Abstract
Due to the recent development of a cell culture model, hepatitis C virus (HCV) can be efficiently propagated in cell culture. This allowed us to reinvestigate the subcellular localization of HCV structural proteins in the context of an infectious cycle. In agreement with previous reports, confocal immunofluorescence analysis of the subcellular localization of HCV structural proteins indicated that, in infected cells, the glycoprotein heterodimer is retained in the endoplasmic reticulum. However, in contrast to other studies, the glycoprotein heterodimer did not accumulate in other intracellular compartments or at the plasma membrane. As previously reported, an association between the capsid protein and lipid droplets was also observed. In addition, a fraction of labeling was consistent with the capsid protein being localized in a membranous compartment that is associated with the lipid droplets. However, in contrast to previous reports, the capsid protein was not found in the nucleus or in association with mitochondria or other well-defined intracellular compartments. Surprisingly, no colocalization was observed between the glycoprotein heterodimer and the capsid protein in infected cells. Electron microscopy analyses allowed us to identify a membrane alteration similar to the previously reported “membranous web.” However, no virus-like particles were found in this type of structure. In addition, dense elements compatible with the size and shape of a viral particle were seldom observed in infected cells. In conclusion, the cell culture system for HCV allowed us for the first time to characterize the subcellular localization of HCV structural proteins in the context an infectious cycle.
Hepatitis C virus (HCV) is a small enveloped virus that belongs to the Hepacivirus genus in the Flaviviridae family (27). Its genome encodes a single polyprotein precursor of ∼3,010 amino acid residues, which is synthesized on endoplasmic reticulum (ER)-associated ribosomes. The polyprotein is cleaved co- and posttranslationally by cellular and viral proteases to yield at least 10 mature products. HCV genome encodes three structural proteins: a capsid protein (C) and two envelope glycoproteins (E1 and E2). These proteins are released from the N-terminal region of the polyprotein by signal peptidase cleavages (15). In addition, processing in the C-terminal region of the capsid protein by a signal peptide peptidase leads to the generation of a mature capsid protein (32).
In the absence of a robust cell culture model for HCV, the analyses of the subcellular localization of HCV proteins have been performed with heterologous expression systems or in the context of HCV replicons (reviewed in references 15 and 33). Transient expression of HCV envelope glycoproteins with heterologous expression systems has shown that HCV envelope glycoproteins E1 and E2 assemble as a noncovalent heterodimer (11). Due to the presence of retention signals in the transmembrane domains of HCV envelope glycoproteins (8, 9), the glycoprotein heterodimer is mainly retained in the ER (17). However, in some expression systems, a fraction of HCV envelope glycoproteins has also been found to be located in the intermediate compartment and the cis-Golgi apparatus (12, 29, 37) and at the plasma membrane (3, 13, 24).
When expressed with heterologous expression systems or in the context of HCV replicons, the subcellular distribution of the capsid protein seems to be complex. Most of the protein is cytoplasmic where it is found both attached to the ER and at the surface of lipid droplets (for a review, see reference 31). The different extents to which the capsid protein is attached either to lipid droplets or membranes may be dependent on the amount of lipid droplets present in various cell types (22). In some conditions, a minor proportion of the capsid protein has also been found to be located in the nucleus (43). More recently, the capsid protein has also been found to colocalize with mitochondrial markers in Huh-7 cells containing a full-length HCV replicon (39).
Very recently, a cell culture model has been developed for HCV (26, 42, 44). This system is based on the transfection of the human hepatoma cell line Huh-7 with genomic RNA derived from a cloned viral genome. This culture system allows the production of virus that can be efficiently propagated in cell culture. Although a large amount of data has been accumulated on most HCV proteins during the past 15 years, the development of a cell culture system for HCV allows reinvestigation of the biological and biochemical properties of HCV proteins in a more relevant context. Here, we analyzed the subcellular localization of HCV structural proteins in Huh-7 cells infected with an infectious HCV clone. Our data show that, in infected cells, HCV glycoprotein heterodimer is retained in the ER and the capsid protein is detected in association with lipid droplets. However, in contrast to previous reports, no other subcellular localization was found for these proteins. In addition, no colocalization was observed between the glycoprotein heterodimer and the capsid protein in HCV-infected cells. Electron microscopy analyses identified membrane alterations in infected cells; however, dense elements compatible with the size and shape of a viral particle were seldom observed in HCV-infected cells.
MATERIALS AND METHODS
Cell culture and HCV production.
Huh-7 human hepatoma cells (35) were grown in Dulbecco modified essential medium (Invitrogen) supplemented with 10% fetal bovine serum. The plasmid pJFH1 containing the full-length cDNA of JFH1 isolate, which belongs to subtype 2a (GenBank accession no. AB047639), has been described previously (42). To generate genomic HCV RNA, the plasmid pJFH1 was linearized at the 3′ end of the HCV cDNA by XbaI digestion. After treatment with mung bean nuclease, the linearized DNA was then purified and used as a template for in vitro transcription with the MEGAscript kit from Ambion. In vitro-transcribed RNA was delivered to cells by electroporation as described previously (25). Viral stocks were obtained by harvesting cell culture supernatants 1 week posttransfection. Secondary viral stocks of ca. 105 50% tissue culture infective doses/ml were obtained by additional amplifications on naive Huh-7 cells. Except for some electron microscopy studies, all of the experiments were done with Huh-7 cells infected with secondary viral stocks. For immunofluorescence studies, HCV-infected cells were treated with trypsin, seeded on coverslips, and grown for 3 to 4 days before processing for immunolabeling. Virus titration was performed as described previously (26) by immunostaining the cells with anti-E2 monoclonal antibody (MAb) 3/11.
Antibodies.
Rat MAb 3/11 (19) was produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Mouse anti-E2 MAb AP33 has been previously described (7). Human anti-E1 MAb 1C4 (Innogenetics hybridoma clone IGH398) was obtained from a patient chronically infected with an HCV subtype 1b strain. This immunoglobulin G1 (IgG1) MAb has been mapped to the V3 region of E1 (amino acids 235 to 240) and has been shown to cross-react with E1 peptides from genotypes 1 to 6 (G. Maertens et al., unpublished data). Anti-capsid ACAP27 (28) and anti-NS3 (486D39) MAbs were kindly provided by J. F. Delagneau (Bio-Rad, France). Human anti-E2 MAb CBH5 (21), mouse anti-ERGIC-53 MAb (38), and anti-capsid MAb 6G7 (23) were kindly provided by S. Foung (Stanford University), H. P. Hauri (University of Basel, Basel, Switzerland) and H. B. Greenberg (Stanford University), respectively. Mouse anti-CD63 MAb TS63 (5) was a gift from E. Rubinstein (INSERM U602, Villejuif, France). Mouse anti-GM130 and anti-LAMP-1 MAbs were purchased from BD Biosciences. Rabbit antibodies to calnexin, calreticulin, and protein disulfide isomerase (PDI) were from Stressgen. Mouse anti-cytochrome c MAb 6H2.B4 was from Pharmingen. Guinea pig polyclonal antibody to human ADRP was purchased from Progen. Alexa 488-conjugated goat anti-rabbit, anti-mouse, anti-rat, or anti-human IgG, and isotype-specific Alexa488-conjugated goat anti-mouse IgG2a and Alexa555-conjugated goat anti-mouse IgG1 were purchased from Molecular Probes. Cy3-conjugated goat anti-mouse, anti-rat, or anti-guinea pig IgG were purchased from Jackson Immunoresearch (West Grove, PA).
Indirect immunofluorescence microscopy.
Infected Huh-7 cells grown on 12-mm glass coverslips were fixed with 3% paraformaldehyde and then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS). Both primary and secondary antibody incubations were carried out in PBS containing 10% goat serum for 30 min at room temperature. For double-label immunofluorescence with primary antibodies from different species, Cy3- and Alexa 488-conjugated secondary antibodies were used. For double-label immunofluorescence with anti-C MAb ACAP27 (IgG2a) and another mouse MAb (IgG1), isotype-specific Alexa 488-conjugated goat anti-mouse IgG2a and Alexa 555-conjugated goat anti-mouse IgG1 were used. Lipid droplets were stained with oil red O as described previously (22). Coverslips were mounted on slides by using Mowiol 4-88 (Calbiochem). Confocal microscopy was performed with an SP2 confocal laser-scanning microscope (Leica) using a ×100/1.4 numerical aperture oil immersion lens. Double-label immunofluorescence signals were sequentially collected by using single fluorescence excitation and acquisition settings to avoid crossover. Images were processed by using Adobe Photoshop software.
Cell surface labeling.
Huh-7 cells infected by JFH1 virus or transfected with the plasmid phCMV-E1E2 (3) or phCMV-E1E2(LAL) (6), as well as control Huh-7 cells were used for cell surface labeling. Cells were incubated for 1 h on ice with the primary antibody. MAb 3/11 was used to detect E2 glycoprotein expressed at the cell surface. Experiments with a rabbit polyclonal anti-calnexin antiserum were carried out as a control for the lack of permeabilization of cell membranes. Cells were then washed three times with cold PBS, fixed with 3% paraformaldehyde. Alexa-488-conjugated goat anti-rat or rabbit secondary antibody incubation was carried out in PBS containing 10% goat serum for 30 min at room temperature.
Endoglycosidase digestions.
Huh-7 cells infected by JFH1 virus or 393T cells transfected with the plasmid phCMV-E1E2 (3) were lysed with 0.5% Igepal CA-630 in TBS (50 mM Tris-Cl [pH 7.5], 150 mM NaCl). Cell lysates were used for immunoprecipitation with anti-E2 MAb AP33 as previously described (16). Immunoprecipitated proteins were eluted from protein-A Sepharose in 30 μl of dissociation buffer (0.5% sodium dodecyl sulfate [SDS] and 1% 2-mercaptoethanol) by boiling for 10 min. The protein samples were then divided into equal portions for digestion with either endo-β-N-acetylglucosaminidase H (endo H) or peptide:N-glycosidase F (PNGase F) and an undigested control. Digestions were carried out for 1 h at 37°C in the buffer provided by the manufacturer. Proteins were then analyzed by Western blotting.
Western blotting.
After separation by SDS-polyacrylamide gel electrophoresis (PAGE), protein preparations were transferred to nitrocellulose membranes (Hybond-ECL; Amersham) by using a Trans-Blot apparatus (Bio-Rad) and revealed with a specific MAb, followed by the addition of goat anti-mouse or anti-rat immunoglobulin conjugated to peroxidase (Jackson Immunoresearch). The proteins of interest were revealed by enhanced chemiluminescence detection (ECL; Amersham) as recommended by the manufacturer.
Electron microscopy.
For ultrastructural analysis, cells were fixed in 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 48 h. Cells were then washed in phosphate buffer, harvested, and postfixed with 1% osmium tetroxide for 1 h. They were then dehydrated in a graded acetone series, and cell pellets were embedded in Epon resin, which was allowed to polymerize for 24 h at 60°C. Ultrathin sections were cut on an ultramicrotome (Reichert, Heidelberg, Germany), collected on copper grids and stained with 1% uranyl acetate-1% lead citrate. The grids were then observed with a 1010 XC electron microscope (JEOL, Tokyo, Japan).
RESULTS
The glycoprotein heterodimer is located in the ER of HCV-infected cells.
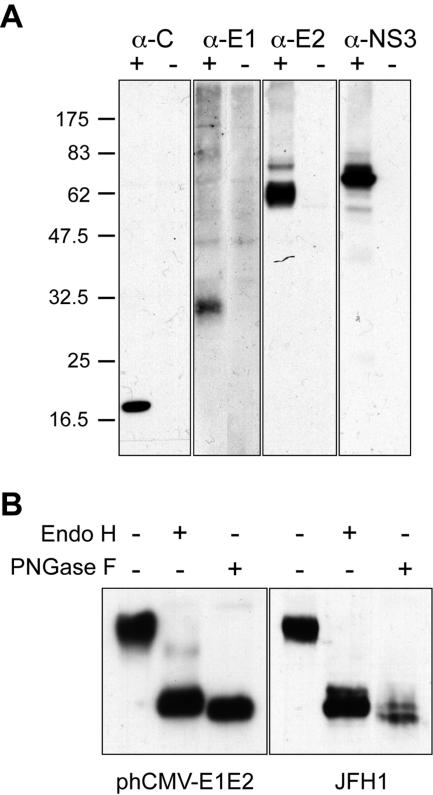
Efficient viral replication of JFH1 isolate was obtained by amplification on naive Huh-7 cells with virus titers of ca. 105 50% tissue culture infective doses/ml. This allowed us to reinvestigate the subcellular localization of HCV structural proteins in the context of an infectious cycle. The subcellular distribution of HCV envelope glycoproteins was examined by confocal immunofluorescence microscopy. To define the intracellular localization of E1 and E2 during HCV infection, we looked for antibodies that recognize the glycoproteins of the JFH1 strain in immunofluorescence analyses. Huh-7 cells were either mock infected or infected with HCV, grown on glass coverslips, fixed with paraformaldehyde, and processed for indirect immunofluorescence with various anti-E1 or anti-E2 antibodies. A large panel of antibodies that were raised against HCV glycoproteins of genotype 1 was screened. However, only anti-E1 MAb 1C4 and anti-E2 MAbs 3/11, AP33, and CBH5 were found to specifically label HCV-infected Huh-7 cells, but not naive cells in indirect immunofluorescence. Immunoblot analysis confirmed that MAb 1C4 recognizes the glycoprotein E1 and that MAbs 3/11 and AP33 recognize the glycoprotein E2 of the JFH1 strain (Fig. 1A and data not shown).
FIG. 1.
Expression of HCV proteins in HCV-infected cells. (A) Analysis of the expression of HCV proteins C, E1, E2, and NS3 by Western blotting. Lysates of naive (−) or HCV-infected (+) cells were separated by SDS-PAGE and revealed by Western blotting with MAbs ACAP27 (anti-C), 1C4 (anti-E1), 3/11 (anti-E2), and 486D39 (anti-NS3). The sizes of protein molecular mass markers are indicated on the right (in kilodaltons). (B) Analyses of the glycans associated with HCV glycoprotein E2. Lysates of HCV-infected cells Huh-7 cells (JFH1) or 293T cells transfected with a plasmid expressing HCV envelope glycoproteins (phCMV-E1E2) were immunoprecipitated with anti-E2 MAb AP33. The immunoprecipitates were treated or not treated with endo H or PNGase F. Proteins were then separated by SDS-PAGE and revealed by Western blotting with the anti-E2 MAb 3/11.
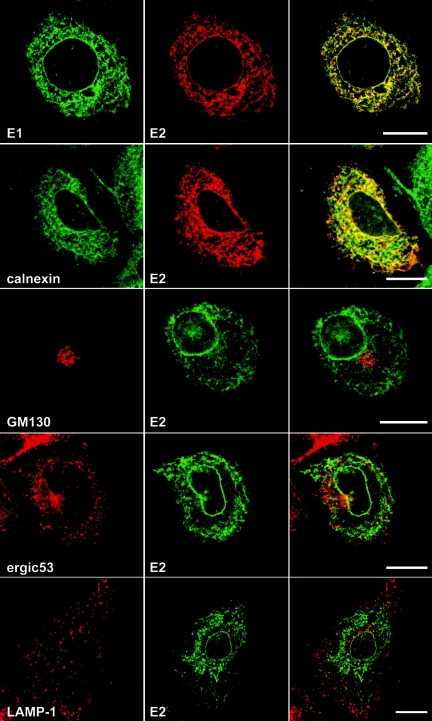
HCV-infected cells stained with MAbs 1C4, 3/11, or AP33 displayed a pattern of specific fluorescence in a network of cytoplasmic membranes and the nuclear envelope (Fig. 2). A very similar pattern of staining was also observed with MAb CBH5 (not shown). As expected, since they assemble as a heterodimer during their folding (11), E1 and E2 glycoproteins colocalized (Fig. 2). Interactions between E1 and E2 of JFH1 strain were confirmed by coimmunoprecipitation (data not shown). To further define the intracellular localization of HCV glycoprotein heterodimer in infected cells, double-label immunofluorescence experiments were carried out with an anti-E2 MAb. HCV-infected cells grown on glass coverslips were incubated with MAb 3/11, together with antibodies specific for various intracellular compartments. Within the secretory pathway, E2 colocalized predominantly with proteins of the ER, including calnexin (Fig. 2, calnexin), calreticulin, and PDI (not shown). In contrast to other studies (29, 37), E2 did not colocalize with the marker of the intermediate compartment ERGIC-53 (Fig. 2, ergic53) or with the Golgi marker GM130 (Fig. 2, GM130). In addition, E2 did not colocalize with markers of the endocytic pathway, such as EEA-1, a protein localized in early endosomes; CD63, a marker of multivesicular endosomes (not shown); or LAMP-1, a marker of late endosomes and lysosomes (Fig. 2, lamp-1). It is worth noting that we did not detect any change in the subcellular localization of HCV glycoprotein heterodimer when analyzed at different times posttransfection (48, 72, and 96 h) or postinfection (48 and 72 h). These findings demonstrate that the glycoprotein heterodimer localizes predominantly to the ER in HCV-infected cells, and they are in agreement with previous data obtained in the context of the expression of the full-length HCV polyprotein (17).
FIG. 2.
Confocal immunofluorescence analysis of the intracellular distribution of HCV glycoproteins. Infected cells grown on coverslips were fixed and processed for double-label immunofluorescence for E1, E2, and the following cellular markers: calnexin, a chaperone of the ER; GM130, a Golgi matrix protein; ERGIC-53, a marker of the ER-to-Golgi intermediate compartment; or LAMP-1, a marker of late endosomes and lysosomes. Anti-E2 mouse MAb AP33 was used for the colocalization with E1. For the other experiments, E2 was revealed with the rat MAb 3/11. Representative confocal images of individual cells are shown with the merge images in the right column. Bar, 20 μm.
The glycoprotein heterodimer is not expressed at the plasma membrane of HCV-infected cells.
Recently, it has been shown that, in some transient expression systems, a fraction of HCV envelope glycoproteins can also be detected at the plasma membrane (3, 13, 24). We therefore investigated whether HCV glycoprotein heterodimer can leave the ER and be exported to the plasma membrane in the context of HCV-infected cells.
To determine whether a fraction of HCV glycoprotein heterodimer has left the ER compartment, we analyzed whether the glycans associated with HCV glycoprotein E2 have been modified by Golgi enzymes by evaluating their sensitivity to endo H treatment. Resistance to digestion with endo H is indicative that glycoproteins have moved from the ER to at least the medial and trans-Golgi, where complex sugars are formed. PNGase F treatment, which removes all types of N-linked glycans, was used as a control of deglycosylation. As shown in Fig. 1B, the glycoprotein E2 remained endo H sensitive in HCV-infected cells, whereas a faint endo H-resistant band was observed for E2 expressed in cells transfected with a plasmid expressing HCV envelope glycoproteins. As previously observed (14), due to the presence of a residual N-acetyl-glucosamine at each glycosylation position after endo H treatment, the endo H-treated E2 migrated slightly more slowly than the PNGase F-treated protein. An additional slightly slower migrating band was detected for both PNGase F- and endo H-treated proteins (Fig. 1B). Such a partial resistance to PNGase F treatment has already been observed for truncated forms of HCV glycoprotein E2, as well as for E1 expressed in HCV pseudotyped particles (36).
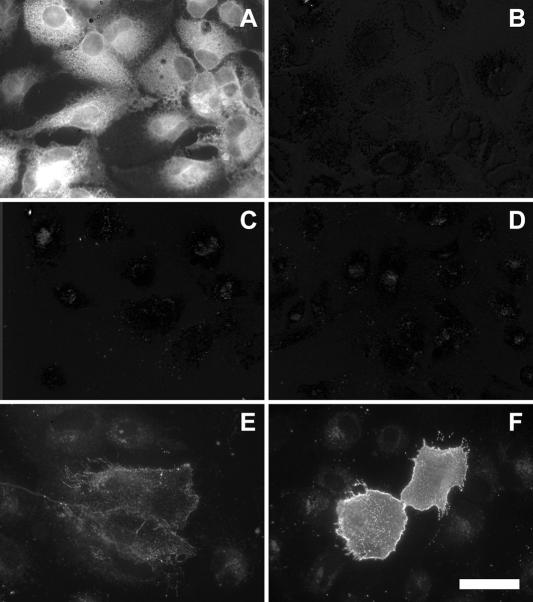
Cell surface expression was determined by surface labeling using the anti-E2 MAb 3/11. Although an E2 staining was clearly detected in HCV-infected cells treated with Triton X-100 (Fig. 3, compare panels A and B), no E2 protein was detected at the cell surface (Fig. 3C). Interestingly, E2 was detected at the plasma membrane of Huh-7 cells transfected with a plasmid expressing HCV envelope glycoproteins of genotype 1a (Fig. 3E). It is worth noting that there was a correlation between the level of expression and cell surface detection (data not shown). In addition, as previously observed (6, 10), the level of expression of E2 at the cell surface was higher when the charged residues in the transmembrane domain of E2 were mutated (Fig. 3F). When expressed from a plasmid, the expression level of HCV envelope glycoproteins of JFH1 isolate was approximately 15 times higher than in the context of HCV-infected cells; however, they were not detected above background at the cell surface (data not shown), indicating that there might be some differences between isolates or subtypes for cell surface expression. Altogether, these data indicate that HCV glycoprotein heterodimer does not accumulate at the plasma membrane in HCV-infected cells.
FIG. 3.
Cell surface expression of HCV glycoprotein E2. Naive (B and D) or infected (A and C) Huh-7 cells grown on coverslips were labeled with the anti-E2 MAb 3/11. A set of cells was fixed with 3% paraformaldehyde and processed for immunofluorescence labeling after permeabilization with Triton X-100 (A and B). Cell surface labeling (C, D, E, and F) was carried out on ice with MAb 3/11 before the fixation with 3% paraformaldehyde and the incubation with Alexa488-labeled secondary antibody. Huh-7 cells transfected with a plasmid expressing wild-type HCV envelope glycoproteins (phCMV-E1E2; panel E) or HCV envelope glycoproteins containing a mutation of the charged residues in the transmembrane domain of E2 [phCMV-E1E2(LAL); panel F] were used as controls of cell surface expression of E2. All images were acquired and processed with the same settings. Bar, 50 μm.
Intracellular localization of the capsid protein in HCV-infected cells.
To detect the capsid protein, we used the 6G7 (23) and ACAP27 (28) antibodies, two mouse MAbs that recognize two different epitopes located at amino acids 29 to 39 and amino acids 40 to 53, respectively. Consistent with the high levels of sequence conservation among core proteins from different genotypes, both MAbs gave positive signals in Huh-7 cells infected with JFH1 virus (Fig. 4 and data not shown). Immunoblot analysis confirmed that they recognize the core protein of the JFH1 strain (Fig. 1 and data not shown). The capsid protein migrated as a single species with an apparent molecular mass of 21 kDa, which most likely corresponded to the processed form of the capsid protein from which the signal sequence of E1 has been removed.
FIG. 4.
Confocal immunofluorescence analysis of the intracellular distribution of the capsid protein. Infected cells grown on coverslips were fixed and processed for double-label immunofluorescence for C (green) and the following cellular markers (red): ERGIC-53, a marker or the ER-to-Golgi intermediate compartment; GM130, a Golgi matrix protein; calnexin, a chaperone of the ER; or ADRP, a marker of the cytosolic pool of lipid droplets. Lipid droplets were stained with oil red O. Representative confocal images of individual cells are shown with the merge images in the right column. Insets display zoomed views of the indicated area. Bar, 20 μm.
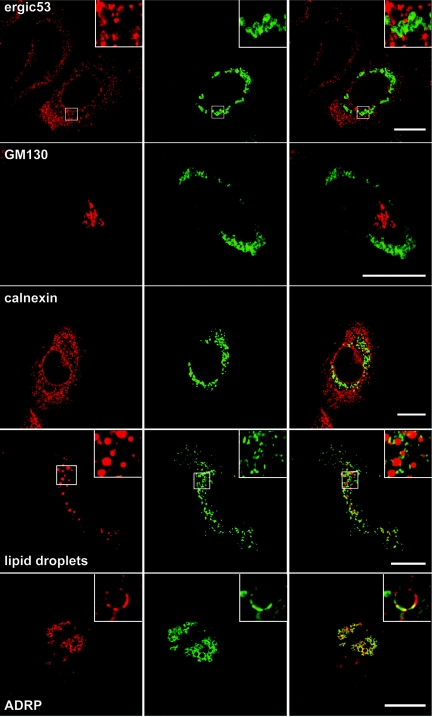
HCV-infected cells incubated with either anti-C protein MAbs displayed very similar patterns of bright fluorescence strictly limited to the cell cytoplasm and frequently concentrated in the perinuclear region (Fig. 4 and data not shown). In contrast, naive Huh-7 cells were not stained, indicating the specificity of the labeling (data not shown). In the perinuclear region, the staining pattern often displayed ring-like structures (Fig. 4). Double-label immunofluorescence experiments were performed in order to define the intracellular localization of the C protein. We did not observe, in contrast to E2, any colocalization between C and markers of the ER, such as calnexin (Fig. 4), calreticulin, or PDI (data not shown). In contrast to another report analyzing the subcellular localization in a cell line inducibly expressing HCV structural proteins (29), the perinuclear area containing the C protein was not localized in the Golgi complex and did not colocalize with the marker of the intermediate compartment (Fig. 4). In addition, no colocalization was observed with markers of the endocytic pathway, such as EEA-1 or Lamp-1 (data not shown).
Expression with heterologous expression systems or in the context of a full-length HCV replicon has shown that HCV capsid protein can associate with lipid droplets (1, 22, 37). We therefore analyzed whether the C protein expressed in HCV-infected cells would colocalize with lipid droplets. When lipid droplets were labeled with oil red O, an association between the C protein and lipid droplets was clearly observable. However, in contrast to previous reports, a fraction of the C protein was not directly associated with the lipid droplets, and the pattern of labeling was consistent with this fraction of the C protein being localized in a membranous compartment that is associated with the lipid droplets (see zoomed inset in Fig. 4). It is worth noting that we did not detect any change in the subcellular localization of the C protein when analyzed at different times posttransfection (48, 72, and 96 h) or postinfection (48 and 72 h).
To confirm the association of C with lipid droplets, double-label immunofluorescence experiments were carried out with an antibody to ADRP, which is a marker of the cytosolic pool of lipid droplets. In most infected cells, C and ADRP displayed an extensive colocalization (Fig. 4). At a higher magnification, C and ADRP were often concentrated on different parts of single lipid droplet-like ring structures (Fig. 4 ADRP, inset). In addition, no competition between both proteins for the association to lipid droplets was observed, as judged by the relative levels of ADRP and C immunoreactive signals in individual cells (not shown).
It has been shown that a fraction of the C protein expressed with heterologous systems can be located in the nucleus (43). However, we did not detect such a subcellular localization in HCV-infected cells (Fig. 4). Other reports have also proposed that a fraction of the C protein could be associated with mitochondria (39) and/or an Apo-AII-positive compartment (1). However, double-label immunofluorescence experiments in HCV-infected Huh-7 cells did not confirm the colocalization of the C protein with Apo-AII or mitochondria (data not shown). The detection of the capsid protein in the nucleus or the mitochondria as observed in other reports is potentially due to protein overexpression, to saturation of fluorescence signals, or to the absence of particle formation.
Altogether, our results show that the capsid protein is localized at the surface of lipid droplets and in a membranous compartment that is associated with the lipid droplets.
Relative intracellular localization of HCV glycoprotein heterodimer, capsid protein, and NS3.
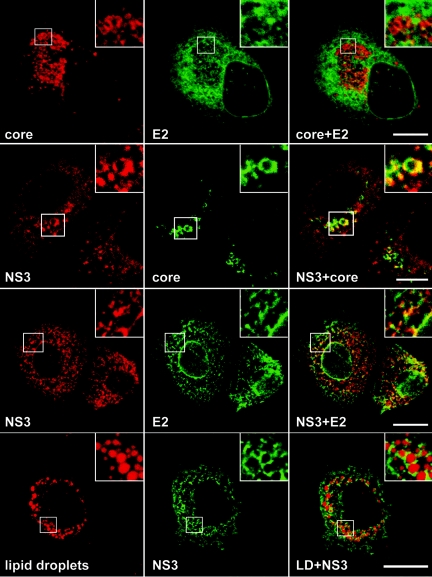
HCV nonstructural proteins NS3 to NS5B are the viral components of HCV replication complex (2), and they have been shown to colocalize in Huh-7 cells containing a subgenomic replicon (20). In addition, due to the presence of NS4B, the expression of HCV nonstructural proteins induces the formation of a virus-induced structure designated the membranous web (18). HCV structural and nonstructural proteins have been shown to colocalize with the membranous web (18), suggesting that the replication and assembly factories might be located within the same virus-induced organelle. We therefore analyzed whether HCV glycoprotein heterodimer would colocalize with the C protein and whether the localization of structural proteins would overlap that of nonstructural proteins. We chose to use an anti-NS3 antibody to detect one of the components of the replication complex. Immunofluorescence with the anti-NS3 MAb was positive on HCV-infected cells (Fig. 5) and negative on control cells (data not shown). Immunoblot analysis confirmed that the anti-NS3 MAb recognizes the NS3 protein of the JFH1 strain (Fig. 1A).
FIG. 5.
Relative intracellular localization of HCV proteins C, E2, and NS3 analyzed by confocal immunofluorescence. Infected cells grown on coverslips were fixed and processed for double-label immunofluorescence for C and E2 (top row), NS3 and C (middle row), or NS3 and E2 (bottom row). Lipid droplets were stained with oil red O. Representative confocal images of individual cells are shown with the merge images in the right column. Insets display zoomed views of the indicated area. Bar, 20 μm.
Double-label immunofluorescence experiments with anti-C and anti-E2 antibodies did not show any colocalization between C and E2 (Fig. 5). This is in agreement with the colocalization of E2 with ER markers and the absence of colocalization between the C protein and ER makers (Fig. 2 and 4). In contrast, a partial colocalization was observed between the C protein and NS3 and between E2 glycoprotein and NS3 (Fig. 5). NS3 displayed an ER-like pattern of immunofluorescence, which colocalized partially with E2. In addition, in a number of cells NS3 labeling was often more intense in the perinuclear area, where it partially colocalized with the C protein (Fig. 5). Interestingly, when lipid droplets were labeled with oil red O, the NS3 protein was also found around lipid droplets (Fig. 5).
Ultrastructural aspects of HCV-infected cells.
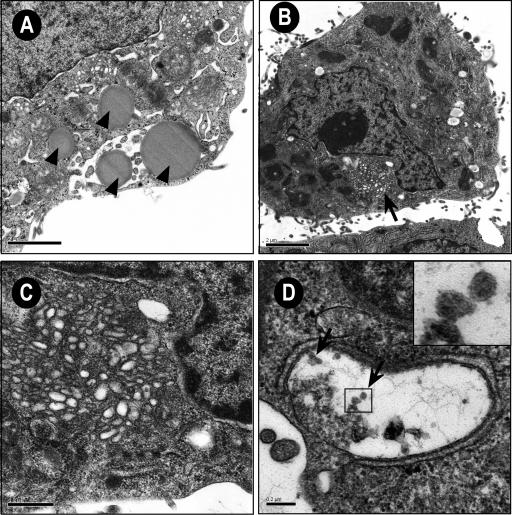
To identify ultrastructural modifications induced by JFH1 replication and the potential presence of viral particles, we investigated Huh-7 cells containing replicative JFH1 genome by electron microscopy. In a first set of experiments, we used Huh-7 cells transfected with JFH1 genomic RNA to have virtually all of the cells containing a replicative virus (Fig. 6). We then confirmed our observations on HCV-infected cells (data not shown). Lipid droplets were often observed in cells containing replicative JFH1 genome (Fig. 6A); however, there is no evidence that lipid droplets accumulate more in HCV-infected cells than in control naive Huh-7 cells. A membranous web composed of small vesicles was also found in many Huh-7 cells containing replicative JFH1 genome (Fig. 6B) but not in naive cells (data not shown). Figure 6C shows a membranous web at higher magnification. This structure was similar to the membranous web previously identified in U-2 OS human osteosarcoma-derived cell lines inducibly expressing the HCV polyprotein and in Huh-7 cells harboring a subgenomic HCV replicon (18, 20). Virus-like particles were never found in the membranous web. Some electron-dense elements compatible in size and shape with virus-like or core particles were sometimes found in small cisternae of Huh7 cells containing replicative JFH1 genome (Fig. 6D). These particles were smaller and more regular than the glycogen particles that are abundantly present in the Huh7 cells, but they were rare and their relationship with viral structures remains to be determined.
FIG. 6.
Membrane alterations in Huh-7 cells containing replicative JFH1 genome visualized by electron microscopy. (A) Cell containing several lipid droplets (arrows). Bar, 2 μm. (B) Low-power overview showing a membranous web (arrow). Bar, 2 μm. (C) Higher magnification of the membranous web. Bar, 0.5 μm. (D) Small cisternae containing electron-dense elements compatible in size and shape with virus-like or core particles (arrows). Bar, 0.2 μm. Inset displays zoomed view of the indicated area.
DISCUSSION
The recent development of a cell culture model for HCV (26, 42, 44) allows the production of virus that can be efficiently propagated in cell culture. This cell culture system has allowed us to reinvestigate the subcellular localization of HCV structural proteins in the context of an infectious cycle. Here, we showed that, in infected cells, HCV glycoprotein heterodimer is retained in the ER and the capsid protein is detected in association with lipid droplets. In contrast to previous reports, the glycoprotein heterodimer and the capsid protein were not detected in other subcellular localizations. Interestingly, electron microscopy analyses identified membrane alterations in infected cells. However, dense elements compatible with viral particles were seldom observed in HCV-infected cells.
HCV glycoprotein heterodimer is retained in the ER in infected cells. This is in agreement with several studies with heterologous expression systems containing HCV envelope glycoproteins or in the context of the full-length HCV polyprotein (11, 14, 17). Indeed, ER localization signals have been identified in the transmembrane domains of E1 and E2 (8, 9). Other studies have, however, shown that a fraction of HCV envelope glycoproteins can also be found in the ERGIC and in the cis-Golgi apparatus (29, 37). The presence of HCV glycoprotein heterodimer in these compartments is potentially due to protein overexpression, to saturation of fluorescence signals, or to the absence of particle formation. Indeed, such localization in the ERGIC and in the cis-Golgi apparatus was not observed in HCV-infected cells.
HCV glycoprotein heterodimer is not exported to the plasma membrane in infected cells. In some conditions of expression, a small fraction of HCV envelope glycoproteins has been shown to accumulate at the plasma membrane, and this led to the development of a system to pseudotype retroviral particles with HCV envelope proteins that has proven crucial for studying HCV entry (3, 13, 24, 36). However, cell surface expression of HCV glycoprotein heterodimer is not observed in HCV-infected cells, and export at the plasma membrane with the heterologous system might be due to the accumulation of small amounts of glycoproteins escaping the ER retention machinery due to saturation of this mechanism. It is worth noting that in the case of JFH1 envelope glycoproteins expressed from a plasmid, the glycoproteins were not detected at the cell surface, suggesting that there might also be some differences between isolates or subtypes for the subcellular localization of the envelope glycoproteins.
The capsid protein is detected in association with lipid droplets in infected cells. This is in agreement with other studies using heterologous expression systems or in the context of a full-length HCV replicon (1, 22, 37). However, in contrast to these studies, a fraction of the C protein was not directly associated with the lipid droplets, and this suggests a localization of the C protein in a membranous compartment that is associated with the lipid droplets, in addition to the localization of another fraction of the C protein at the surface of lipid droplets. Lipid droplets consist of a core of triglycerides and cholesterol esters surrounded by a monolayer of phospholipids and a proteinaceous coat (34). The most likely mechanism by which triglycerides and cholesterol esters end up in cytosolic lipid particles is that they first accumulate between the leaflets of the ER lipid bilayer and, after reaching a critical size, a lipid droplet would bud off the cytosolic side of the ER membrane (40). However, some droplets can remain physically connected to the ER membrane (4). From our immunolocalization study, it can be suggested that the C protein localizes at the site of lipid droplet formation. Although the C protein was not found to colocalize with the markers of ER membranes in HCV-infected cells, its localization at the potential site of lipid droplet formation suggests that the C protein does not associates with the rough ER but might attach to a subcompartment of the ER, which is potentially the smooth ER. We can also not exclude that this subcompartment corresponds to cytoplasmic raft microdomains that have been recently reported to contain the C protein (30).
The absence of colocalization between the C protein and classical markers of the ER compartment contrasts with some reports indicating that the C protein interacts with rough ER membranes (for a review, see reference 31). Interestingly, it has been reported that the traffic between rough ER membranes, the site of capsid protein synthesis, and lipid droplets is regulated by signal peptide peptidase cleavage in the C-terminal region of the capsid protein (32). It is therefore likely that in the context of HCV-infected cells, transport of the C protein to the site of lipid droplet assembly is rapid due to rapid cleavage by the signal peptide peptidase. It has also been previously reported that the different extents to which the capsid protein is associated with lipid droplets or rough ER membranes may be dependent on the amount of lipid droplets present in various cell types (22). Since HCV-infected cells accumulate lipid droplets, they provide the conditions for a shift toward accumulation of the C protein in association with lipid droplets.
The significance of the association of HCV capsid protein to a lipid droplet related compartment is not understood. Since the capsid protein is a major component of the viral particle, one might expect that this interaction plays a role in some step of virion morphogenesis. It is possible that the sites of lipid droplet formation provide a platform for nucleocapsid assembly by concentrating viral and/or cellular components that are necessary for the assembly of the nucleocapsid and/or by excluding those that inhibit this process. Interestingly, NS3 appears to partially colocalize with C in this compartment.
The glycoprotein heterodimer and the capsid protein do not colocalize in HCV-infected cells. Since the glycoprotein heterodimer and the capsid protein are the protein components of HCV particle, one would expect that these proteins would accumulate at the same site for virion assembly. Indeed, production of infectious virus particles likely requires spatially and temporally coordinated interactions of components that make up an infectious virion. The absence of colocalization between the glycoprotein heterodimer and the capsid protein would suggest that particle assembly is a two-step process involving nucleocapsid assembly, followed by a budding process occurring in two separate compartments. However, in the absence of sufficient data, it would be premature to generate a model of HCV particle assembly. To have a better idea of HCV morphogenesis, one would need to analyze the subcellular localization of HCV structural proteins in living cells. This would help elucidate the dynamics of the protein interactions between different cellular compartments.
Some viruses require membrane surfaces on which to assemble their replication complex. Such interactions have been well documented for positive-strand RNA viruses (41). In the case of HCV, a structure called membranous web has been previously identified in cell lines inducibly expressing the HCV polyprotein (18). In addition, such structures have also been shown to contain HCV RNA replication complex in cells harboring subgenomic replicons (20). Interestingly, a membranous web composed of small vesicles was also found in HCV-infected cells. However, these structures did not contain any virus-like particles, suggesting that HCV particle assembly does not occur in the membranous web.
The recent development of a cell culture model for HCV has allowed us to investigate for the first time the subcellular localization of HCV structural proteins in the context of a replicative and assembly competent virus. Interestingly, our data indicate that investigating the properties of HCV proteins expressed in heterologous systems do not necessarily reflect those that they have in the context of an infectious cycle. In conclusion, the cell culture system for HCV warrants reinvestigation of the biochemical and biological properties of HCV proteins in order to better understand their functional significance in the context of active viral replication and morphogenesis.
Acknowledgments
We thank André Pillez, Sophana Ung, and Sylvie Trassard for technical assistance. We are grateful to J. F. Delagneau, S. Foung, H. P. Hauri, H. B. Greenberg, E. Rubinstein, F. L. Cosset, and B. Bartosch for providing reagents. The data presented here were generated with the help of the Imaging Core Facility of the Calmette Campus and the Electron Microscopy Facility of the University of Tours.
This study was supported by EU grant QLRT-2001-01329 and grants from the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales (ANRS) and INSERM ATC-Hépatite C. Some of the authors were supported by fellowships from the French Ministry of Research (F.H.), the ANRS (D.D. and E.B.), and the CNRS (C.V.). T.W. was partly supported by grants from the Ministry of Health, Labor, and Welfare of Japan; the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO); and Research on Health Sciences focusing on Drug Innovation of the Japan Health Sciences Foundation. J.D. is an international scholar of the Howard Hughes Medical Institute.
REFERENCES
- 1.Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C pseudo-particles containing functional E1E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette-Mackie, E. J., N. K. Dwyer, T. Barber, R. A. Coxey, T. Takeda, C. M. Rondinone, J. L. Theodorakis, A. S. Greenberg, and C. Londos. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid. Res. 36:1211-1226. [PubMed] [Google Scholar]
- 5.Charrin, S., F. Le Naour, M. Oualid, M. Billard, G. Faure, S. M. Hanash, C. Boucheix, and E. Rubinstein. 2001. The major CD9 and CD81 molecular partner: identification and characterization of the complexes. J. Biol. Chem. 276:14329-14337. [DOI] [PubMed] [Google Scholar]
- 6.Ciczora, Y., N. Callens, C. Montpellier, B. Bartosch, F. L. Cosset, A. Op De Beeck, and J. Dubuisson. 2005. Contribution of the charged residues of HCV glycoprotein E2 transmembrane domain to the functions of E1E2 heterodimer. J. Gen. Virol. 86:2793-2798. [DOI] [PubMed] [Google Scholar]
- 7.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocquerel, L., S. Duvet, J.-C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel, L., J.-C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a key role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Martynoff, G., A. Venneman, and G. Maertens. 1997. Analysis of post-translational modifications of HCV structural proteins by using the vaccinia virus expression system, p. 39-44. In M. Rizzetto, R. H. Purcell, and H. Gerin (ed.), Viral hepatitis and liver disease. Minerva Medica, Turin, Italy.
- 13.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 14.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell. Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 16.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 18.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, H. H., M. Donets, H. B. Greenberg, and S. M. Feinstone. 1993. Characterization of hepatitis C virus structural proteins with a recombinant baculovirus expression system. Hepatology 17:763-771. [PubMed] [Google Scholar]
- 24.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 26.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 28.Maillard, P., K. Krawczynski, J. Nitkiewicz, C. Bronnert, M. Sidorkiewicz, P. Gounon, J. Dubuisson, G. Faure, R. Crainic, and A. Budkowska. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 75:8240-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martire, G., A. Viola, L. Iodice, L. V. Lotti, R. Gradini, and S. Bonatti. 2001. Hepatitis C virus structural proteins reside in the endoplasmic reticulum as well as in the intermediate compartment/cis-Golgi complex region of stably transfected cells. Virology 280:176-182. [DOI] [PubMed] [Google Scholar]
- 30.Matto, M., C. M. Rice, B. Aroeti, and J. S. Glenn. 2004. Hepatitis C virus core protein associates with detergent-resistant membranes distinct from classical plasma membrane rafts. J. Virol. 78:12047-12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 32.McLauchlan, J., M. K. Lemberg, R. G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moradpour, D., R. Gosert, D. Egger, F. Penin, H. E. Blum, and K. Bienz. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antivir. Res. 60:103-109. [DOI] [PubMed] [Google Scholar]
- 34.Murphy, D. J., and J. Vance. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24:109-115.10203758 [Google Scholar]
- 35.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 36.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweizer, A., J. A. M. Fransen, T. Bächi, L. Ginsel, and H.-P. Hauri. 1988. Identification, by a monoclonal antibody, of a 53-kD protein associated with tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J. Cell Biol. 107:1643-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwer, B., S. Ren, T. Pietschmann, J. Kartenbeck, K. Kaehlcke, R. Bartenschlager, T. S. Yen, and M. Ott. 2004. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J. Virol. 78:7958-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Meer, G. 2001. Caveolin, cholesterol, and lipid droplets? J. Cell Biol. 152:29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villanueva, R., Y. Rouillé, and J. Dubuisson. 2005. Interactions between virus proteins and host cell membranes. Int. Rev. Cytol. 245:171-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]