Abstract
The Arabidopsis genome contains many gene families that are not found in the animal kingdom. One of these is the multidrug and toxic compound extrusion (MATE) family, which has homology with bacterial efflux transporters. Arabidopsis has at least 54 members of this family, which often are found in tandem repeats. Analysis of ALF5, one member of this Arabidopsis family, suggests that its function is required for protection of the roots from inhibitory compounds. Loss of ALF5 function results in the sensitivity of the root to a number of compounds, including a contaminant of commercial agar. Moreover, expression of the Arabidopsis ALF5 cDNA in yeast confers resistance to tetramethylammonium. These phenotypes are consistent with a role for ALF5 as an efflux transporter. Both transcriptional and translational fusions of ALF5 to the β-glucuronidase reporter gene show that ALF5 is expressed strongly in the root epidermis, a tissue in direct contact with the external environment. The distinct requirement for ALF5 function is remarkable because of the large number of MATE gene family members in Arabidopsis, one of which is adjacent to ALF5 and 83% identical to ALF5 at the amino acid level.
INTRODUCTION
Plants grow in varied environments in which their roots are exposed to toxic or inhibitory chemical substances (Wink, 1997). Toxins can be pollutants, herbicides, or the xenobiotic products of neighboring plants or endemic microorganisms. Such exogenous toxins are detoxified and removed from the cytoplasm of plant cells in a manner related to the vacuolar storage of endogenous metabolites (Wink, 1997; Dixon et al., 1998).
Both toxins and secondary metabolites are removed from the plant cell cytoplasm and stored in the vacuole (Wink, 1997). In the best characterized mode of vacuolar sequestration, the toxin or metabolite is conjugated to glutathione, and the glutathione-tagged compound is then transported to the vacuole (Sandermann, 1992). Accumulation of anthocyanin pigment in the maize vacuole requires the activity of a glutathione S-transferase encoded by the Bronze-2 gene (Marrs et al., 1995). This system in plants is analogous to both the excretion of glutathione conjugates from mammalian cells and the vacuolar sequestration of the Cd(II)(glutathione)2 complex in yeast. Arabidopsis AtMRP, which is capable of vacuolar concentration of glutathione-conjugated compounds, was identified on the basis of homology with the ATPase-driven efflux pumps responsible for glutathione conjugate transport in mammalian cells (MRP1) and yeast (YCF1) (Lu et al., 1997; Sanchez-Fernandez et al., 1998; Tommasini et al., 1998).
The sequestration of metabolites in the vacuole of plants and yeast also is mediated by active secondary transport systems (Ohsumi and Anraku, 1981; Wink, 1997) that require the vacuolar H+-ATPase and vacuolar pyrophosphatase for maintenance of a proton gradient across the tonoplast (Luettge and Ratajczak, 1997; Zhen et al., 1997). The genes encoding the vacuolar H+ antiporters for metal cations such as calcium (CAX1) and sodium (AtNHX1) from Arabidopsis facilitate the vacuolar sequestration of calcium and sodium ions, respectively (Hirschi et al., 1996; Apse et al., 1999; Gaxiola et al., 1999; Hirschi, 1999). The alkaloids reticuline and scoulerine and the malonylated derivative of 1-aminocyclopropane-1-carboxylic acid, the precursor of the plant hormone ethylene, also are concentrated in plant vacuoles. The in vitro uptake behavior of these metabolites in isolated plant vacuoles suggests that their accumulation is facilitated by proton antiport systems (Deus-Neumann and Zenk, 1986; Bouzayen et al., 1989). In yeast, the fact that resistance to the toxins staurosporine, vanadate, and hygromycin is dependent on vacuolar acidification suggests that these toxins are exported from the cytosol via vacuolar secondary transporters (Yoshida and Anraku, 2000).
A family of putative secondary transporters unique to plants and microbes has been termed the multidrug and toxic compound extrusion (MATE) family (Brown et al., 1998). Hydropathy analysis suggests that proteins of the MATE family have a common topology consisting of 12 transmembrane (TM) domains. The MATE family is divided into three groups on the basis of sequence relatedness. Members of the two more related groups are found in prokaryotes, whereas the third group is composed exclusively of proteins from eukaryotes. Members of this third group of MATE proteins are present in the genomes of both Saccharomyces cerevisiae and Schizosaccharomyces pombe and are abundant in the Arabidopsis genome. No MATE gene sequences have been identified in the genomic sequence of any member of the animal kingdom.
On the basis of the analysis of two bacterial family members, NorM and YdhE (Morita et al., 1998), the MATE family is thought to encode efflux pumps. Expression of the Vibrio parahaemolyticus norM gene in Escherichia coli increases resistance to the antibiotic norfloxacin as well as to another quinoline antibiotic, ciprofloxacin, and to the structurally unrelated antimicrobial agents ethidium, streptomycin, and kanamycin. Expression of ydhE, which encodes an E. coli ortholog of NorM, from a multicopy plasmid confers on E. coli resistance to a group of antimicrobial agents that are similar to but not overlapping with those to which NorM confers resistance. The significant reduction in norfloxacin accumulation in E. coli cells attributed to NorM expression can be eliminated rapidly by dissipation of the proton gradient across the plasma membrane, suggesting that NorM, and presumably other MATE protein family members, functions as a proton antiporter (Morita et al., 1998).
There is little evidence for the function of the MATE genes in eukaryotes beyond the sequence similarities with other family members. The function of the yeast ERC1 MATE gene has been inferred from the ethionine resistance phenotype conferred when ERC1 is expressed from a multicopy plasmid (Shiomi et al., 1988). The resistance to ethionine, a toxic analog of methionine, is thought to be a consequence of the sequestration of S-adenosylethionine in the vacuole. This explanation is based on the observation in yeast that external methionine is converted to S-adenosylmethionine (SAM) and stored in the vacuole (Thomas and Surdin-Kerjan, 1997). In support of this model, strains overexpressing ERC1 in the presence of exogenous methionine or ethionine accumulate high levels of SAM or S-adenosylethionine, respectively, in the vacuole (Shiomi et al., 1991). Early studies describe a saturable transport system capable of concentrating SAM in isolated yeast vacuoles, which could reflect ERC1 function (Schwencke and de Robichon-Szulmajster, 1976).
Here we describe alf5, a mutant that lacks the function of one of the MATE gene family members. The Arabidopsis alf5 loss-of-function phenotype results in sensitivity to compounds that fail to affect the growth and development of wild-type plants. The phenotype of the alf5 mutant, the localization of ALF5 expression, and the function of ALF5 in yeast imply a direct role for ALF5 in the extrusion of inhibitors in the Arabidopsis root and raise the possibility of the use of it and related MATE genes to engineer plants that are resistant to environmental toxins.
RESULTS
Root Growth of the alf5 Mutant Is Inhibited by Bacto Agar
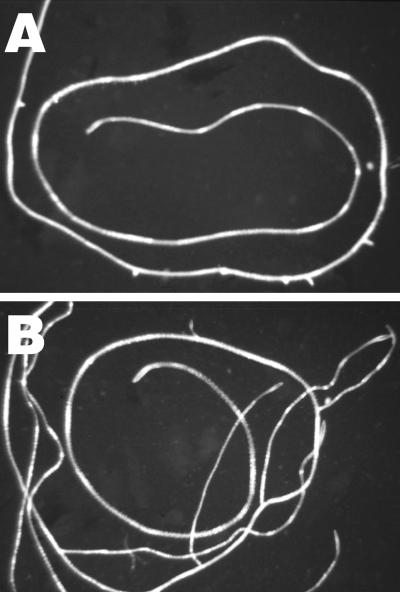
alf5 was identified on the basis of its aberrant lateral root formation phenotype observed in seedlings grown on Petri plates. The newly formed lateral roots of the mutant ceased to elongate soon after emergence from the primary root (Figure 1A), whereas wild-type lateral roots elongated normally (Figure 1B). No obvious morphological defects in the cellular organization of the arrested alf5 lateral root meristems were observed with light microscopy.
Figure 1.
Primary Roots Grown on Plant Nutrient Medium Solidified with Bacto Agar.
(A) alf5.
(B) Wild-type.
The primary roots were coiled on an agar surface for ease of photography.
The defect in alf5 lateral root growth was dependent on a water-soluble compound present in Bacto agar. When grown in soil, the appearance of the alf5 plants, including the growth of lateral roots, was normal, suggesting that the inhibition of lateral roots was a response to culturing on nutrient medium in solidified agar. If the alf5 plants were grown on medium with Bacto agar that had been washed extensively with water or on a purer commercial agar (Noble), the alf5 plants had no defect in lateral root growth and were indistinguishable from wild-type plants. Addition of the Bacto agar supernatant wash to Noble agar plates resulted in the arrest of lateral root growth in alf5 plants. None of the components of the synthetic medium conferred alf5-specific growth inhibition. Therefore, the alf5 mutation conferred sensitivity to a soluble contaminant present in Bacto agar.
alf5 Root Growth Is Hypersensitive to Commercial Polyvinylpyrrolidone and Pyrrolidinone
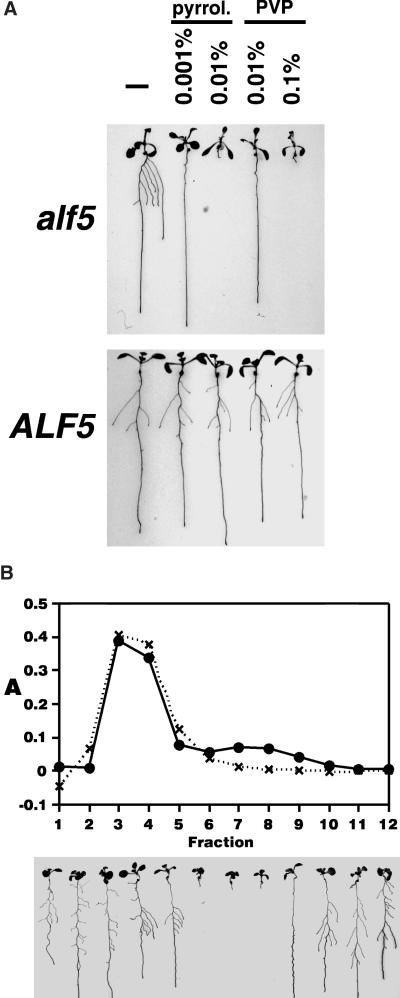
The growth of alf5 roots also was sensitive to commercial polyvinylpyrrolidone (PVP) at concentrations that had no effect on the growth of wild-type roots. When added to Noble agar at a concentration of 0.01% (w/v), PVP strongly inhibited alf5 lateral root growth (Figure 2A). Moreover, all root growth of the alf5 mutant, including that of the primary root, was inhibited at a 10-fold higher concentration (Figure 2A). The root growth of wild-type plants was not affected by either concentration of PVP.
Figure 2.
Commercial PVP and Pyrrolidinone Inhibit alf5 Root Growth.
(A) alf5 (top) and ALF5 (bottom) plants grown on Noble agar–solidified nutrient medium alone (left) or containing either pyrrolidinone (pyrrol.) or PVP (w/v).
(B) Gel filtration chromatography of PVP. A 5% (w/v) PVP sample was fractionated by gel filtration with 50 mM sodium phosphate, pH 7.0, as an eluant. The UV light absorbance of each fraction was measured at 240 nm (closed circles), and the UV light absorbance of the PVP-quercetin complex activity of each fraction was measured at 350 nm (hatches). Below each chromatogram fraction is the response of alf5 seed sown on PN Noble agar (all assays are described in Methods) containing that fraction. The root systems were stained with safranin O dye and splayed immediately before photography.
The inhibition of alf5 roots in the presence of commercial PVP was caused by a minor contaminant in commercial PVP and not by the predominant polymeric material. A 5% solution of 10-kD PVP was fractionated on a desalting column with 50 mM sodium phosphate, (pH 7.0), as the eluant. The eluted peak of alf5-inhibitory activity was separated clearly from the predominant peak of UV light absorbance and the peak of PVP-quercetin complex activity (Figure 2B). The elution of the alf5-inhibitory activity in fractions that followed the elution of the high-molecular-mass PVP (Figure 2B) implied that the alf5-inhibitory substance had a relatively low molecular mass.
Some likely chemical contaminants of PVP were not the alf5-inhibitory substance. There is no alf5-specific root growth inhibition by 1-vinyl-2-pyrrolidinone, γ-aminobutyric acid, γ-hydroxybutyric acid lactone, ammonium chloride, or N-methylpyrrolidinone. Commercial pyrrolidinone inhibited alf5 lateral root growth at 0.001% and both primary and lateral root growth at 0.01%. However, the alf5-inhibitory activity could be separated from pyrrolidinone. Pyrrolidinone eluted from the desalting column with water as the eluant, whereas the alf5-inhibitory activity failed to elute from the column and was not present in any fractions containing pyrrolidinone.
alf5 Is a Recessive Mutation on Chromosome 3
The alf5 mutation behaved as a single Mendelian recessive trait located on chromosome 3 between the visible markers gl1 and axr2 (Wilson et al., 1990). The triply heterozygous GL1/gl1 ALF5/alf5 axr2/AXR2 mutant was constructed by crossing the gl1/gl1 alf5/alf5 mutant and the axr2/axr2 dominant mutant. Segregation of these alleles in the recombinant offspring placed alf5 between gl1 and axr2, 11 cM from gl1 and 2 cM from axr2.
Positional Cloning of the alf5 Locus
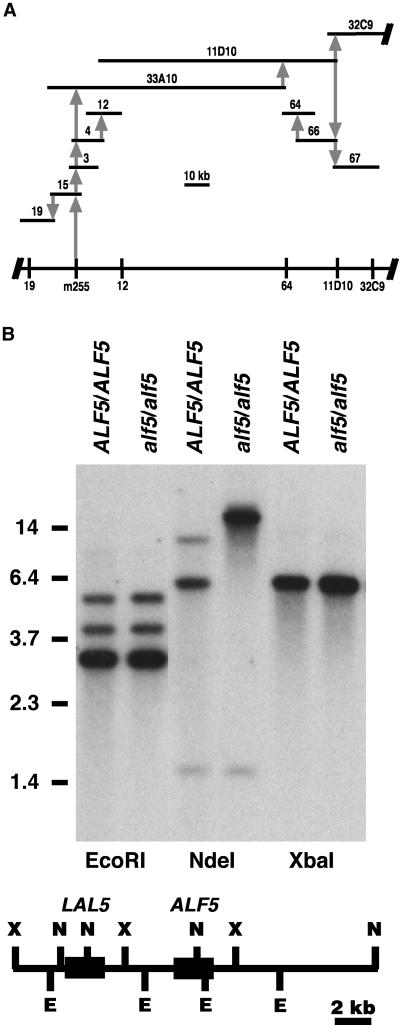
The alf5 locus was contained within a contiguous genomic sequence on chromosome 3 by a chromosome walk. We created four cleaved-amplified polymorphic sequence (CAPS) markers from two cloned genes (ArLIM15 and ABI3) and two restriction fragment length polymorphism markers (g4711 and m255) located approximately between gl1 and axr2 (Wilson et al., 1990; Giraudat et al., 1992; Klimyuk and Jones, 1997). The m255 CAPS marker was found to be tightly linked to the alf5 mutation and was used to initiate a physical map of overlapping genomic fragments, both λ and bacterial artificial chromosome (BAC) library clones. The assembled physical map depicted in Figure 3A extends over 200 kb. During construction of the physical map, several CAPS markers (Figure 3A) that span 135 kb of this contig were created to locate alf5.
Figure 3.
Positional Cloning of alf5.
(A) Physical genomic map of the alf5 region. The span of each Arabidopsis genomic clone (horizontal bars) is proportional to the 10-kb scale, and its position is relative to its genome coverage. Clones 33A10, 11D10, and 32C9 are Texas A&M University BACs; clones 3, 4, 12, 15, 19, 64, 66, or 67 are from λ libraries. The positions of CAPS makers (λ19, m255, λ12, λ64, 11D10, and 32C9) along chromosome 3 are depicted at the bottom. The initial association of each cloned segment to the contig is represented by a gray vertical arrow: the sequence used as a hybridization probe is the base of the arrow, and the sequence identified by the hybridization probe is at the arrow point.
(B) ALF5 and alf5 genomic DNA. Genomic DNA of ALF5 and alf5 plant lines was digested with the restriction enzymes EcoRI, NdeI, and XbaI. The DNA gel blot was hybridized to the ALF5 cDNA. The mobilities of standard restriction fragments are indicated at left with their sizes in kb. At bottom, the expected restriction map was compiled from the BAC MDB19 genomic sequence. E, EcoRI; N, NdeI; X, XbaI. Black boxes represent the ALF5 and LAL5 coding regions.
An F2 mapping population generated from an alf5 Columbia (Col) ecotype and a Wassilewskija (Ws) ecotype hybrid cross gave recombinant offspring whose breakpoints confined the alf5 mutation to a 35-kb interval. It should be noted that a similar cross of alf5 (Col) × Landsberg erecta failed to yield any (0 of 930) alf5/alf5 plants with a crossover between the alf5 mutation and any marker in our physical contig (separated by as much as 135 kb). After scoring 300 recombinant F2 plants from the Col × Ws cross, the genotype of a single alf5/alf5 plant was found to be heterozygous (Col/Ws) at the λ64 CAPS marker and a single alf5/alf5 plant was found to be heterozygous at the 32C9 CAPS marker. Three genomic clones, λ64, λ66, and λ67, spanned the junction of the λ64 and 32C9 CAPS markers.
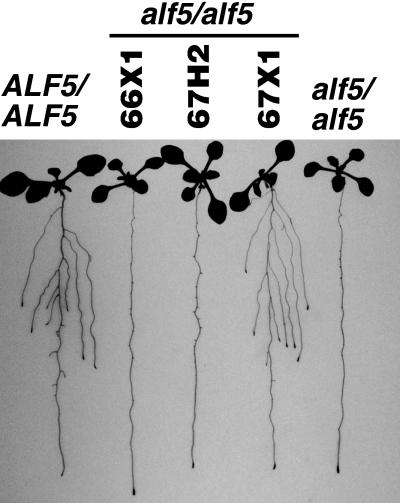
alf5 plants were transformed with overlapping fragments of λ64, λ66, and λ67 genomic clones to test the ability of each of these segments to suppress the Alf5− phenotype. Of three transgenic plants (66X1, 67H2, and 67X1) shown in Figure 4, only the plant harboring the 67X1 transgene suppressed the alf5 defect. Moreover, in subsequent generations, the Alf5+ phenotype cosegregated with the kanamycin resistance marker used to select for the T-DNA transformation. DNA gel blot analysis confirmed that multiple copies of the 67X1 transgene are present in the transgenic alf5 plant.
Figure 4.
Complementation of the alf5 Mutation.
Plants were sown on a Noble agar plate supplemented with 0.02% PVP and grown for 10 days. ALF5/ALF5 (left) and alf5/alf5 (right) plants flank three alf5/alf5 plants, each harboring a transgene (66X1, 67H2, or 67X1). The roots were dyed and splayed before photography.
The alf5 Mutant Results from a Deletion of 29 bp
The alf5 allele has a 29-bp deletion within a coding sequence identified in the genomic region covered by the 67X1 transgene, shown in Figure 5. After sequencing the 6.1-kb 67X1 genomic clone, only a single coding sequence within 67X1 could be inferred by homology with available GenBank sequence entries. All clones isolated from a cDNA library by hybridization to the 67X1 transgene cross-hybridized to our largest ALF5 cDNA clone, suggesting that they were representatives of a single gene transcript in 67X1. The predicted gene, MDB19.4, within the genomic sequence (GenBank accession number AB023036) submitted by the Arabidopsis Genome Initiative (AGI) was identical to ALF5. MDB19.4/ALF5 is the only open reading frame (ORF) that appears in the AGI sequence annotation of the region covered by 67X1. From polymerase chain reaction (PCR) sequencing, we identified a 29-bp deletion in the alf5 mutant not found in a related wild-type ALF5 reference plant (see Methods). An NdeI endonuclease restriction site was present within the 29-bp sequence deleted in the alf5 mutant (Figure 5). The resulting NdeI restriction fragment length polymorphism was detected by DNA gel blot analysis (Figure 3B), confirming the sequence difference between alf5 and ALF5 plants. The deletion in alf5 likely results in the complete loss of ALF5 function, because the deletion disrupts the reading frame before half of the conserved coding sequence can be translated.
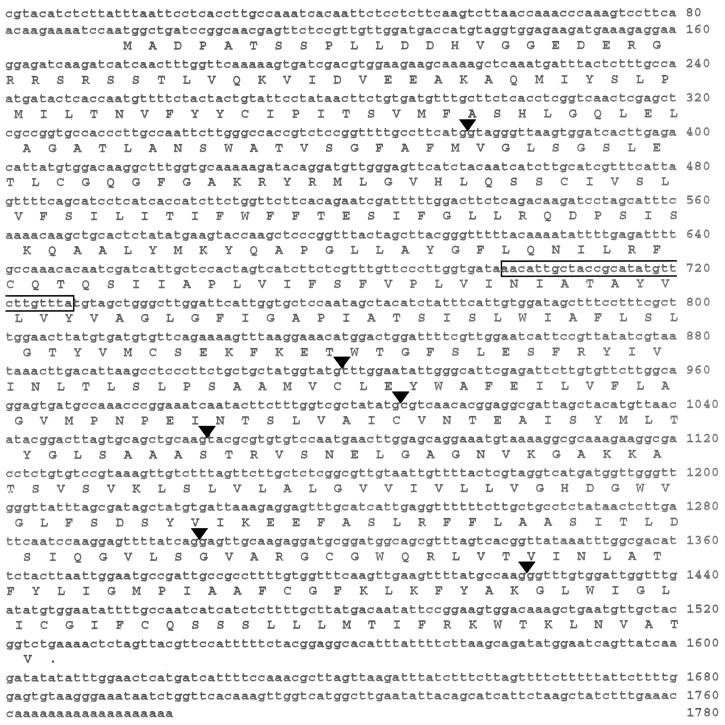
Figure 5.
cDNA of the ALF5 Transcript.
The 1780-nucleotide sequence is in lowercase letters, with the number of the rightmost nucleotide given at the right of each row. The amino acid sequence of the largest ORF is given below the nucleotide sequence in uppercase letters. The single letter designation for an amino acid is centered below the corresponding codon. Triangles above the nucleotide sequence indicate the positions of the six introns in the genomic sequence. The box indicates the 29-bp sequence deleted from alf5.
ALF5 Is a Member of the MATE Protein Family
ALF5 encodes a member of the MATE family of integral membrane proteins. An alignment of the deduced polypeptide sequence of ALF5 with related MATE protein sequences is shown in Figure 6. Three of these MATE sequences are derived from annotations of genomic sequences produced by the AGI. For each MATE polypeptide in the alignment, we predicted the location of TM domains using a computer algorithm (see Methods). As many as 12 putative TM domains in each MATE polypeptide localize approximately to equivalent regions in each MATE member, shown in Figure 6. We found that this algorithm (and others) failed to consistently identify the fourth and tenth potential TM domains. The deletion in the alf5 mutant terminates the normal reading frame of ALF5 within the middle of the fifth TM domain. Homology searches (using BLAST) with the nearly completed Arabidopsis genomic sequence in GenBank identified at least 54 possible MATE ORFs.
Figure 6.
Amino Acid Alignment of ALF5 with Other MATE Family Sequences.
The ALF5 sequence is shown aligned with the sequences of Arabidopsis F13P17.20, Arabidopsis F7H19.220, Arabidopsis T6A23.29, Saccharomyces cerevisiae Erc1p, and Vibrio parahaemolyticus NorM. Amino acids are indicated by the standard single letter designation, and dashes indicate gaps. Residues are highlighted in white letters on black if three or more MATE sequences have an identical residue at the aligned position. Gray shading indicates the positions of 12 potential transmembrane domains, TM1 through TM12.
LAL5, a Duplicate Copy of ALF5 Adjacent to ALF5
An ORF designated Like ALF5 (LAL5) that shares 88% nucleotide identity to ALF5 and encodes a polypeptide with 83% identity to ALF5 is located immediately downstream of ALF5. LAL5 is equivalent to MBD19.3 in AGI sequence annotation. LAL5 and ALF5 have the same orientation on chromosome 3, and no intervening coding sequence is apparent between them. The predicted exon/intron structures of ALF5 and LAL5 are virtually identical. Although we do not know whether LAL5 is transcribed, the putative exon sequences of ALF5 and LAL5 display more conservation (between 85 and 90% nucleotide identity) than the noncoding intervening intron sequences (between 45 and 71% identity). The gross structure of the LAL5 gene is intact in the alf5 mutant, as determined by comparison of DNA gel blot hybridizations with restriction fragment lengths estimated from AGI sequencing (Figure 3B).
Yeast Strains Expressing ALF5 Are Resistant to Tetramethylammonium Chloride
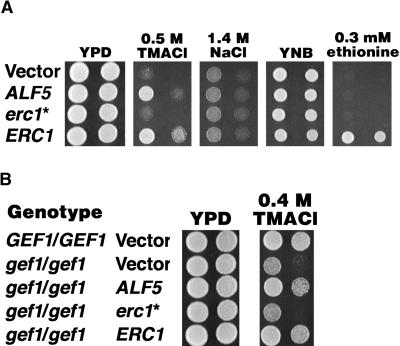
Because the ALF5 amino acid sequence and the sensitivity of alf5 mutants to inhibitors suggest that this gene is related to membrane extrusion pumps, we expressed ALF5 in yeast to determine whether it altered sensitivity to toxic cations. Sensitivity to tetramethylammonium chloride (TMACl) has been used to assess the ability of yeast cells to sequester the toxic cation tetramethylammonium (Ferrando et al., 1995; Gaxiola et al., 1998). To permit high expression of ALF5 in yeast, we fused the Arabidopsis ALF5 cDNA or S. cerevisiae ERC1 coding sequence to the yeast ADH1 transcriptional promoter and terminator sequences on a multicopy plasmid. As shown in Figure 7A, a yeast strain expressing either ALF5 or ERC1 is resistant to growth-inhibitory concentrations of TMACl, compared with a strain harboring the control plasmid or expressing the truncated erc1 allele (see Methods). However, the yeast strain expressing ALF5 or ERC1 failed to suppress the growth-inhibitory effects of either 1.4 M NaCl (Figure 7A) or 1.7 M KCl (data not shown).
Figure 7.
High Copy Expression of ADH1::ALF5 in Yeast Confers Resistance to TMACl.
(A) The growth of serial dilutions of yeast on rich medium (YPD) without added salt or rich medium containing 0.5 M TMACl or 1.4 M NaCl, synthetic medium alone (YNB), or synthetic medium containing 0.3 mM ethionine. Each strain has a high copy plasmid, either the empty vector or a coding sequence (the ALF5 cDNA or S. cerevisiae erc1 or ERC1), expressed by the S. cerevisiae ADH1 promoter and terminator sequences. The erc1 gene (*) in the yeast strain designated as wild type has a frameshift that renders it nonfunctional. We corrected the frame to make it functional and designated it ERC1 (see Methods). YNB, synthetic yeast nitrogen base plus 2% dextrose and 200 μM uracil.
(B) The growth of serial dilutions of the diploid gef1/gef1 yeast on rich medium alone (YPD) or rich medium containing 0.4 M TMACl. Each strain has a high copy plasmid as in (A).
ALF5 expression also suppressed the enhanced TMACl sensitivity of the diploid gef1/gef1 strain (Figure 7B). The S. cerevisiae GEF1 gene encodes a chloride channel family member. Diploid gef1/gef1 strains have increased sensitivity to various cations, including tetramethylammonium (Ferrando et al., 1995; Gaxiola et al., 1998). As shown in Figure 7B, the growth of the gef1/gef1 mutant was inhibited by concentrations of TMACl that did not appreciably affect the growth of the wild-type GEF1/GEF1 strain. Expression of either ALF5 or ERC1 enhanced the growth of the gef1/gef1 mutant strain on 0.4 M TMACl. Thus, the TMACl resistance conferred by ALF5 is independent of the intrinsic GEF1 yeast cation detoxification pathway.
Yeast strains that express high levels of ALF5 did not show resistance to ethionine. In contrast, we found that high copy expression of ERC1, the yeast ortholog of ALF5, conferred significant resistance to 0.3 mM ethionine (Figure 7A), as reported previously (Shiomi et al., 1988).
ALF5::GUS Fusions Are Expressed in the Root Epidermis
Analysis of transgenic plants containing ALF5 fused to the β-glucuronidase (GUS) reporter gene indicates that this gene is highly expressed in the root epidermis and cortex. The same pattern of ALF5::GUS expression was obtained from plants transformed with either a transcriptional or a translational fusion of ALF5. In the transcriptional fusion, the complete intergenic region upstream of ALF5, presumably including the 5′ transcriptional promoter, was fused to the GUS reporter gene. In the translational fusion, the first 467 amino acids encoded by ALF5, including all 12 putative TM domains, were fused in frame to the GUS reporter gene. This fusion removed just 10 C-terminal nonconserved residues from ALF5. Because this construct is derived from the complementing 67X1 transgene, the translational fusion retained the same 5′ untranslated region that provided expression for complementation of the alf5 defect.
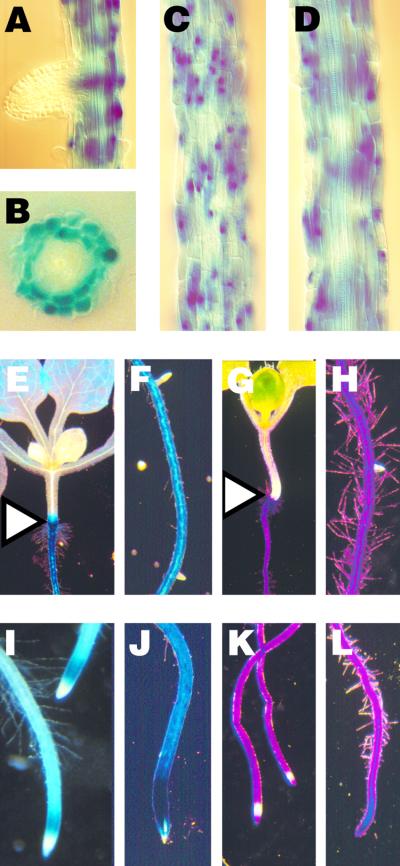
The strongest ALF5::GUS reporter gene expression, as shown in Figure 8, was detected in the outer root layers (the epidermis and cortex; Figures 8B, 8C, and 8D). The pattern of expression at the root apex appeared to be contingent on the age of the root. No GUS activity was observed in newly formed lateral root primordia (Figures 8A and 8F) The strongest expression was observed consistently in the elongation zone of young roots (Figure 8I), but activity was still absent from the meristematic region. In older roots, the GUS activity spread into the meristematic region so that there was strong expression over the entire root tip (Figure 8J). The hypocotyl/root junction shown in Figure 8E was clearly delineated by the presence of GUS activity in the root and collet and its absence in the hypocotyl.
Figure 8.
ALF5::GUS Reporter Gene Expression and Dye-Stained Roots.
(A), (B), (C), (D), (E), (F), (I), and (J) Reporter gene expression of the ALF5::GUS translational fusion.
(B) A cross-section of the root shows that ALF5::GUS is strongly expressed in the outer epidermal layer and cortex. Focus on the surface image of the root (C) or on the central root vasculature (D) shows that intense ALF5::GUS staining is present in the outer cell layers. (E) ALF5::GUS expression in the root clearly delineates the hypocotyl/root junction (triangle). (I) ALF5::GUS expression is excluded from young meristematic regions, including the lateral root primordia (F). (J) Strong expression at the meristem of older roots appears to be on the surface (the tear in the root tip reveals underlying white, nonexpressing tissue).
(G), (H), (K), and (L) Arabidopsis seedlings stained with toluidine blue O dye.
(G) The hypocotyl/root junction (triangle) is delineated by dye staining of the root and collet but no staining of the hypocotyl. (K) Dye is excluded from young meristematic regions, including the lateral root primordia (H). (L) The entire tip of older roots is darkly stained by dye.
Coincident Staining Pattern of Dyes and ALF5::GUS
The brief dye staining of Arabidopsis roots has a pattern strikingly similar to that of GUS activity encoded by the ALF5::GUS transgene. Lipophilic basic dyes, such as toluidine blue O and safranin O, readily stain Arabidopsis roots. Although entire plants were submerged completely in dye solution, toluidine blue O readily stained only the root epidermis and not the aerial parts of the plant, as shown in Figure 8G. Because dye is not absorbed by the hypocotyl as it is by the root, the hypocotyl/root junction was demarcated by dye staining similar to the ALF5::GUS staining shown in Figure 8A. Dye also was restricted from the newly formed lateral root primordium (Figure 8H; cf. Figure 8F). The lateral roots that elongated from the primary root (Figure 8K) were readily stained in the elongation zone and along its mature length. The restriction of stain retreated in the meristematic region in more mature roots until the root tip was covered completely by the most intense dye staining (Figure 8L).
DISCUSSION
We found that ALF5 protects roots from growth inhibition by a number of compounds contaminating commercial preparations. The alf5 loss-of-function allele revealed the sensitivity of Arabidopsis roots to a contaminant found in commercial Bacto agar, PVP, and pyrrolidinone. In the absence of these compounds, the growth of alf5 plants was indistinguishable from the growth of wild-type plants.
Our data suggest that ALF5 plays a direct role in either the vacuolar sequestration or the cellular efflux of these toxins. The cloned ALF5 gene encodes a member of the MATE protein family, which is represented by the bacterial multidrug efflux transporters NorM and YdhE. The MATE proteins share sequence homology and a common topology. The presence of 12 potential TM domains is typical of the architecture observed in integral membrane permeases (Paulsen et al., 1996). High copy expression of the ALF5 gene in yeast confers a phenotype similar to overexpression of ERC1, a yeast MATE family member. In both cases, the overexpressing strains have increased resistance to TMACl.
The spatial expression of ALF5 in Arabidopsis roots is consistent with a direct role in toxin resistance. The ALF5::GUS reporter gene is expressed maximally in the root epidermis and cortex, the tissues that come into immediate contact with the soil and the soluble toxic compounds within it. The localization of ALF5::GUS expression is coincident with the susceptibility of root cells to dye staining. This localization is not a result of limited penetration of 5-bromo- 4-chloro-3-indoxyl-b-D-glucuronic acid because the same protocol used to visualize ALF5::GUS localizes another reporter construct (SMT1::GUS) to the root vasculature and meristem (Diener et al., 2000). The shoot and the hypocotyl fail to stain with dyes, probably because they are covered with a hydrophobic cuticle (Wink, 1997).
The mechanism of ALF5-mediated toxin resistance is unresolved, although the sequence similarity of the Alf5 protein to other efflux pumps suggests that the plant protein either extrudes compounds from the cell or sequesters them in the vacuole. Localization of ALF5 to either the plasma membrane or the vacuole would reveal its site of action, and identification of the toxic compounds would permit a direct biochemical test of its function in transport.
The Arabidopsis genome contains at least 54 possible MATE genes. The Arabidopsis Membrane Protein Library (http://www.cbs.umn.edu/arabidopsis/) catalogs 2018 coding sequences with 4 or more putative TM domains. The MATE sequences compiled by the Arabidopsis Membrane Protein Library are estimated to have between 8 and 13 TM domains. The MATE sequences account for more than 6% of the Arabidopsis sequences with between 8 and 13 TM domains.
The tandem arrangement of the ALF5 and LAL5 genes is representative of the organization of some MATE genes. Twenty-five of 54 Arabidopsis MATE sequences (46%) are constituents of tandem arrays. These arrays are clusters of two to five ORFs with a common orientation on the chromosome. The higher degree of homology within these clusters, compared with MATE genes elsewhere in the Arabidopsis genome, suggests that the arrays arose by gene duplication. The tandem duplication of genes provides a mechanism for amplifying gene expression. Analysis of bacteria selected for increased antibiotic resistance has shown that they often contain tandem duplicated copies of an antibiotic resistance gene that is present in low copy number in unselected bacteria (Wiebauer et al., 1981). By analogy with bacteria, the tandem repetition of the MATE genes could provide a mechanism for increasing the resistance of plants to toxic compounds in the soil.
If LAL5 is functional in the root, there are at least two explanations for our ability to detect a phenotype for alf5. One possibility is that the two MATE genes have distinct activities despite their close relatedness. Diversity of function within an array appears to explain the phenotypes of mutations in a single member of the leucine-rich repeat resistance genes, which are present in tandem arrays similar to those of the MATE genes (Ellis and Jones, 1998). Despite the apparent gene duplications, the loss-of-function allele of an individual member of this resistance gene array can generate sensitivity to a specific pathogen. According to this interpretation, the phenotypes of the alf5 mutant would be a consequence of the loss of those functions not shared with LAL5. A second possibility is that ALF5 and LAL5 genes have an identical function. If this is the case, then the phenotypes of the alf5 mutant could result from a reduction in the level of that function. Alternately, ALF5 and LAL5 could be expressed and required in different tissues. The resolution of these possibilities will require a detailed analysis of LAL5, including the isolation of strains lacking a functional LAL5, so that the phenotypes of lal5 and lal5 alf5 strains can be compared.
The identification of ALF5 as a component of the detoxification system in roots suggests the possibility of engineering plants that overexpress particular MATE proteins to achieve useful phenotypes. For example, these transgenic plants might grow in soils containing chemicals that would inhibit the growth of wild-type plants.
METHODS
Chemicals
Unless indicated otherwise, chemicals were purchased from Sigma/Aldrich: polyvinylpyrrolidone (PVP) with average molecular masses of 10, 40, or 360 kD (PVP-10, PVP-40, or PVP-360), PVP with an average molecular mass of 10 kD (85,645-2), Kollidon 17 PF or 25 (BASF Corp., Ludwigshafen, Germany; 010750-01 or 009,961), 1-vinyl-2-pyrrolidinone (V340-9), 1-ethyl-2-pyrrolidinone (14,635-8), 2-imidazolidone (I-60-1), 2-pyrrolidinone (P7,437-0), γ-amino-n-butyric acid (A5835), and γ-hydroxybutyric acid lactone (H7629). The catlogue number for each chemical appears in parentheses.
Plant Lines and Growth Conditions
Plants were grown under continuous white light. Seed were sterilized by dispersion in 10% bleach and 0.01% Triton X-100 detergent for 10 min, washed several times with water, and sown on aseptic agar plates. For as long as 3 weeks after sowing, plants were grown in growth chambers on a chemically defined plant nutrient (PN) agar medium: 5 mM KNO3, 2.5 mM potassium phosphate, pH 5.5, 2 mM Ca(NO3)2, 2 mM MgSO4, 50 μM Fe·EDTA plus micronutrients, and 7.5 g/L Bacto agar or Noble agar (Difco Laboratories, Detroit, MI) with or without a 0.5% sugar supplement, either glucose (PNG) or sucrose (PNS). Micronutrient stock (×1000) was 70 mM H3BO3, 14 mM MnCl2, 0.5 mM CuSO4, 1 mM ZnSO4, 0.2 mM Na2MoO4, 10 mM NaCl, and 0.01 mM CoCl2. Seed were selected for kanamycin resistance by sowing on PNG plates supplemented with 20 mg/L kanamycin A. Plants also were sown on soil, or 1- to 2-week-old plantlets were transplanted to soil (Metro-Mix 200 Growing Medium, Scotts-Sierra Horticultural Products, Marysville, OH), and grown in a greenhouse with lamp-supplemented sunlight for continuous illumination. Seed were stratified for 1 to 3 days at 4°C. axr2 seed were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). alf5 was a spontaneous recessive mutation segregating among the progeny of a gl1/gl1 trp1-100/trp1-100 parent in the Columbia (Col) ecotype (Rose et al., 1992). Comparison of mutant (alf5) and wild-type (ALF5) sequences was performed with lines homozygous for either ALF5/ALF5 (BF4.9) or alf5/alf5 (BF4.8) but derived from the same heterozygous parent.
Plant Transformation
Plant transformation was mediated by the Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998). GV3101 was transformed by electroporation (Mattanovich et al., 1989). The pBIN19-based binary plasmids (Clontech, Palo Alto, CA) were selected on Luria-Bertani plates with 25 mg/L kanamycin A (Olszewski et al., 1988), and pPZP200-based plasmids were selected with 100 mg/L spectinomycin (Hajdukiewicz et al., 1994). Transgenic plants were selected on PN agar supplemented with 20 mg/L kanamycin A.
5-Bromo-4-Chloro-3-Indolyl-β-Glucuronic Acid and Dye Staining
ALF5::GUS transgenic plants were submerged, vacuum infiltrated in β-glucuronidase (GUS) staining buffer plus 0.2 mg/L 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Rose Scientific Ltd., Alberta, Canada), and then incubated overnight at 36°C (Jefferson et al., 1987). After rinses with water, tissue was fixed in 90, 70, and then 50% ethanol and left in 50% glycerol for photography. Arabidopsis seedlings were stained briefly (∼5 min) in either 0.5% toluidine blue O or 0.5% safranin O (Eastman Organic Chemicals, Rochester, NY) and then rinsed with tap water. Dye-stained roots were photographed immediately.
Nucleic Acids
Unless indicated otherwise, all molecular biology protocols were derived from standard protocols (Ausubel et al., 1997). Plant genomic DNA was prepared using a protocol for DNA extraction provided by Qiagen (Valencia, CA). Oligonucleotides were purchased from and DNA sequencing was performed by Research Genetics (Huntsville, AL). Plasmid DNA was prepared by the Qiagen column method. A polymerase chain reaction (PCR) kit for DNA amplification was purchased from Perkin-Elmer. DNA probes for hybridization were radiolabeled using the Prime-It kit (Stratagene, La Jolla, CA). DNA hybridization buffer consisted of 7% SDS, 1% BSA, 1 mM EDTA, and 250 mM sodium phosphate, pH 7.2.
Arabidopsis bacterial artificial chromosome (BAC) genomic clones were isolated from the Texas A&M University library and purchased from the Arabidopsis Biological Resource Center. λ bacteriophage clones were isolated from two libraries: one library was constructed from atr1 mutant DNA (Bender and Fink, 1998), and the other library, a gift from Dr. Ronald Davis (Stanford University, Palo Alto, CA), was constructed from sheared Col DNA and the λGEM11 vector. ALF5 cDNA clones were isolated from a λgt10 cDNA library (a gift from Christian Luschnig, Whitehead Institute, Cambridge, MA) derived from young plants (Col ecotype). DNA fragments were subcloned into the pBluescript II SK+ vector (Stratagene). For transgenic complementation, genomic subclones were inserted into the polylinker of the pBIN19 vector (Clontech). The full length ALF5 λ cDNA clone was removed by XmaI digestion, subcloned into the XmaI site of pBluescript II SK+ vector (BS/ALF5) for sequencing, and subcloned into the XmaI site of the yeast 2-μm plasmid pAD4M in an orientation that yields ALF5 expression in yeast driven by the ADH1 promoter (Ballester et al., 1989).
The HindIII–EcoRI T-DNA fragment of pBI101 or pBI101.2 (Clontech) was subcloned into a polylinker of pPZP212 (Hajdukiewicz et al., 1994) to generate the pPZP212/GUS1 or the pPZP212/GUS2 binary GUS expression vector, respectively. The translational ALF5::GUS fusion was constructed by subcloning the 3.4-kb XbaI–BspEI fragment of pBS/67X1 between the XbaI and XmaI polylinker sites of pPZP212/GUS2. For the transcriptional ALF5::GUS fusion, DNA of the ALF5 5′ promoter was amplified by PCR using primers AD357 (5′-GCGCTAGCCATTGGATTTTTTTGTTG-3′) and AD358 (5′-GCGCTAGCGTTTGGGTGGCGCACGA-3′). Each primer is chimeric and contains an 5′ NheI restriction site fused to the genomic sequence. The 3.5-kb PCR product was digested with NheI and subcloned in the XbaI polylinker site of pPZP212/GUS1 in the proper orientation with the GUS reporter gene. The junction of the PCR-amplified Arabidopsis sequence and the GUS reporter gene was confirmed by single strand sequencing.
The erc1 coding sequence from the S288C yeast background was amplified by PCR and subcloned into the pAD4M 2-μm plasmid. To make the ERC1 coding sequence (described by Shiomi et al., 1991), a single A/T base pair from a mononucleotide repeat (underlined in the primers) was removed from the wild-type erc1 coding sequence by in vitro plasmid mutagenesis using the primers AD454 (5′-ACGTGG-TCAAGGGAGACAAAAAATAGGTGGGTACATCAAC-3′) and AD455 (5′-GTTGATGTACCCACCTATTTTTTGTCTCCCTTGACCACGT-3′).
DNA Marker Genotyping
A crude leaf preparation of plant tissue was made using a protocol modified by John Celenza (Boston University, Boston, MA) (Klimyuk et al., 1993). A 4-mm2 piece of leaf was placed in a 1.5-mL microcentrifuge tube placed on dry ice, and ground with a Teflon pestle. The ground tissue was resuspended in 10 μL of 0.5 M NaOH, vacuum infiltrated for 30 sec, heated at 100°C for 45 sec, and then diluted with 100 μL of 0.2 M Tris-HCl, pH 8.0, and 1 mM EDTA. One microliter of leaf preparation was added to a PCR mixture with a volume ≥ 20 μL. Primers for the simple sequence length polymorphism markers were purchased from Research Genetics (Bell and Ecker, 1994). The generation and analysis of cleaved-amplified polymorphic sequence (CAPS) markers has been described (Konieczny and Ausubel, 1993). Details of CAPS markers generated for this work can be obtained from the Arabidopsis database World Wide Web site (http://www.arabidopsis.org/).
Gel Filtration Chromatography
The sample was dissolved in eluant, either reverse osmosis–treated water or 50 mM sodium phosphate (diluted from a stock of 0.5 M sodium phosphate, pH 7.0). Prepacked desalting NAP-25 gel filtration columns were purchased from Pharmacia LKB Biotechnology. A 1-mL sample was added to the column, and 1-mL fractions were collected. Absorbance was measured with a Hitachi (Tokyo, Japan) U-2000 spectrometer. The absorbance of a 200-μL fraction diluted into 800 μL of the elution buffer was measured at 250 nm. Standard curves were made with sample dilutions of 0.1, 0.25, 0.5, 1, and 2%. PVP and polyphenolic compounds, such as quercetin, form a noncovalent association that shifts the absorbance spectrum of the aromatic polyphenol. To measure the change in the absorbance of quercetin, 100 μL of a fraction was diluted into 900 μL of 20 ppm quercetin in 50 mM sodium phosphate, pH 7.0. The mixture was left at room temperature for 1 hr, and the change in A340 was measured. To assess the alf5-inhibiting activity, 200 μL of each fraction was diluted into 10 mL of PN Noble agar. Seed of alf5 and ALF5 were sown on the test agar and scored after 2 weeks.
Yeast Strains
Yeast strains were grown under standard conditions (Guthrie and Fink, 1991). Strain 10556-24D (MATa ura3-1 leu2-3,112) was transformed with the variants of the pAD4M 2-μm plasmid by selection for Leu+ colonies. The diploid GEF1/GEF1 and gef1/gef1 strains have been described by Gaxiola et al. (1998).
Resistance Assay for Yeast Strains
A fivefold dilution series (in water) from a saturated liquid culture grown overnight was added (5-μL drops) in parallel to agar plates, either YPD (yeast extract [1%], peptone [2%], and dextrose [2%]), to which 1.4 M NaCl or 0.4 M or 0.5 M tetramethylammonium chloride (TMACl) was added, or synthetic yeast nitrogen base plus 2% dextrose and 200 μM uracil, to which 0.3 mM ethionine was added. After ∼2 days at 30°C, yeast growth on the plates was photographed.
Sequence Alignment and Prediction
Sequence data were analyzed with DNAStar software (Madison, WI). The transmembrane (TM) domains in the multidrug and toxic compound extrusion genes were predicted using SOUSI (Hirokawa et al., 1998).
Photography
Photography was performed with a Nikon (Tokyo, Japan) FE2 35-mm camera and Elite Chrome 100 slide film (Eastman Kodak). Slide images were captured with a SprintScan 35 slide scanner (Polaroid Corporation, Cambridge, MA). Autoradiographs were captured with a Lacie Silverscanner III (LaCie Ltd., Hillsboro, OR). The exposure, brightness, contrast, and cropping of images were manipulated using Photoshop (Adobe Systems, Mountain View, CA), Canvas (Deneba Systems Inc., Miami, FL), and/or Freehand (Macromedia Inc., San Francisco, CA) software.
GenBank Accession Numbers
The GenBank accession numbers are as follows: Arabidopsis F13P17.20, AAC27412; Arabidopsis F7H19.220, CAA19819; Arabidopsis T6A23.29, AAC67367; Saccharomyces cerevisiae Erc1p, 382,954; and Vibrio parahaemolyticus NorM, BAA31456.
Acknowledgments
A.C.D. is grateful for the technical assistance offered by John Celenza, Amir Sherman, and Bonnie Bartel. A.C.D. thanks fellow members of the Fink laboratory and both Kendal Hirschi and Julie Stone, who gave critical readings of the manuscript. This research was supported by National Science Foundation Grant MCB 9317175.
References
- Apse, M.P., Aharon, G.S., Snedden, W.A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1997). Current Protocols in Molecular Biology. (New York: John Wiley and Sons).
- Ballester, R., Michaeli, T., Ferguson, K., Xu, H.P., McCormick, F., and Wigler, M. (1989). Genetic analysis of mammalian GAP expressed in yeast. Cell 59, 681–686. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bender, J., and Fink, G.R. (1998). A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 5655–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzayen, M., Latche, A., Pech, J.-C., and Marigo, G. (1989). Carrier-mediated uptake of 1-(malonylamino)cyclopropane-1-carboxylic acid in vacuoles isolated from Catharanthus roseus cells. Plant Physiol. 91, 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M.H., Paulsen, I.T., and Skurray, R.A. (1998). The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31, 393–395. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Deus-Neumann, B., and Zenk, M.H. (1986). Accumulation of alkaloids in plant vacuoles does not involve an ion-trap mechanism. Planta 166, 250–260. [DOI] [PubMed] [Google Scholar]
- Diener, A.C., Li, H., Zhou, W.-X., Whoriskey, W.J., Nes, W.D., and Fink, G.R. (2000). STEROL METHYLTRANSFERASE 1 controls the level of cholesterol in plants. Plant Cell 12, 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, D.P., Cummins, L., Cole, D.J., and Edwards, R. (1998). Glutathione-mediated detoxification systems in plants. Curr. Opin. Plant. Biol. 1, 258–266. [DOI] [PubMed] [Google Scholar]
- Ellis, J., and Jones, D. (1998). Structure and function of proteins controlling strain-specific pathogen resistance in plants. Curr. Opin. Plant Biol. 1, 288–293. [DOI] [PubMed] [Google Scholar]
- Ferrando, A., Kron, S.J., Rios, G., Fink, G.R., and Serrano, R. (1995). Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol. Cell. Biol. 15, 5470–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola, R.A., Yuan, D.S., Klausner, R.D., and Fink, G.R. (1998). The yeast CLC chloride channel functions in cation homeostasis. Proc. Natl. Acad. Sci. USA 95, 4046–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola, R.A., Rao, R., Sherman, A., Grisafi, P., Alper, S.L., and Fink, G.R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 96, 1480–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G.R. (1991). Guide to Yeast Genetics and Molecular Biology. (San Diego, CA: Academic Press).
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOUSI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379. [DOI] [PubMed] [Google Scholar]
- Hirschi, K.D. (1999). Expression of Arabidopsis CAX1 in tobacco: Altered calcium homeostasis and increased stress sensitivity. Plant Cell 11, 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi, K.D., Zhen, R.G., Cunningham, K.W., Rea, P.A., and Fink, G.R. (1996). CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 8782–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk, V.I., and Jones, J.D. (1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Klimyuk, V.I., Carroll, B.J., Thomas, C.M., and Jones, J.D. (1993). Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 3, 493–494. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Lu, Y.P., Li, Z.S., and Rea, P.A. (1997). AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: Isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc. Natl. Acad. Sci. USA 94, 8243–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luettge, U., and Ratajczak, R. (1997). The physiology, biochemistry and molecular biology of the plant vacuolar ATPase. In The Plant Vacuole, R.A. Leigh and D. Sanders, eds (San Diego, CA: Academic Press), pp. 253–297.
- Marrs, K.A., Alfenito, M.R., Lloyd, A.M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. [DOI] [PubMed] [Google Scholar]
- Mattanovich, D., Ruker, F., Machado, A.C., Laimer, M., Regner, F., Steinkellner, H., Himmler, G., and Katinger, H. (1989). Efficient transformation of Agrobacterium spp. by electroporation. Nucleic Acids Res. 17, 6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, Y., Kodama, K., Shiota, S., Mine, T., Kataoka, A., Mizushima, T., and Tsuchiya, T. (1998). NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi, Y., and Anraku, Y. (1981). Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 256, 2079–2082. [PubMed] [Google Scholar]
- Olszewski, N.E., Martin, F.B., and Ausubel, F.M. (1988). Specialized binary vector for plant transformation: Expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 16, 10765–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, I.T., Brown, M.H., and Skurray, R.A. (1996). Proton-dependent multidrug efflux systems. Microbiol. Rev. 60, 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A.B., Casselman, A.L., and Last, R.L. (1992). A phosphoribosylanthranilate transferase gene is defective in blue fluorescent Arabidopsis thaliana tryptophan mutants. Plant Physiol. 100, 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez, R., Ardiles-Diaz, W., Van Montagu, M., Inze, D., and May, M.J. (1998). Cloning and expression analyses of AtMRP4, a novel MRP-like gene from Arabidopsis thaliana. Mol. Gen. Genet. 258, 655–662. [DOI] [PubMed] [Google Scholar]
- Sandermann, H., Jr. (1992). Plant metabolism of xenobiotics. Trends Biochem. Sci. 17, 82–84. [DOI] [PubMed] [Google Scholar]
- Schwencke, J., and de Robichon-Szulmajster, H. (1976). The transport of S-adenosyl-l-methionine in isolated yeast vacuoles and spheroplasts. Eur. J. Biochem. 65, 49–60. [DOI] [PubMed] [Google Scholar]
- Shiomi, N., Fukuda, H., Morikawa, H., Fukuda, Y., and Kimura, A. (1988). Cloning of a gene for S-adenosylmethionine synthesis in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 29, 302–304. [Google Scholar]
- Shiomi, N., Fukuda, H., Fukuda, Y., Murata, K., and Kimura, A. (1991). Nucleotide sequence and characterization of a gene conferring resistance to ethionine in yeast Saccharomyces cerevisiae. J. Ferment. Bioeng. 71, 211–215. [Google Scholar]
- Thomas, D., and Surdin-Kerjan, Y. (1997). Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61, 503–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini, R., Vogt, E., Fromenteau, M., Hortensteiner, S., Matile, P., Amrhein, N., and Martinoia, E. (1998). An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J. 13, 773–780. [DOI] [PubMed] [Google Scholar]
- Wiebauer, K., Schraml, S., Shales, S.W., and Schmitt, R. (1981). Tetracycline resistance transposon Tn1721: recA-dependent gene amplification and expression of tetracycline resistance. J. Bacteriol. 147, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222, 377–383. [DOI] [PubMed] [Google Scholar]
- Wink, M. (1997). Compartmentation of secondary metabolites and xenobiotics in plant vacuoles. In The Plant Vacuole, R.A. Leigh and D. Sanders, eds (San Diego, CA: Academic Press), pp. 141–169.
- Yoshida, S., and Anraku, Y. (2000). Characterization of staurosporine-sensitive mutants of Saccharomyces cerevisiae: Vacuolar functions affect staurosporine sensitivity. Mol. Gen. Genet. 263, 877–888. [DOI] [PubMed] [Google Scholar]
- Zhen, R.-G., Kim, E.J., and Rea, P.A. (1997). The molecular and biochemical basis of pyrophosphate-energized proton translocation at the vacuolar membrane. In The Plant Vacuole, R.A. Leigh and D. Sanders, eds (San Diego, CA: Academic Press), pp. 298–339.