Abstract
Factors that restrict a virus from establishing productive infection in a new host species are important to understand because cross-species transmission events are often associated with emergent viral diseases. To determine the evolutionary pressures on viruses in new host species, we evaluated the molecular evolution of a feline immunodeficiency virus derived from a wild cougar, Puma concolor, during infection of domestic cats. Analyses were based on the coding portion of genome sequences recovered at intervals over 37 weeks of infection of six cats inoculated by either intravenous or oral-nasal routes. All cats inoculated intravenously, but only one inoculated orally-nasally, became persistently viremic. There were notable accumulations of lethal errors and predominance of G-to-A alterations throughout the genome, which were marked in the viral polymerase gene, pol. Viral structural (env and gag) and accessory (vif and orfA) genes evolved neutrally or were under purifying selection. However, sites under positive selection were identified in reverse transcriptase that involved residues in the nucleotide binding pocket or those contacting the RNA-DNA duplex. The findings of extensive G-to-A alterations in this cross-species infection are consistent with the recently described editing of host cytidine deaminase on lentivirus genomes. Additionally, we demonstrate that the primary site of hypermutation is the viral pol gene and the dominant selective force acting on this feline immunodeficiency virus as it replicates in a new host species is on key residues of the virus polymerase.
Viruses responsible for many new human and animal diseases are themselves not new but are resident in wild animal populations (8, 20). A cross-species transmission event may result in infection, and possibly in disease, in an exposed individual, but transmission of the virus between members of the new species does not necessarily occur. For example, human diseases caused by viruses such as Sin Nombre (Bunyaviridae) and Nipah (Paramyxoviridae) viruses result from exposure to infected reservoir species but infected humans do not transmit the virus effectively to other humans (10, 27, 29, 32, 33). In contrast, human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2, respectively), which derive from simian immunodeficiency viruses (13, 15) (all lentiviruses in the family Retroviridae), are readily transmitted from human to human and are effectively maintained in the human population. Despite the success of HIV-1 and HIV-2 in their new host, there is evidence that, in regions where contact among primates and humans occurs, humans are infected with, but do not transmit, a variety of primate retroviruses (55). Thus, barriers imposed by the new host individual may present a suboptimal environment for virus production that curtails subsequent spread in the population.
The ability of lentiviruses to establish infection in a new host species is of substantial interest for several reasons. Lentiviruses infect many mammalian species and, despite high mutation and recombination rates (18, 38, 43), are considered to be highly host specific. There is no apparent disease associated with endemic lentivirus infections (1, 2, 4, 5, 31), but the consequences of lentivirus transmission to a new host species can be devastating (30). Lentiviruses establish persistent infections in their hosts in the face of robust immune responses, which suggests that components of the acquired immune system are not exclusive barriers to successful cross-species transmission. Finally, lentiviruses are distinguished from other members of the retrovirus family by carrying accessory genes that modulate virus infectivity. Although lentiviruses differ in the type of accessory genes that they carry, most have a vif gene (25). vif protects the virus from host RNA editing enzymes that cause extensive G-to-A hypermutation of the viral genome and can render a virus defective (11, 14, 23, 28, 41, 42, 59, 60). The discovery that vif is necessary for productive viral infection highlights the importance of innate responses in controlling lentivirus infections (39). In accord with the premise that lentiviruses are host specialists, the protection conferred by viral vif is specific for the editing enzymes of the primary host species (3, 21, 40, 44, 56). Given the extent of the current HIV pandemic, there is a significant incentive to understand features that constrain lentiviruses from establishing infection in a new host.
Lentiviruses can infect new host species by experimental exposure (26, 47) in spite of the formidable barriers imposed by host defenses. In some cases, viremia is transient and virus extinction is attributed to host immune defenses, although the actual mechanism of immune clearance is not always determined. For example, domestic cats are host to a newly described feline immunodeficiency virus (FIV) but can be experimentally infected with FIVpco, which is derived from cougars (Puma concolor) (47, 49-51). Of interest, a recent study demonstrated that cats inoculated orally-nasally (o.n.) were less likely to sustain infection than cats inoculated intravenously (i.v.) (47). In this experimental cross-species infection, there were no features of host immune responses that correlated with viral persistence or clearance.
The goal of this study was to investigate mechanisms that account for successful or abortive lentiviral infection in a new host. Our data are based on samples from FIVpco infection of domestic cats. This model system of cross-species feline lentivirus infection is well established (49-51), and longitudinal studies on immunological profiles and virus distribution in these FIVpco-infected cats have been reported (47). Here, we evaluated the molecular evolution of FIVpco isolate PLV-1695 in domestic cats to determine (i) the evolutionary forces on the virus in this new host and (ii) viral features that distinguish successful from abortive virus infection in the new host.
MATERIALS AND METHODS
Samples.
Four cats were inoculated by oral-nasal (ON1 to ON4) or intravenous (IV1 to IV4) routes as described previously (47). Cats IV2 and ON3 were not evaluated in this study. Peripheral blood mononuclear cells (PBMC) were obtained at weeks 4, 11, 19, and 37. Virus could not be amplified after week 11 from cats ON1 and ON2 and after week 19 from ON4.
Limiting dilutions of genomic DNA template from each time point for all cats were amplified, and where possible, amplicons from two different PCRs were cloned and sequenced. Only one clone of the 5′ half-genome was obtained from the week 11 sample for cat ON2 and of the 3′ half-genome from the week 19 samples for IV3 and IV4 and the week 37 sample for IV1. Additional clones were evaluated from the week 4 samples of IV1 (five clones of the 5′ half-genome and six clones of the 3′-half genome) and ON1 (four clones of the 5′ half-genome) to optimize heteroduplex mobility assay (HMA) conditions.
PLV-1695 amplification and cloning.
The source of the virus used in this study was the biological isolate puma lentivirus 1695 (PLV-1695), derived from concanavalin A-stimulated PBMC from an infected cougar (51). The virus stock used to inoculate cats was produced in MYA-1 cells (47). Virus was recovered from a 2-ml aliquot of MYA-1 supernatant by centrifugation at 100,000 × g for 1.5 h. The virus pellet was treated with DNase I (Roche Applied Science, Indianapolis, IN) for 1 h. Viral RNA was purified using the QIAGEN QiaAmp viral RNA kit (Valencia, CA). cDNA was synthesized using 2 pmol of a virus-specific primer, CoLTR (CTCAGGCAGATGTCAGGGTTCAAT), in a reaction with SuperScript II RNase H-minus reverse transcriptase (RT) (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Three clones of the 5′ half-genome and four clones of the 3′ half-genome of the inoculum virus were derived and sequenced as described below. In addition, two clones of both 5′ and 3′ half-genomes were derived from the original infected cougar PBMC (PLV-PB). The ancestral full-length clone was derived a year prior to this experiment from PLV-1695 passaged in 3201 cells.
The proviral sequences evaluated in this study were derived from genomic DNA prepared from 1 × 106 PBMC of each infected cat using the QIAamp DNA blood minikit (QIAGEN, Valencia, CA). The PLV-1695 genome was amplified using nested PCR in two overlapping half-genome fragments. The 5′ half-genome spanned from gag through the first 30 bp of dUTPase and was 3.5 kb in length. First-round primers for the 5′ half-genome were 5PLV7F (TCCTTTTCAGCGGCGTTCGT) and 5PLV6R (TGGGCTACTTTCTGTCCTTGTTCG), and second-round primers were 5PLV9F (CGAGGGACGTGCGAACA) and 5PLV8R (CTGCTGCTACCAAATCATAACCTG). Conditions for the first-round 5′ half-genome amplification were 94°C for 3 min, then 35 cycles of 94°C for 30 s and 53°C for 30 s, and extension at 70°C for 4 min. Second-round PCR conditions were 94°C for 3 min, then 35 cycles of 94°C for 30 s and 54°C for 30 s, and extension at 70°C for 3.5 min. For the 3′ half-genome, which was 4.3 kb and included dUTPase through the end of env, first-round primers were 3PLV11aF (GCCGGAACCTACAGACCCCTT) and 3PLV12mR (ACTTTGTTCTGCCCATTCTCCTA) and second-round primers were 3PLV13F (GCAGGTTATGATTTGGTAGCA) and 3PLV14bR (CCCATTCTCCTATTGTAGCACTT). Conditions for the first-round 3′ half-genome amplification were 94°C for 3 min, then 35 cycles of 94°C for 30 s and 53°C for 30 s, and extension at 70°C for 4.5 min. Second-round PCR conditions were 94°C for 3 min, then 35 cycles of 94°C for 30 s and 51°C for 30 s, and extension at 70°C for 4.3 min. Takara Mirus Bio (Madison, WI) ExTaq DNA polymerase was used for all PCRs. PCR products were gel isolated, purified with the QIAGEN QIAquick gel extraction kit, and cloned into the pDrive vector using the QIAGEN PCR cloning kit. One Shot TOP10 chemically competent Escherichia coli cells (Invitrogen Corporation, Carlsbad, CA) were transformed and plated on LB agar plates with antibiotic selection. Plasmid DNA was recovered from overnight liquid cultures using a QIAGEN Spin Miniprep kit and sequenced. The PLV genomic sequence has been deposited in GenBank (DQ192583). Composite data sets of the 5′ and 3′ half-genome sequences are available from the authors upon request.
HMA.
Viral diversity and evolution studies are often conducted on multiple sequences derived from an individual at several time points. The location of the sequence fragment for evaluation is based on viral genes presumed or known to be important to infection or pathogenesis; for example, HIV-1 env carries determinants for viral entry and neutralization. In this analysis, we made no assumptions as to the regions of the viral genome that would be key for establishing infection or would be under selection. Therefore, we evaluated the complete coding region of the PLV-1695 genome derived from infection of six cats. However, the increased length of the sequenced product precluded analyses of multiple fragments from each cat. We developed a modified heteroduplex mobility assay to determine how representative the sequenced clones were of the genetic diversity within each PCR-amplified sample. To determine the type of migration pattern to expect for a given combination of sequences, the insert was excised from a sequenced clone. DNA was digested with AseI (5′ fragment) or AvaII (3′ fragment), and the digested fragments were purified. Each isolated band was mixed with the digested insert of a different clone to determine the patterns of hetero- and homoduplexes that would arise due to sequence or restriction fragment size differences. Digests were mixed with annealing buffer consisting of 100 mM NaCl, 10 mM Tris HCl, pH 7.8, and 2 mM EDTA. Reaction mixtures were denatured at 95°C for 3 min, cooled for 4 min at 4°C, heated to 55°C for 4 min, and chilled again for 4 min at 4°C. Products were resolved on a 5% polyacrylamide TBE Ready gel (Bio-Rad, Hercules, CA) for 60 min at 170 V. Ethidium bromide-stained gels were visualized with a 520-nm excitation laser on a Fujifilm FLA 3000G fluorescent image analyzer by using Fujifilm Image reader V4.22.
All PCR products derived from infected cats were evaluated using this modified HMA and compared to the patterns determined from clone sequences. No novel HMA patterns were discerned in PCR products.
Phylogenetic analysis.
Sequences were aligned using ClustalW (48) with manual adjustment. The complete 5′ (3,477 bp) and 3′ (4,316 bp) half-genome fragments from the virus stock used to inoculate the cats, from PLV-PB, and from all infected cats were subjected to phylogenetic analysis. The optimal model of evolution based on the hierarchical likelihood ratio test was determined in Modeltest (36) to be a Tamura-Nei model including invariant sites and rate variation among sites (TrN+I+G). Phylogenetic trees were estimated in PAUP* v4.0b10 (45) using maximum likelihood under the TrN+I+G model, and 100 bootstrap replicates were obtained using the fast-heuristic search algorithm. Mutation rate estimates and 95% confidence intervals were determined using maximum likelihood based on serial samples and the optimized model of evolution for the data set as implemented in PEBBLE (12).
Frequency distribution of codons under neutral evolution.
A phylogenetic tree was estimated for the ancestral PLV sequence and sequences from the last time point for all cats that sustained infection (all i.v. inoculated cats and ON4). The appropriate model of evolution was determined for a sequence data set of FIVpco genomes, which included the ancestral PLV-1695 sequence, using Modeltest (36) to account for the normal base frequencies and substitution rates in these feline lentiviruses. The expected frequency distributions of codons occurring under the null hypothesis of selectively neutral evolution were obtained from 1,000 simulation replicates across a tree of each gene containing the ancestral PLV sequence and the last sequences from cats that sustained the infection (week 37 sequences from IV1, IV2, and IV4 and week 19 sequences from ON4) by using Seq-Gen (37). The frequency of each codon was summed across evolved sequences at the tips of the tree, and the cumulative frequency distribution was determined for 1,000 replicates. Then the observed codon frequency for each of the sequences from infected cats was computed, and its position on the cumulative frequency distribution was determined. A matrix was generated that contained the number of sequences that fell in the <5%, 5 to 95%, and >95% regions of the frequency distribution. This matrix of expected and observed codon frequencies was analyzed using a Fisher exact test for each of the 13 pairs of codons that contain A or G in the third position. If the frequency of any codon from the PLV inoculum sequences did not fall in the 5 to 95% region, the codon was excluded from the analysis. This analysis was repeated on the PRT portion of pol from 10 individual FIVpco isolates (DQ308412 to -21) in order to compare codon usage of FIVpco in the natural cougar host to those detected during experimental infection of domestic cats.
Frequency of substitutions by position.
The frequency of each type of mutation (e.g., A to C, C to A, A to G, etc.) was determined for each sequence position for each gene. This analysis was performed using phylogenetic trees estimated as described above from all FIVpco and PLV sequences using the model of evolution identified by Modeltest (36) as most appropriate. First, the ancestral sequence at each of the interior nodes was estimated using the BASEML module of PAML (57) under the GTR+G model of evolution. Then, the frequency of each type of substitution at each gene position was summed across all branches in the clade of infected cat and PLV inoculum sequences, from the basal node to the terminal nodes. This analysis was repeated on the protease and RT (PRT) portion of pol from nine individual FIVpco isolates described above and on a previously described data set consisting of FIVpco pol fragments (4) in order to compare substitutions that arose during FIVpco infection of the natural cougar host to those detected during experimental infection of domestic cats.
Distribution of substitutions.
The frequency with which multiple substitutions at a site were observed was compared with the frequency obtained when substitutions are distributed randomly. This analysis was conducted only for each of the common mutations (A to G, G to A, and T to C). To generate the frequency distribution for substitutions in each gene, the frequency of sites with 0, 1, 2, … substitutions was determined for each gene/substitution type. Using the number of substitutions and sequence length for each combination of gene/substitution type, the appropriate number of substitutions was distributed at random and the frequency of sites with 0, 1, 2, … substitutions was recorded. This was repeated 1,000 times for gag, 5′ and 3′ pol, and env to determine the proportion of sites expected to have a given number of substitutions under a model of random substitution. In each gene, multiple substitutions were rare under random expectation, reaching the greatest expected frequency of only 0.055 for G-to-A mutations in PRT. Significant deviation from random distribution was assessed using a chi-square test among genes and among substitution types.
To determine whether substitution sites were randomly distributed along the sequence, the observed frequency distribution (Fo) of distances between substitution sites of the common mutations (A to G, G to A, and T to C) in each of four genes was compared with the distribution for randomly placed substitutions in 1,000 replicates. For each replicate i, the number of observed substitution sites was distributed at random on a sequence of appropriate length, and the frequency distribution (fi) of the distances between substitution sites was computed. These 1,000 instances of fi were summed to give an overall frequency distribution Fs. Each distribution, fi, Fo, and Fs, was converted to a cumulative frequency distribution, ci, Co, and Cs, respectively. The Kolmogorov-Smirnov D statistic was used to compare these cumulative frequency distributions. This statistic is the absolute value of the greatest difference between two cumulative frequency distributions, f1(X) and f2(X), for any given X. The sampling variation of D for random substitutions was estimated by computing Di for each ci against Cs. The difference between the observed distribution and the distribution for random substitutions was summarized by computing Do for the comparison of Co against Cs. The probability of the observed value of Do, or one more extreme, was inferred by comparison with the empirical distribution of Di. The distribution of A-to-G, G-to-A, and T-to-C mutations was assessed at all substitution sites and for just those sites having multiple (≥2) substitutions.
Selective profiles.
The selection pressures were determined for each gene in the PLV genome except for rev. A maximum likelihood tree was produced for each gene based on the optimized model of evolution determined in Modeltest (36). The sequence alignment and tree were submitted to the Hyphy package available from DataMonkey (http://www.datamonkey.org), and the single-likelihood ancestor counting (SLAC) and fixed effects likelihood (FEL) methods using the REV (general time reversible) model of evolution were used to assess selection (34, 35). In addition, several models of codon-based substitution available in the CODEML module of PAML (57, 58) were considered that allowed different values for the parameter ω, the ratio of the rates of nonsynonymous and synonymous substitutions. Model M0 assesses the fit of a single value of ω to the data. Model M1 is contrasted with model M2, which includes an estimate of the proportion of sites having a value of ω > 1, to give a test for positive selection. A second such test is achieved by contrasting M7, in which a beta distribution is used to model the frequency of sites with ω ≤ 1, with M8, which has an additional site category with ω > 1. A likelihood ratio test with the Bonferroni correction for multiple tests was used to determine the model that best fit the data. The Bayes empirical Bayes (BEB) calculation of posterior probabilities for site classes was used to calculate the probabilities of sites under positive selection (58).
Nucleotide sequence accession number.
The PLV genomic sequence has been deposited in GenBank (DQ192583).
RESULTS
Phylogenetic relationship of sequences.
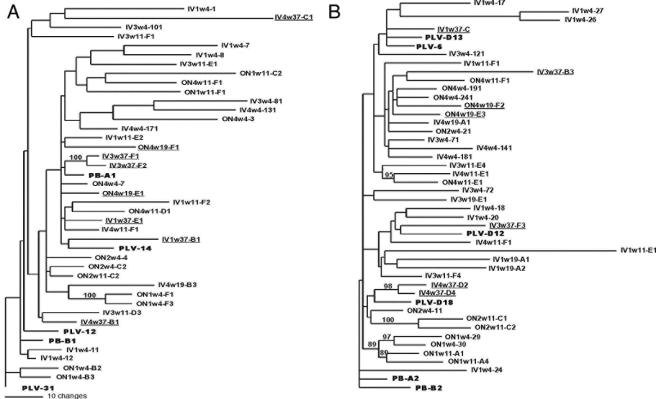
We evaluated the phylogenetic relationship among sequences obtained from infected cats to determine if there were common genetic features that distinguished viruses that persisted or were cleared. The maximum likelihood estimates of evolutionary rate for the 5′ and 3′ half-genomes were similar, 0.26% (0.012 to 0.475%) and 0.37% (0.167 to 0.567%) per site per year, respectively. There were no significant differences between rate estimates for i.v. group and o.n. group half-genomes. The branch lengths of sequences from both the 5′ and 3′ half-genomes derived from experimentally infected cats are in general longer than those of the inoculum PLV sequences and those derived from PBMC of the infected cougar (shown in boldface in Fig. 1). Despite evidence of diversification in PLV sequences in infected cats, there was weak phylogenetic signal in the 5′ portion of the genome (Fig. 1A), which spanned through the end of the viral polymerase. Only the branch containing sequences from the week 4 ON1 and week 37 IV3 samples had support. However, there was strong support for phylogenetic affiliation of 3′ genome sequences (Fig. 1B) obtained from both time points from ON1 and the final samples from ON2 (week 11) and IV4 (week 37). There was no relationship between inter-time point sequences in any of the cats except ON1.
FIG. 1.
Phylogenetic relationship of sequences obtained over 37 weeks from six cats inoculated with PLV-1695. (A) 5′ half-genome. (B) 3′ half-genome. Clone names indicate route of inoculation, cat identification number, date on which the sample was obtained in weeks postinoculation, and a clone number. Trees were generated by maximum likelihood using a TrN+I+G model of evolution. Branches with bootstrap values greater than 80% are shown. Sequences from the PLV inoculum or PBMC of the infected cougar from which PLV-1695 was obtained are boldface in the figure. The final sequences obtained from the cats that sustained infection (IV1, IV3, IV4, and ON4) are underlined.
Viral diversity.
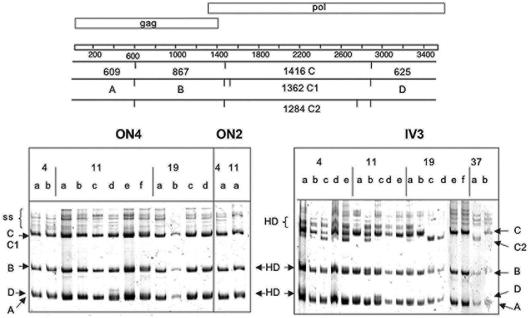
Because it was cost-prohibitive to sequence multiple full-length clones from an animal at each time point, we used an HMA to determine how well the sequenced clones represented the overall viral diversity in each sample. The 5′ and 3′ half-genomes obtained from PCR of serial dilutions of genomic DNA were digested with AseI and AvaII, respectively, as described in Materials and Methods. The diversity within the 5′ half-genome was due to both point mutations and the acquisition of new AseI sites in pol (Fig. 2). There was no evidence of other major variants in any of the 5′ PCR products, indicating that our clones were representative of the diversity within a cat. The 3′ half-genome PCR products were more variable in all infected cats. The AvaII sites were conserved in all sequences in the alignment. Thus, the majority of the HMA diversity arose from length variation of up to 15 bp at the 5′ region of orfA (shown schematically in Fig. 3C and D). Again, the 3′ half-genome clones appeared to represent the diversity seen in the PCR products for all animals.
FIG. 2.
A representative HMA of the 5′ half-genome of ON2, ON4, and IV3. PCR products from different weeks postinfection were digested with AseI and prepared for HMA as described in Materials and Methods. The AseI sites and resultant fragment sizes are shown schematically at the top of the figure in relation to the genomic structure. Each fragment is given a letter designation to indicate its position on the gel. Heterogeneity at the AseI site that flanked fragment C in pol resulted in different fragment sizes and is indicated as C to C2. Heteroduplex formation occurred in fragments B and D in o.n. inoculated cats, and AseI fragment C was the most common. In contrast, sequences from IV3 had variants C and C2, which were identified at each time point. Heteroduplex formation in all fragments was more noticeable in all IV3 PCRs except at the week 37 time point. The numbers at the top of the gel indicate the week of the sample, and the lowercase designation represents an individual PCR product.
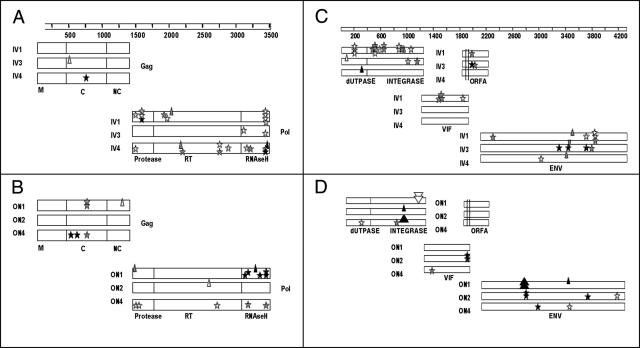
FIG. 3.
Distributions of insertions, deletions, and stop codons in PLV-1695-infected cats. Panels A and B are the 5′ half-genome. Panels C and D represent the 3′ half-genome. Panels A and C are from intravenously inoculated cats, and panels B and D are from orally-nasally inoculated cats. Stars represent stop codons, and small upright and inverted triangles represent single nucleotide deletions and insertions, respectively. The larger triangles represent a 21-bp deletion in ON4 integrase, a 66-bp insertion in ON1 integrase, and a 63-bp deletion in ON1 envelope. White, gray, and black symbols indicate sequences from the first, middle, and final samples, respectively. ON1 and ON2 did not have detectable virus after week 11, and ON4 did not have virus after week 19. Therefore, these dates represent the final samples (in black) for those animals. The genes are displayed to represent their position in the genome and are labeled on the figure as follows: NC, nucleocapsid; C, capsid; M, matrix. The parallel lines in orfA in panels C and D represent a region of length variation.
Distribution of lethal mutations.
We determined the distribution of mutations that would lead to a frameshift or stop codon within coding regions and render the virus defective (Fig. 3). The number of lethal mutations in sequences from all cats was remarkably high and was not evenly distributed across the genome. There were no lethal mutations in matrix (M) and only one in nucleocapsid (NC) in the 38 sequences evaluated. In contrast, lethal mutations were common in pol, which is represented in both the 5′ (PRT) and 3′ (dUTPase and integrase [UI]) half-genomes. Stop codons were numerous in sequences from all cats and clustered in the RNase H region. Cat IV1 sustained a particularly high number of stops in pol despite the fact that this cat maintained infection throughout the 37-week experiment. Larger insertions or deletions occurred only in sequences from ON1 and ON4 and were located in the integrase region of pol and in env. The length variation detected in orfA in most infected cats maintained the coding frame.
Molecular evolutionary parameters.
Because the distribution of lethal mutations appeared to be concentrated in certain areas of the genome, we determined the base frequencies for each gene based on the optimized model of evolution from Modeltest (36). There were no differences between base frequencies of genes derived from o.n. and i.v. inoculated cats. Similar to other retroviruses, there was a high prevalence of adenosine (A) residues in these sequences but the relative proportion of A was significantly higher in 5′ pol than in other coding regions (Student t test, P = 0.03; Table 1). The ratio of transitions to transversion was also significantly higher in 5′ pol.
TABLE 1.
Evolutionary genetics parameters for each PLV gene obtained from infected cats
| Parameter | Gag | PRTa | UIb | Vif | ORFe A | Env |
|---|---|---|---|---|---|---|
| Freqf A | 0.3859 | 0.4368 | 0.4065 | 0.3679 | 0.3498 | 0.3773 |
| Freq C | 0.1825 | 0.1406 | 0.1471 | 0.1503 | 0.1326 | 0.1632 |
| Freq G | 0.2113 | 0.1870 | 0.2090 | 0.2157 | 0.2470 | 0.2001 |
| Freq T | 0.2203 | 0.2355 | 0.2374 | 0.2661 | 0.2705 | 0.2594 |
| Ts/Tvc | 23.7904 | 45.0098 | 18.4540 | 16.5405 | 25.9121 | 23.9770 |
| Selectiond | M1, M7 | M2, M8 | M2, M8 | M0 | M0 | M1, M7 |
5′ region of pol containing protease and reverse transcriptase.
3′ region of pol containing dUTPase and integrase.
Transition-to-transversion ratio.
Model of selection determined in PAML.
ORF, open reading frame.
Freq, frequency.
Codon bias.
The nucleotide frequency bias towards A in HIV-1 can be attributed, in part, to the recently described action of a host cytidine deaminase, APOBEC3G (42, 60). During reverse transcription, this enzyme deaminates cytidine on the single-stranded DNA produced as the RNA template is degraded and results in incorporation of A instead of G in the proviral plus-sense DNA strand. Recently, the action of a feline cytidine deaminase with homology to human APOBC3F has been reported on feline foamy viruses (19). We reasoned that, if there was an increase in G-to-A mutations resulting from the action of domestic cat cytidine deaminase on a foreign lentivirus, then codons that were degenerate for A or G at the third position would be represented by a higher frequency of codons with third-position A. We tested the null hypothesis that these codons would be distributed randomly based on a frequency distribution generated from the sequence data. The observed frequency of 6 of the 13 codons that are represented by NNR (where N is any nucleotide and R is either A or G; Table 2) was significantly different than expected from the frequency distribution. In all six cases, the frequency of NNA fell in the 95% tail of the distribution. The codon bias was not equally distributed across all genes, as four of the six codons biased for third-position A were found in pol.
TABLE 2.
Codons with a synonymous third-position A that fell in the 95% tail of a codon frequency distribution
| Codon | Gene | Amino acid |
|---|---|---|
| TTA | Gag and PRTa | Leu |
| TCA | Env | Ser |
| GTA | Env | Val |
| GAA | PRT | Glu |
| AAA | PRT | Lys |
| AGA | PRT and UIb | Arg |
5′ region of pol containing protease and reverse transcriptase.
3′ region of pol containing dUTPase and integrase.
To ascertain that this codon bias was due to replication of the cougar-derived PLV isolate in domestic cats, we utilized the same method described above to evaluate the codon frequency in the PRT region of nine FIVpco isolates. The only codon in PRT from FIVpco that occurred at a higher-than-expected frequency was GGG, which encodes Gly.
Substitution frequencies.
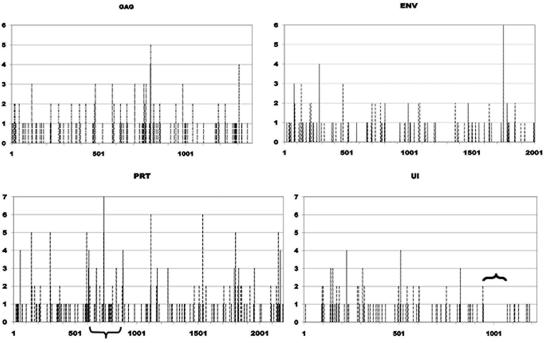
As a second test of the randomness of substitutions, we determined the frequency of each nucleotide state change for each gene along branches of the phylogenetic tree (see Materials and Methods). G-to-A mutations represented 41.5% (range, 33.0 to 51.4%) of all substitutions. There was no correlation between the relative numbers of G and the location of G-to-A mutations in any of the genes. Notably, the 124-bp region in UI devoid of G-to-A mutations (Fig. 4) contains five GG and eight GA dinucleotides, which are the preferred target for APOBEC3 modification of HIV-1 (59). Of significance, this site is immediately 3′ of the central polypurine tract (cPPT), which also is lacking G-to-A mutations.
FIG. 4.
Distribution of G-to-A mutations across the PLV genome. The frequency of G-to-A mutations was determined for each sequence position for gag, 5′ pol (PRT), 3′ pol (UI), and env as described in Materials and Methods. The nucleotide position for each gene is given on the x axis, and the y axis shows the number of sequences in which a G-to-A mutation arose at that position in the gene. The bracket in the panel showing the UI gene is immediately 3′ of the cPPT. The inverted bracket in PRT shows the region evaluated for nucleotide substitution in sequential FIVpco isolates.
We evaluated the nucleotide substitution frequency in the FIVpco PRT sequences used above to ascertain that the high frequency of G-to-A mutations was not an intrinsic property of cougar lentivirus replication. G-to-A changes comprised 13.6% of all substitutions while 30% of all changes were A to G. We also evaluated a previously described data set (4) of sequences that span the region of PRT with the highest incidence of G-to-A changes (shown by an inverted bracket in Fig. 4). The sequences were derived from between two and four sequential PBMC samples of six individual cougars (Table 3). Whereas purine transitions comprise between 50 and 82% of all point substitutions that occurred in this fragment of the FIVpco genome, the number of A-to-G changes was equal to or greater than the number of G-to-A mutations in five of the six animals. Thus, the frequency of G-to-A changes reported here for PLV replicating in a new host relates to the host environment and not to intrinsic features of FIVpco replication.
TABLE 3.
Nucleotide substitution in a fragment of pol in sequential FIVpco samples from naturally infected cougars
| Animal | Intervala | Nucleotidesb | Mutationc | No. of mutations
|
|
|---|---|---|---|---|---|
| G to A | A to G | ||||
| SF617 | 2 (3 yr) | 11,040 | 32 (0.29) | 11 | 10 |
| SF618 | 2 (1.5 yr) | 11,040 | 22 (0.20) | 5 | 13 |
| SF620 | 4 (2 yr) | 11,500 | 27 (0.23) | 16 | 5 |
| SF623 | 2 (1 yr) | 9,660 | 13 (0.13) | 4 | 4 |
| YF125 | 4 (4 yr) | 8,100 | 10 (0.12) | 3 | 3 |
| YM131 | 2 (2 yr) | 6,480 | 8 (0.12) | 1 | 3 |
Number of sequential samples from which sequences were derived and the interval over which the samples were collected in years.
Total number of nucleotides evaluated. Fragment size varied from 460 to 540 bp.
Number of single nucleotide substitutions in the alignment. Heterogeneous sites were not included in the total count. The number in parentheses is the percentage of the total number of sites that sustained a substitution.
Distribution of substitutions.
To determine if some sites are more likely to have multiple substitutions than others (e.g., whether are there “hotspots” for the location or type of substitution), we compared the frequency with which multiple substitutions were observed with the frequency obtained when substitutions are distributed randomly. Multiple substitutions were rare for randomized substitutions with the greatest expected frequency of only 0.055 for G-to-A mutations in PRT. Within our sequences, there was significant heterogeneity observed among genes in the frequency of multiple substitutions for A to G, P < 0.005, and G to A, P < 0.017, but not T to C. G-to-A mutations had the most extreme nonrandom distribution in all genes, but this was especially pronounced in PRT. In addition, the highest incidence of multiple substitutions (0.58) occurred for G-to-A mutations within PRT.
We also determined if there was clustering of substitution sites in each gene by determining if sites acquiring substitutions were distributed randomly on the sequence. A-to-G and T-to-C mutation sites were randomly distributed in all genes. However, there was a nonrandom pattern of G-to-A mutation at single hit sites for both gag and PRT (P < 0.001 and P < 0.0001, respectively). When only sites with multiple substitutions were considered, then PRT was the only gene to have a nonrandom distribution (P < 0.02). Thus, there are hotspots within PRT for G-to-A mutations, and these sites tend to cluster.
Selection profiles.
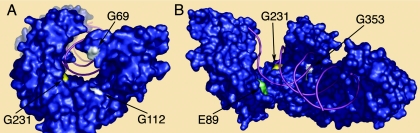
We utilized several methods to determine the primary evolutionary forces on each of the viral genes and to identify sites in each gene under positive selection. From the PAML analysis, the model of selection that best fit the data was the same for the i.v. and o.n. group data sets for all genes (Table 1). The accessory genes vif and orfA were evolving under a purifying model of evolution. For both gag and env, approximately 65 to 75% of sites were neutrally evolving with the remainder under purifying selection. Whereas the likelihood ratio test indicated that the UI region was under positive selection, the estimated proportion of sites under positive selection was zero under the M2 model and Bayes empirical Bayes identified only one selected site under M8. No sites in UI were identified to be under positive selection by FEL or SLAC. The only gene with convincing evidence for positive selection was 5′ pol. Three sites that fell in the region encoding RT were identified by all methods (BEB, SLAC, and FEL) to be under positive selection. Two sites in RT and two in the cleaved leader sequence to protease were identified by BEB and FEL, and an additional site in RT was identified only by BEB. All of the positively selected sites in RT fell at residues important for enzyme-ligand contact (Fig. 5). Two of these sites fell within the nucleotide binding domain, and they correspond to HIV-1 RT residues G112 and S68 or T69 (the FIV RT sequence is one amino acid shorter in this region than the HIV-1 RT). Three sites under selection were at positions that contact the RNA template, corresponding to E89, G231 (in the “primer grip”), and K353 in the HIV-1 RT sequence (9, 16, 17) (Fig. 5). The amino acids at these sites in inoculum PLV sequences were G69, E89, G112, G231, and R353. Thus, the residues at positions 89, 112, and 231 are conserved between primate and feline lentiviruses. Mutations identified in sequences derived from PLV-infected cats were R, K, or E at site 69; K at site 89; R or E at site 112; R or E at site 231; and K at site 353. All amino acid changes resulted from G-to-A mutations.
FIG. 5.
Model of reverse transcriptase showing the locations of positively selected sites in PLV-1695 RT. The model is based on the crystal structure of HIV-1 RT (1rOa). Sites under positive selection are indicated in white (supported by BEB, SLAC, and FEL), yellow (supported by BEB and FEL), and pale green (supported by BEB). The amino acid residue in the parental PLV-1695 sequence and the position relative to the crystal are shown. Only the p66 chain is shown for clarity. (A) Nucleotide binding site. (B) Longitudinal view with polymerase catalytic domain to the left and RNase H domain to the right.
DISCUSSION
This report provides the first detailed analysis of the evolutionary genetics of a feline lentivirus during in vivo infection of a new host species. Our data indicate that there are significant accumulations of fatal mutations in the PLV-1695 genome, as this cougar-derived feline immunodeficiency virus replicates in domestic cats and the most common mutation throughout the genome was G to A. Our previous report on the molecular evolution of FIVpco (4) and data presented herein demonstrate that a high frequency of lethal mutation and G-to-A bias were not properties of FIVpco in the natural cougar host. There are three mechanisms that could account for these data. An imbalance in the pyrimidine nucleoside pool of infected cells can enhance G-to-A mutation frequency (24, 53). Thus, the target cell supporting replication of a virus in a new host could affect the nature and frequency of nucleotide substitutions. Second, the most common error made during reverse transcription of HIV-1 in single replication cycle experiments is a G-to-A mutation (22). This finding suggests that intrinsic properties of the viral RT can be responsible for a nucleotide bias. Most recently, editing of viral minus-strand DNA by a host cytidine deaminase, APOBEC3G, has been reported for HIV-1 (14, 42, 59, 60). The HIV-1 vif protein is essential to mitigate the effect of host editing enzymes by targeting them for degradation (7, 23), although additional mechanisms may also be involved (11, 46). In the absence of vif, APOBEC3G is packaged into virions. In the next round of infection, this enzyme deaminates cytidine to uracil during first-strand DNA synthesis, which leads to incorporation of A instead of G at the edited site in the plus-sense DNA strand. In vitro, vif is specific to the editing enzymes from the host of origin, which may provide one explanation for the host specificity of lentiviruses (3, 21, 40, 56). It is likely that all three mechanisms contribute to the marked G-to-A hypermutation observed in some HIV-1 and other lentivirus infections. In the cross-species feline lentivirus infection discussed herein, the data are most consistent with the significant impact of the innate defense of host editing on a new virus.
The distribution of errors and the sites of G-to-A mutations were not uniform over the genome. The pol gene had the highest base pair frequency of A, the highest transition-to-transversion ratio, the most stop codons, and the highest frequency of sites associated with G-to-A changes and was the only gene in which the distribution of G-to-A mutations exhibited a nonrandom pattern. The action of host cytidine deaminases has only recently been described, and there are few reports to indicate differential susceptibility of sites on the lentivirus genome in natural (52) or experimental (59) infections. Indeed, we would not have detected this phenomenon if we had not evaluated the entire coding region of the PLV-1695 genome from these cats. Yet, the replication strategy of lentiviruses provides an explanation for susceptibility of pol. The process of reverse transcription converts the viral single-stranded RNA genome into double-stranded DNA. During synthesis of the minus-strand DNA, the viral RNA genome is degraded by RNase H activity in the RT but RNA primers are retained to initiate second-strand DNA production. There are at least two polypurine tracts in the lentivirus genome that provide the primer templates for second-strand synthesis. In both FIV and HIV-1, there is a polypurine tract near the 3′ long terminal repeat and a cPPT in the integrase coding region at the end of pol (6, 54). The preferred initiation site for second-strand synthesis in HIV-1 is the PPT. However, if the cPPT is used, the 3′ end of the genome including the end of pol, accessory genes, and env will be the first genes to be converted to double-stranded DNA. Thus, these genes should sustain the fewest mutations from a host cytidine deaminase because it acts only on single-stranded DNA (59). In support of this thesis, the region of UI that is devoid of G-to-A mutations (Fig. 4) lies directly 3′ of the cPPT described for FIV (54). This region is replete with the dinucleotide repeats that are the targets identified for human cytidine deaminases. Protection of the region at, and 3′ of, the cPPT has also been reported for a hypermutated HIV-1 group O isolate (52) and near the PPT of HIV-1 clone NL4-3 under experimental conditions (59). To complete second-strand synthesis, there is a strand transfer to the 5′ portion of the genome and gag and finally pol are duplexed. pol may be the most susceptible gene in the lentiviral genome because it is exposed as single-stranded DNA the longest. Thus, the patterns of G-to-A mutation observed in our PLV genomes during infection in the new cat host are in accord with those predicted from host cytidine deaminase editing. It is also noteworthy that, across the entire coding region of the PLV genome, five of the seven sites most strongly supported to be under positive selection fell in RT and were located at highly conserved areas of lentiviral RT. The effect of these substitutions on RT function will require further study. However, these data suggest that RT residues contacting the RNA template and in the nucleotide binding pocket may be important in the evolution of a lentivirus in a changing environment.
One of the objectives of this study was to determine if the virus evolutionary history could provide insight as to why infections initiated by intravenous routes were successful, while three cats infected orally-nasally were only transiently infected. Our analysis of selection profiles demonstrated that few sites in the PLV genome were under positive selection and most were located at functional positions in the RT-encoding regions of pol. Thus, there was no apparent signature of immune selection on the virus obtained from either i.v. or o.n. inoculated cats, which is consistent with immunological findings (47). Clearly there were no marked differences in the number or distribution of lethal mutations in the genome that correlated with viral fitness in these infections. However, the phylogenetic profiles may indicate that there was a similar trend towards virus extinction in both i.v. and o.n. inoculated cats that operated at different rates. For example, the sequences with the highest intrasample relatedness were obtained during the first 12 weeks of infection from the two cats with abortive infection, ON1 and ON2, and from the week 37 samples of two of the four cats evaluated with persistent viremia, IV3 and IV4. Proviral loads were 10-fold higher in IV1 to IV4 and ON4 at the onset of detectable infection cells and declined in all cats at week 11 by at least an order of magnitude, and infected cells were not detectable thereafter in ON1 and ON2 (47). Viral genomes sampled over time in cats with higher proviral loads would not appear to be related, even if they derived from the same infected cell, because the genomes of each progeny would be differentially edited during the next cycle of infection. The higher relatedness among sequences from cats with low viral loads may reflect the increased probability of sampling a remnant population of cells that do not contribute substantially to the virus pool. Thus, the additional burden of lethal mutations imposed on the virus from host editing enzymes increases the threshold of viable particles needed to establish a successful cross-species infection. These data demonstrate that, even under optimized experimental infection conditions, host innate defenses can provide important barriers to cross-species infections. However, our data also highlight the resilient nature of lentiviruses, which can be host specific and still adapt quickly to a changing environment.
Acknowledgments
This work was supported in part by National Institutes of Health Allergy and Infectious Disease grants AI 052055 and AI 054303, NSF grant 0346458, Morris Animal Foundation grant D01ZO-111, and a grant from the Wilburforce Foundation. Funding for the Molecular Computational Core Facility, Center for Structural and Functional Neuroscience, was derived from NIH P20 RR15583-01 and the NSF EPS-0091995.
We acknowledge summer students at the New Zealand Bioinformatics Institute for help with computer analysis, E. Burkala for critical reading of the manuscript, Jennifer Yactor for collection of cat blood samples, and several anonymous reviewers for helpful comments.
REFERENCES
- 1.Beer, B., J. Denner, C. R. Brown, S. Norley, J. zur Megede, C. Coulibaly, R. Plesker, S. Holzammer, M. Baier, V. M. Hirsch, and R. Kurth. 1998. Simian immunodeficiency virus of African green monkeys is apathogenic in the newborn natural host. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18:210-220. [DOI] [PubMed] [Google Scholar]
- 2.Biek, R., A. G. Rodrigo, D. Holley, A. Drummond, C. R. Anderson, Jr., H. A. Ross, and M. Poss. 2003. Epidemiology, genetic diversity, and evolution of endemic feline immunodeficiency virus in a population of wild cougars. J. Virol. 77:9578-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broussard, S. R., S. I. Staprans, R. White, E. M. Whitehead, M. B. Feinberg, and J. S. Allan. 2001. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter, M. A., and S. J. O'Brien. 1995. Coadaptation and immunodeficiency virus: lessons from the Felidae. Curr. Opin. Genet. Dev. 5:739-745. [DOI] [PubMed] [Google Scholar]
- 6.Charneau, P., and F. Clavel. 1991. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J. Virol. 65:2415-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 8.Daszak, P., A. A. Cunningham, and A. D. Hyatt. 2000. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287:443-449. [DOI] [PubMed] [Google Scholar]
- 9.Ding, J., K. Das, C. Tantillo, W. Zhang, A. D. Clark, Jr., S. Jessen, X. Lu, Y. Hsiou, A. Jacobo-Molina, K. Andries, et al. 1995. Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution. Structure 3:365-379. [DOI] [PubMed] [Google Scholar]
- 10.Escutenaire, S., and P. P. Pastoret. 2000. Hantavirus infections. Rev. Sci. Technol. 19:64-78. [DOI] [PubMed] [Google Scholar]
- 11.Goncalves, J., and M. Santa-Marta. 2004. HIV-1 Vif and APOBEC3G: multiple roads to one goal. Retrovirology 1:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goode, M., and A. G. Rodrigo. 2004. Using PEBBLE for the evolutionary analysis of serially sampled molecular sequences, p. 6.8.1-6.8.26. In A. D. Baxevanis (ed.), Current protocols in bioinformatics. Wiley, Hoboken, N.J. [DOI] [PubMed]
- 13.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 14.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, E. C. 2001. On the origin and evolution of the human immunodeficiency virus (HIV). Biol. Rev. Camb. Philos. Soc. 76:239-254. [DOI] [PubMed] [Google Scholar]
- 16.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 17.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, et al. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lochelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y. B. Kim, U. Truyen, U. Rosler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Munk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahy, B. W., and C. C. Brown. 2000. Emerging zoonoses: crossing the species barrier. Rev. Sci. Technol. 19:33-40. [DOI] [PubMed] [Google Scholar]
- 21.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 22.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 24.Martinez, M. A., J. P. Vartanian, and S. Wain-Hobson. 1994. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc. Natl. Acad. Sci. USA 91:11787-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, R. J., J. S. Cairns, S. Bridges, and N. Sarver. 2000. Human immunodeficiency virus and AIDS: insights from animal lentiviruses. J. Virol. 74:7187-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morin, T., F. Guiguen, B. A. Bouzar, S. Villet, T. Greenland, D. Grezel, F. Gounel, K. Gallay, C. Garnier, J. Durand, T. Alogninouwa, L. Mselli-Lakhal, J. F. Mornex, and Y. Chebloune. 2003. Clearance of a productive lentivirus infection in calves experimentally inoculated with caprine arthritis-encephalitis virus. J. Virol. 77:6430-6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mounts, A. W., H. Kaur, U. D. Parashar, T. G. Ksiazek, D. Cannon, J. T. Arokiasamy, L. J. Anderson, and M. S. Lye. 2001. A cohort study of health care workers to assess nosocomial transmissibility of Nipah virus, Malaysia, 1999. J. Infect. Dis. 183:810-813. [DOI] [PubMed] [Google Scholar]
- 28.Navarro, F., and N. R. Landau. 2004. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16:477-482. [DOI] [PubMed] [Google Scholar]
- 29.Nunes-Araujo, F. R., S. D. Nishioka, I. B. Ferreira, A. Suzuki, R. F. Bonito, and M. S. Ferreira. 1999. Absence of interhuman transmission of hantavirus pulmonary syndrome in Minas Gerais, Brazil: evidence from a serological survey. Clin. Infect. Dis. 29:1588-1589. [DOI] [PubMed] [Google Scholar]
- 30.Ogunbodede, E. O. 2004. HIV/AIDS situation in Africa. Int. Dent. J. 54:352-360. [PubMed] [Google Scholar]
- 31.Packer, C., S. Altizer, M. Appel, E. Brown, J. Martenson, S. J. O'Brien, M. Roelke-Parker, R. Hofmann-Lehmann, and H. Lutz. 1999. Viruses of the Serengeti: patterns of infection and mortality in African lions. J. Anim. Ecol. 68:1161-1178. [Google Scholar]
- 32.Parashar, U. D., L. M. Sunn, F. Ong, A. W. Mounts, M. T. Arif, T. G. Ksiazek, M. A. Kamaluddin, A. N. Mustafa, H. Kaur, L. M. Ding, G. Othman, H. M. Radzi, P. T. Kitsutani, P. C. Stockton, J. Arokiasamy, H. E. Gary, Jr., and L. J. Anderson. 2000. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. J. Infect. Dis. 181:1755-1759. [DOI] [PubMed] [Google Scholar]
- 33.Pini, N., S. Levis, G. Calderon, J. Ramirez, D. Bravo, E. Lozano, C. Ripoll, S. St. Jeor, T. G. Ksiazek, R. M. Barquez, and D. Enria. 2003. Hantavirus infection in humans and rodents, northwestern Argentina. Emerg. Infect. Dis. 9:1070-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 35.Pond, S. L., and S. D. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 36.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 37.Rambaut, A., and N. C. Grassly. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13:235-238. [DOI] [PubMed] [Google Scholar]
- 38.Rhodes, T., H. Wargo, and W. S. Hu. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrofelbauer, B., Q. Yu, and N. R. Landau. 2004. New insights into the role of Vif in HIV-1 replication. AIDS Rev. 6:34-39. [PubMed] [Google Scholar]
- 42.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 43.Shriner, D., A. G. Rodrigo, D. C. Nickle, and J. I. Mullins. 2004. Pervasive genomic recombination of HIV-1 in vivo. Genetics 167:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, J. H., D. L. Miller, R. A. Fouchier, M. A. Soares, K. W. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swofford, D. L. 1999. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4.3 ed. Sinauer Associates, Sunderland, Mass.
- 46.Takeuchi, H., S. Kao, E. Miyagi, M. A. Khan, A. Buckler-White, R. Plishka, and K. Strebel. 2005. Production of infectious SIVagm from human cells requires functional inactivation but not viral exclusion of human APOBEC3G. J. Biol. Chem. 280:375-382. [DOI] [PubMed] [Google Scholar]
- 47.Terwee, J. A., J. K. Yactor, K. S. Sondgeroth, and S. Vandewoude. 2005. Puma lentivirus is controlled in domestic cats after mucosal exposure in the absence of conventional indicators of immunity. J. Virol. 79:2797-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VandeWoude, S., C. A. Hageman, S. J. O'Brien, and E. A. Hoover. 2002. Nonpathogenic lion and puma lentiviruses impart resistance to superinfection by virulent feline immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 29:1-10. [DOI] [PubMed] [Google Scholar]
- 50.VandeWoude, S., S. O'Brien, and E. Hoover. 1997. Infectivity of lion and puma lentiviruses for domestic cats. J. Gen. Virol. 78:795-800. [DOI] [PubMed] [Google Scholar]
- 51.VandeWoude, S., S. J. O'Brien, K. Langelier, W. D. Hardy, J. P. Slattery, E. E. Zuckerman, and E. A. Hoover. 1997. Growth of lion and puma lentiviruses in domestic cat cells and comparisons with FIV. Virology 233:185-192. [DOI] [PubMed] [Google Scholar]
- 52.Vartanian, J. P., M. Henry, and S. Wain-Hobson. 2002. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J. Gen. Virol. 83:801-805. [DOI] [PubMed] [Google Scholar]
- 53.Vartanian, J. P., U. Plikat, M. Henry, R. Mahieux, L. Guillemot, A. Meyerhans, and S. Wain-Hobson. 1997. HIV genetic variation is directed and restricted by DNA precursor availability. J. Mol. Biol. 270:139-151. [DOI] [PubMed] [Google Scholar]
- 54.Whitwam, T., M. Peretz, and E. Poeschla. 2001. Identification of a central DNA flap in feline immunodeficiency virus. J. Virol. 75:9407-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfe, N. D., W. M. Switzer, J. K. Carr, V. B. Bhullar, V. Shanmugam, U. Tamoufe, A. T. Prosser, J. N. Torimiro, A. Wright, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. Heneine. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932-937. [DOI] [PubMed] [Google Scholar]
- 56.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 58.Yang, Z., W. S. Wong, and R. Nielsen. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107-1118. [DOI] [PubMed] [Google Scholar]
- 59.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]