-like sequences CCGCAGA and CCATTGT, respectively, which were also accompanied by a TATA-motif 25 nts upstream (Fig. 2A). The sequences of surrounding the other two sites showed no significant similarity to INR but were preceded by sequences with some similarity with the TATA-motif or TFIIB recognition element G/CG/CG/ACGCC (BRE) (17, 26) 30 nts upstream (Fig. 2B). However, we could not obtain evidence for the transcript from a CAGT motif located 4 nts downstream of the transcription start site +1 for pe38 in HeLa14 cells though it was also accompanied by a TATA-motif 30 nts upstream. This suggested that the typical eukaryotic RNA polymerase II promoter structure was not sufficient to initiate transcription in the AcMNPV genome.
We happened to observe the upregulation of β-actin expression in AcMNPV-inoculated mammalian cells in an analysis using a NPV DNA microarray containing a human β-actin DNA spot as a control. Actin is essential for the transport of NPV to the nucleus in insect and mammalian cells (29) and is also necessary not only for constitution of the virogenic stroma but also for budding of the viruses in insect cells (6). In insect cells, AcMNPV infection induces overexpression of cellar actin and it inhibits the polyhedron synthesis and polyhedral formation, although factors stimulating actin expression are not clear (30). In order to investigate if the upregulation is related to the viral transcripts in HeLa14 cells, effects of UV-inactivated viruses on β-actin expression were examined. UV-inactivated viruses which were expected to be impaired in the fusion capability of envelope protein by the denaturation of GP64 (1) were able to induce an upregulation of β-actin expression compared to untreated control viruses, suggesting that the upregulation was caused by events occurring before the internalization of viral components. Thus, the significance of the AcMNPV transcript to the physiology of mammalian cells remains to be solved.
In conclusion, the results of our investigation provided evidence that AcMNPV is capable of expressing some viral genes at least at the transcription level in mammalian cells through the usual pathway of infection. These results emphasize the significance of studying the molecular details of baculovirus-mammalian cell interactions to reinforce the inability of AcMNPV to replicate in mammalian cells for facilitating the use of baculoviruses in the agrobiological, pharmaceutical, and medical fields.
REFERENCES
- 1.Abe, T., H. Hemmi, H. Miyamoto, K. Moriishi, S. Tamura, H. Takaku, S. Akira, and Y. Matsuura. 2005. Involvement of the toll-like receptor 9 signaling pathway in the induction of innate immunity by baculovirus. J. Virol. 79:2847-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe, T., N. Miyake, Y. Nishijima, R. Fujita, K. Sahara, S. Asano, and H. Bando. 2005. Enhancement of cauliflower mosaic virus 35S promoter in insect cells infected with baculovirus. Virus Res. 112:38-41. [DOI] [PubMed] [Google Scholar]
- 3.Abe, T., H. Takahashi, H. Hamazaki, N. Miyano-Kurosaki, Y. Matsuura, and H. Takaku. 2003. Baculovirus induces an innate immune response and confers protection from lethal influenza virus infection in mice. J. Immunol. 171:1133-1139. [DOI] [PubMed] [Google Scholar]
- 4.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 5.Beck, N. B., J. S. Sidhu, and C. J. Omiecinski. 2000. Baculovirus vectors repress phenobarbital-mediated gene induction and stimulate cytokine expression in primary cultures of rat hepatocytes. Gene Ther. 7:1274-1283. [DOI] [PubMed] [Google Scholar]
- 6.Blissard, G. W. 1996. Baculovirus-insect cell interactions. Cytotechnology 20:73-93. [DOI] [PubMed] [Google Scholar]
- 7.Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcription mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170:537-555. [DOI] [PubMed] [Google Scholar]
- 8.Blissard, G. W., and G. F. Rohrmann. 1990. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 35:127-155. [DOI] [PubMed] [Google Scholar]
- 9.Blissard, G. W., H. K. Philip, W. Rosalind, and G. F. Rohmann. 1992. A synthetic early promoter from a baculovirus: roles of the TATA box and conserved start site CAGT sequence in basal levels of transcription. Virology 190:783-793. [DOI] [PubMed] [Google Scholar]
- 10.Boyce, F. M., and N. L. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 93:2348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chisholm, G. E., and D. J. Henner. 1988. Multiple early transcripts and splicing of the Autographa californica nuclear polyhedrosisi virus IE-1 gene. J. Virol. 62:3193-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai, X., L. G. Willis, I. Huijskens, S. R. Palli, and D. A. Theilmann. 2004. The acidic activation domains of the baculovirus transactivators IE1 and IE0 are functional for transcriptional activation in both insect and mammalian cells. J. Gen. Virol. 85:573-582. [DOI] [PubMed] [Google Scholar]
- 13.Gronowski, A. M., D. M. Hilbert, K. C. F. Sheehan, G. Garotta, and R. D. Schreiber. 1999. Baculovirus stimulates antiviral effect in mammalian cells. J. Virol. 73:9944-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornberg, R. D. 1999. Eukaryotic transcriptional control. Trends Cell Biol. 9:M46-M49. [PubMed] [Google Scholar]
- 15.Kost, T. A., and J. P. Condreay. 2002. Recombinant baculoviruses as mammalian cell gene-delivery vectors. Trends Biotechnol. 20:173-180. [DOI] [PubMed] [Google Scholar]
- 16.Kovacs, G. R., L. A. Guarino, B. L. Graham, and M. D. Summers. 1991. Identification of spliced baculovirus RNAs expressed late in infection. Virology 185:633-643. [DOI] [PubMed] [Google Scholar]
- 17.Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg, and R. H. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence specific DNA binding by transcription factor IIB. Genes Dev. 12:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehtolainen, P., K. Tyynela, J. Kannasto, K. J. Airenne, and S. Yla-Herttuala. 2002. Baculovirus exhibits restricted cell type specificity in rat brain: a comparison of baculovirus-and adenovirus-mediated intracerebral gene transfer in vivo. Gene Ther. 9:1693-1699. [DOI] [PubMed] [Google Scholar]
- 19.Maeda, S., T. Kawai, M. Obinata, H. Fujiwara, T. Horiuchi, Y. Saeki, Y. Sato, and M. Furusawa. 1985. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature 315:592-594. [DOI] [PubMed] [Google Scholar]
- 20.Mans, R. M. W., and D. K. Mörsdorf. 1998. In vitro transcription of pe38/polyhedron hybrid promoters reveals sequences essential for recognition by the baculovirus-induced RNA polymerase and for the strength of very late viral promoters. J. Virol. 72:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuyama, T., S. Asano, K. Sahara, and H. Bando. 2003. Functional analysis of an immediate early gene, ie1, of Bombyx mori nucleopolyhedrovirus in mammalian cells. J. Insect Biotechnol. Sericol. 72:82-94. [Google Scholar]
- 22.Morris, T. D., and L. K. Miller. 1994. Mutational analysis of a baculovirus major late promoter. Gene 140:147-153. [DOI] [PubMed] [Google Scholar]
- 23.Murges, D., A. Kremer, and D. Knebel-Morsdörf. 1997. Baculovirus transactivator IE1 is functional in mammalian cells. J. Gen. Virol. 78:1507-1510. [DOI] [PubMed] [Google Scholar]
- 24.Ooi, B. G., C. Rankin, and L. K. Miller. 1989. Downstream sequences augment transcription from the essential initiation site of a baculovirus polyhedron gene. J. Mol. Biol. 210:721-736. [DOI] [PubMed] [Google Scholar]
- 25.Pullen, S. S., and P. D. Friesen. 1995. The CAGT motif functions as an initiator element during early transcription of the baculovirus transregulator ie-1. J. Virol. 69:3575-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qureshi, S. A., and S. P. Jackson. 1998. Sequence-specific DNA binding by the S-shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell 1:389-400. [DOI] [PubMed] [Google Scholar]
- 27.Smale, S. T. 1997. Transcription initiation from TATA-less promoters within eukaryotic protein-coding gene. Biochim. Biophys. Acta 1351:73-88. [DOI] [PubMed] [Google Scholar]
- 28.Tjia, S. T., G. M. zu Altenschildesche, and W. Doefler. 1983. Autographa californica nuclear polyhedrosis virus (AcMNPV) DNA does not persist in mass cultures of mammalian cells. Virology 231:192-200. [DOI] [PubMed] [Google Scholar]
- 29.van Loo, N. D., E. Fortunati, E. Ehlert, M. Rabelink, F. Grosveld, and B. J. Scholte. 2000. Baculovirus infection of nondividing mammalian cells: mechanisms of entry and nuclear transport of capsids. J. Virol. 75:961-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkman, L., K. Storm, V. Aivazachvili, and D. Oppenheimer. 1996. Overexpression of actin in AcMNPV-infected cells interferes with polyhedrin synthesis and polyhedra formation. Virology 225:369-376. [DOI] [PubMed] [Google Scholar]
- 31.Wu, X., S. Stewart, and D. A. Theilmann. 1993. Alternative transcriptional initiation as a novel mechanism for regulating expression of a baculovirus trans activatior. J. Virol. 67:5833-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamagishi, J., R. Isobe, T. Takebuchi, and H. Bando. 2003. DNA microarrays of baculovirus genomes: differential expression of viral genes in two susceptible insect cell lines. Arch. Virol. 148:587-597. [DOI] [PubMed] [Google Scholar]
- 33.Yoo, S., and L. A. Guarino. 1994. The Autographa californica nuclear polyhedrosis virus ie2 gene encodes a transcriptional regulator. Virology 202:746-753. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, Y. J., Y. Z. Yi, Z. F. Zhang, J. L. He, Y. X. Zhang, and X. F. Wu. 2003. Promoter activities in the baculovirus envelope glycoprotein gp64 gene. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35:18-26. (In Chinese.) [PubMed] [Google Scholar]