Abstract
APOBEC3G (A3G) and related cytidine deaminases, such as APOBEC3F (A3F), are potent inhibitors of retroviruses. Formation of infectious human immunodeficiency virus type 1 (HIV-1) requires suppression of multiple cytidine deaminases by Vif. Whether HIV-1 Vif recognizes various APOBEC3 proteins through a common mechanism is unclear. The domains in Vif that mediate APOBEC3 recognitions are also poorly defined. The N-terminal region of HIV-1 Vif is unusually rich in Trp residues, which are highly conserved. In the present study, we examined the role of these Trp residues in the suppression of APOBEC3 proteins by HIV-1 Vif. We found that most of the highly conserved Trp residues were required for efficient suppression of both A3G and A3F, but some of these residues were selectively required for the suppression of A3F but not A3G. Mutant Vif molecules in which Ala was substituted for Trp79 and, to a lesser extent, for Trp11 remained competent for A3G interaction and its suppression; however, they were defective for A3F interaction and therefore could not efficiently suppress the antiviral activity of A3F. Interestingly, while the HIV-1 Vif-mediated degradation of A3G was not affected by the different C-terminal tag peptides, that of A3F was significantly influenced by its C-terminal tags. These data indicate that the mechanisms by which HIV-1 Vif recognizes its target molecules, A3G and A3F, are not identical. The fact that several highly conserved residues in Vif are required for the suppression of A3F but not that of A3G suggests a critical role for A3F in the restriction of HIV-1 in vivo.
APOBEC3G (A3G) and the related cytidine deaminase APOBEC3F (A3F) are potent inhibitors of a wide range of retroviruses and retroelements (2, 5-7, 9, 12, 13, 15, 18, 20-22, 26, 28, 30, 31, 35, 36, 39, 41, 42, 45, 46). In the absence of Vif, A3G and A3F are incorporated into budding virions, where, upon infection of new target cells, they induce cytidine deamination in the minus-strand viral DNA (9, 13, 20, 22, 36, 42, 45), resulting in abortive infection. Human immunodeficiency virus type 1 (HIV-1) Vif recruits the Cullin5-ElonginB-ElonginC E3 ubiquitin ligase (11, 16, 19, 24, 43, 44) to target the cellular antiviral proteins A3G and A3F for degradation (4, 16, 17, 23, 24, 32, 34, 43). HIV/simian immunodeficiency virus (SIV) Vif binds ElonginC through a virus-specific BC box (16, 19, 24, 43, 44). Primate lentiviral Vif proteins also use a highly conserved H-X5-C-X17-18C-X3-5-H motif spanning 30 amino acids upstream of the BC box (Fig. 1) to mediate Cul5 interaction (19).
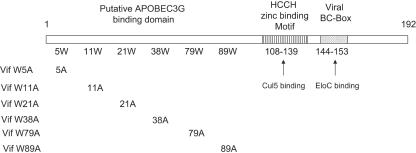
FIG. 1.
Structural features of HIV-1 Vif. The highly conserved SLQYLA motif (amino acids 144 to 153) in Vif forms a BC box that interacts with ElonginC (EloC) (16, 19, 24, 43, 44). Amino acids 108 to 139 in Vif form a highly conserved H-X5-C-X17-18C-X3-5-H motif that mediates the interaction with Cul5 (19). The N-terminal region of HIV-1 Vif has been implicated in the interaction with A3G (23). This region is unusually rich in tryptophans. The indicated mutants were constructed to evaluate the functions of conserved Trp residues.
Vif is a substrate receptor in the Cul5-based E3 ubiquitin ligase complex that mediates target protein selection. The regions of Vif that are involved in binding to target proteins such as A3G and A3F are poorly defined. A long-recognized and striking feature of HIV-1 Vif is the high concentration of tryptophans in its N-terminal region (14). This region has also been implicated in mediating the interaction with A3G (23). Whether any of these highly conserved tryptophans mediates target protein interaction is an intriguing question.
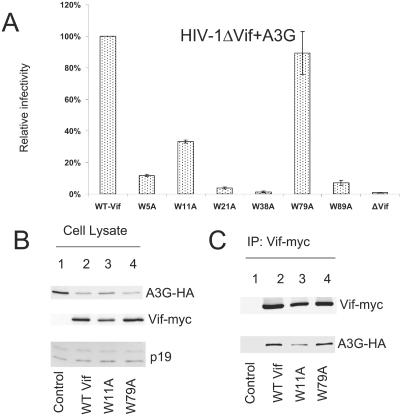
To determine whether these highly conserved Trp residues are important for Vif function, we generated a series of HIV-1 Vif mutant constructs in which individual Trp residues were replaced with Ala (Fig. 1). 293T cells were transfected with HIV-1ΔVif and an A3G expression vector plus wild-type (WT) or mutant Vif expression vectors as indicated (Fig. 2). Virus was produced from transfected 293T cells and tested for infectivity in a standard Magi assay as previously described (38). WT Vif suppressed A3G and maintained HIV-1ΔVif infectivity (Fig. 2A, first column), and this level of infectivity in the presence of WT Vif was considered as 100% infectivity (Fig. 2A). As expected, A3G dramatically reduced the infectivity of HIV-1ΔVif in the absence of Vif (Fig. 2A, eighth column). Most of the conserved Trp residues, including Trp5, -21, -38, and -89, were important for Vif activity against A3G (Fig. 2A), since mutation of these residues destroyed the ability of these Vif mutants to suppress A3G. However, mutation of Trp79 (Fig. 2A, sixth column) and, to a lesser extent, of Trp11 (Fig. 2A, third column) did not significantly affect mutant Vif function against A3G. VifW79A maintained ∼90% of the WT Vif activity (Fig. 2A, sixth column), and VifW11A maintained ∼40% (Fig. 2A, third column).
FIG. 2.
Effect of Vif mutations on A3G suppression. (A) Effect of A3G on infectivity of HIV-1ΔVif in the presence of WT or mutant Vif. HIV-1 viruses were produced in 293T cells coexpressing A3G in the presence of WT or mutant Vif as indicated. Virus infectivity was assessed using Magi indicator cells, with the virus infectivity in the presence of WT Vif set to 100%. Error bars represent the standard deviations from triplicate wells. (B) Immunoblotting of lysates of 293T cells cotransfected with A3G plus a control vector (lane 1) or plus WT Vif (lane 2), VifW11A (lane 3), or VifW79A (lane 4) expression vector. Ribosome P19 protein was used as the sample loading control. (C) Coimmunoprecipitation of A3G with WT or mutant Vif molecules. The expression vector for A3G-HA was cotransfected with a control vector (lane 1) or with WT Vif (lane 2), VifW11A (lane 3), or VifW79A (lane 4) expression vector into 293T cells. At 48 h after transfection, cell lysates were used for immunoprecipitation (IP) of Vif-myc with an anti-myc antibody conjugated to agarose beads. The interaction between A3G-HA and myc-tagged WT or mutant Vif molecules was detected by immunoblotting of immunoprecipitated samples using an antibody against HA to detect A3G-HA. Immunoprecipitated Vif-myc was detected using an anti-Vif antiserum.
We also examined the effect of WT and mutant Vif molecules on A3G expression (Fig. 2B). 293T cells were transfected with an A3G expression vector plus a control vector (Fig. 2B, lane 1) or vector expressing WT Vif-myc (lane 2), VifW11A-myc (lane 3), or VifW79A-myc (lane 4). In agreement with the infectivity data, we observed that the intracellular level of A3G was reduced more by HIV-1 WT Vif (lane 2) than by the vector control (lane 1). Mutants VifW11A (Fig. 2B, lane 3) and VifW79A (lane 4) also showed reduced A3G expression compared to the vector control (lane 1).
The interaction of WT and mutant Vif molecules with A3G was also evaluated by coimmunoprecipitation analysis using the same cell lysates. The Vif-myc proteins were immunoprecipitated from the cell lysates with a monoclonal antibody against the myc tag; coprecipitation of A3G with various Vif molecules was examined by immunoblotting using an antibody against hemagglutinin (HA) to detect A3G-HA.
WT Vif (Fig. 2C, lane 2) and VifW79A (lane 4) efficiently coimmunoprecipitated A3G-HA. A3G-HA was not coimmunoprecipitated in the absence of Vif (Fig. 2C, lane 1), indicating the specificity of the assay. VifW11A also interacted with A3G (lane 3), but with a subtle decrease in reactivity compared to the WT Vif (lane 2). Therefore, Trp79 and to a lesser extent Trp11 in HIV-1 Vif appear to be dispensable for the A3G interaction or its suppression. This result was unexpected, since both Trp11 and Trp79 are highly conserved among various HIV-1 isolates.
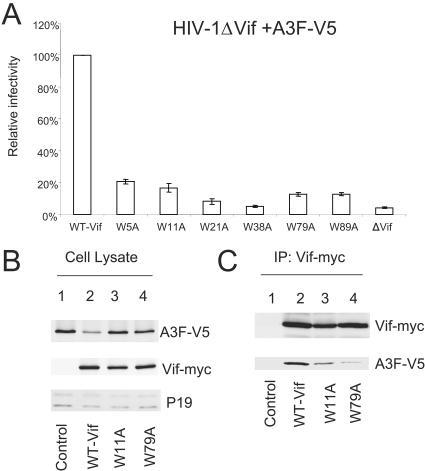
We next examined the effect of Vif Trp mutations on their activities against another target molecule, A3F. In this case, all the Vif mutants, including VifW11A and VifW79A, were essentially ineffective against A3F compared to WT Vif (Fig. 3A). The ability of WT and mutant Vif molecules to affect A3F expression was also examined (Fig. 3B). For this purpose, 293T cells were transfected with an A3F expression vector plus a control vector (Fig. 3B, lane 1) or with vector expressing WT Vif-myc (lane 2), VifW11A-myc (lane 3), or VifW79A-myc (lane 4). In contrast to the results obtained with A3G, mutants VifW11A (Fig. 3B, lane 3) and VifW79A (lane 4) did not significantly reduce A3F expression compared to WT Vif (lane 2). Interaction of WT and mutant Vif molecules with A3F was also evaluated by coimmunoprecipitation analysis. The Vif proteins were immunoprecipitated from cell lysates, and coprecipitation of A3F was examined by immunoblotting. A3F was efficiently coimmunoprecipitated with WT Vif (Fig. 3C, lane 2), and this interaction was specific, since A3F was not detected in the absence of Vif (Fig. 3C, lane 1). Even though the levels of A3F detected in cells expressing VifW11A (Fig. 3B, lane 3) or VifW79A (Fig. 3B, lane 4) were higher than those expressing WT Vif (Fig. 3B, lane 2), significantly less A3F was coimmunoprecipitated with VifW11A (Fig. 3C, lane 3) or VifW79A (Fig. 3C, lane 4). The impaired recognition of A3F by VifW11A and VifW79A correlated with their reduced ability to suppress the antiviral activity of A3F (Fig. 3A) and their impaired ability to reduce the intracellular expression of A3F (Fig. 3B) compared to the WT Vif. Trp5, -21, -38, and -89 were important for the suppression of both A3G (Fig. 2A) and A3F (Fig. 3A), suggesting that these residues may be involved in binding to both target molecules. Alternatively, these residues may be important for the proper folding of the Vif molecule.
FIG. 3.
Effect of Vif mutations on A3F suppression. (A) Effect of A3F on infectivity of HIV-1ΔVif in the presence of WT or Vif mutants. HIV-1 viruses were produced in 293T cells with coexpressing A3F-V5 in the presence of WT or mutant Vif and assayed for infectivity as described for Fig. 2. (B) Immunoblotting of lysates of 293T cells cotransfected with A3F-V5 plus a control vector (lane 1) or plus WT Vif (lane 2), VifW11A (lane 3), or VifW79A (lane 4) expression vector. Ribosome P19 protein was used as the sample loading control. (C) Coimmunoprecipitation of A3F-V5 with WT or mutant Vif molecules. Expression vector for A3F-V5 was cotransfected with a control vector (lane 1) or with WT Vif (lane 2), VifW11A (lane 3), or VifW79A (lane 4) expression vector into 293T cells. At 48 h after transfection, cell lysates were immunoprecipitated (IP) with agarose-conjugated anti-myc antibody, and the interaction of A3F-V5 with myc-tagged WT or mutant Vif molecules was detected by immunoblotting of immunoprecipitated samples with anti-V5 antibody and anti-Vif antiserum.
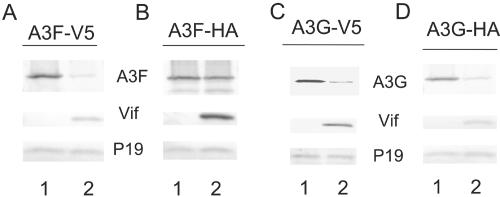
Interaction between A3G and HIV-1 Vif has been mapped to the N-terminal region of A3G (4). The regions in A3F that are important for Vif recognition have not been defined. Thus, it is particularly interesting that we observed that HIV-1 Vif-induced degradation of A3F was quite sensitive to the C-terminal tag peptides. V5-tagged A3F was very sensitive to HIV-1 Vif-induced degradation (Fig. 4A, lane 2) compared to the vector control (Fig. 4A, lane 1). On the other hand, HA-tagged A3F was rather resistant to HIV-1 Vif-induced degradation (Fig. 4B, lane 2) compared to the vector control (Fig. 4B, lane 1). In contrast to A3F, both V5-tagged A3G (Fig. 4C) and HA-tagged A3G (Fig. 4D) were very sensitive to Vif-induced degradation.
FIG. 4.
Influence of C-terminal tag peptides on A3G and A3F expression in the presence or absence of HIV-1 Vif. (A) Immunoblotting of lysates of 293T cells cotransfected with A3F-V5 plus a control vector (lane 1) or an expression vector for WT Vif (lane 2). Ribosome P19 protein was used as the sample loading control. (B) Immunoblotting of lysates of 293T cells cotransfected with A3F-HA plus a control vector (lane 1) or an expression vector for WT Vif (lane 2). (C) Immunoblotting of lysates of 293T cells cotransfected with A3G-V5 plus a control vector (lane 1) or an expression vector for WT Vif (lane 2). (D) Immunoblotting of lysates of 293T cells cotransfected with A3G-HA plus a control vector (lane 1) or an expression vector for WT Vif (lane 2).
Several lines of evidence suggest that HIV-1 Vif may recognize A3G and A3F through distinct interfaces. (i) We have found that some of the highly conserved Trp residues in HIV-1 Vif are differentially required for the recognition of A3F and A3G. Trp79 and Trp11 of HIV-1 Vif were critical for interaction with A3F but less important for A3G recognition. Simon et al. recently also reported that some single-amino-acid mutants of HIV-1 Vif retained selective neutralizing activity against APOBEC3F but not against APOBEC3G, and vice versa (33). (ii) While C-terminal tag modifications of A3F significantly influenced its ability to be recognized by HIV-1 Vif, C-terminal tag modifications of A3G were less influential. (iii) A critical residue (amino acid 128) in the first linker region of A3G influences its recognition by primate lentiviral Vif molecules. A similar residue in A3F is not important. In the case of human A3G and AGM-A3G, a single amino acid at position 128 mediates their species-specific recognition by the Vif protein of the lentivirus in its natural host (3, 21, 22, 29, 40). Altering the negatively charged Asp at position 128 in human A3G to the positively charged Lys found in AGM-A3G makes the mutated human A3G (D128K) resistant to HIV-1 Vif but sensitive to SIVagm Vif (3, 21, 22, 29, 40). Position 128 of human A3F also contains a negatively charged amino acid, Glu. Unlike the situation in human A3G, modification of amino acid 128 in human A3F does not change its recognition by HIV-1 Vif or SIVagm Vif (16).
Collectively, our data support the argument that the N-terminal region of HIV-1 Vif mediates the binding of target molecules such as A3G and A3F. Our data also suggest that Vif may have evolved distinct protein interfaces in order to interact with both A3G and A3F. Thus, HIV-1 Vif has at least four distinct protein-protein interaction interfaces: the BC box with ElonginC, the HCCH motif with Cul5, and two separate interfaces with A3F and A3G.
Although Trp79 is not critical for A3G suppression, this residue is highly conserved among various HIV-1 strains. Trp79 is, however, critical for the suppression of A3F by HIV-1 Vif. Therefore, suppression of A3F by HIV-1 Vif is likely to be important for the survival of the virus in vivo (1, 8, 10, 25, 27, 37). This argument would be consistent with the in vivo observation of HIV-1 G-to-A mutation patterns. A3G primarily mediates GG-to-GA mutations, whereas A3F mostly generates GA-to-AA mutations (1, 8, 10, 25, 27, 37). GA-to-AA mutations are highly represented in viral sequences recovered from HIV-1-infected individuals (15). The highly conserved residues in Vif that are required for the suppression of A3F but not of A3G suggest that A3F represents a major selection force against HIV-1 in vivo.
Acknowledgments
We thank T. Sarkis, K. Luo, and B. Liu for advice and technical assistance; M. Malim and N. Landau for critical reagents; and D. McClellan for editorial assistance. The following reagents were obtained through the AIDS Research Reagents Program, Division of AIDS, NIAID, NIH: MAGI-CCR5 cells and the Vif polyclonal antibody.
W.Z. was supported by the National Science Fund of Zhejiang Province (Y204074). This work was supported by a grant from the NIH (AI062644) and funding from the National Science Foundation of China (NSFC-30425012) and Cheung Kong Scholars Program Foundation of the Chinese Ministry of Education to X-F.Y.
REFERENCES
- 1.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 5.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281-288. [DOI] [PubMed] [Google Scholar]
- 6.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 8.Goff, S. P. 2004. Retrovirus restriction factors. Mol. Cell 16:849-859. [DOI] [PubMed] [Google Scholar]
- 9.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 10.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi, M., A. Takaori-Kondo, Y. Miyauchi, K. Iwai, and T. Uchiyama. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-elongin B-elongin C complex is essential for Vif function. J. Biol. Chem. 280:18573-18578. [DOI] [PubMed] [Google Scholar]
- 12.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 14.Lee, T. H., J. E. Coligan, J. S. Allan, M. F. McLane, J. E. Groopman, and M. Essex. 1986. A new HTLV-III/LAV protein encoded by a gene found in cytopathic retroviruses. Science 231:1546-1549. [DOI] [PubMed] [Google Scholar]
- 15.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 16.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, B., X. Yu, K. Luo, Y. Yu, and X. F. Yu. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lochelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y. B. Kim, U. Truyen, U. Rosler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Munk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, K., Z. Xiao, E. Ehrlich, Y. Yu, B. Liu, S. Zheng, and X. F. Yu. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. USA 102:11444-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 21.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 22.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 23.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 24.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navarro, F., and N. R. Landau. 2004. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16:477-482. [DOI] [PubMed] [Google Scholar]
- 26.Noguchi, C., H. Ishino, M. Tsuge, Y. Fujimoto, M. Imamura, S. Takahashi, and K. Chayama. 2005. G to A hypermutation of hepatitis B virus. Hepatology 41:626-633. [DOI] [PubMed] [Google Scholar]
- 27.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10:291-297. [DOI] [PubMed] [Google Scholar]
- 28.Rosler, C., J. Kock, M. H. Malim, H. E. Blum, and F. von Weizsacker. 2004. Comment on “Inhibition of hepatitis B virus replication by APOBEC3G.” Science 305:1403. [DOI] [PubMed] [Google Scholar]
- 29.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 32.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 33.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 35.Suspene, R., D. Guetard, M. Henry, P. Sommer, S. Wain-Hobson, and J. P. Vartanian. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 102:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suspene, R., P. Sommer, M. Henry, S. Ferris, D. Guetard, S. Pochet, A. Chester, N. Navaratnam, S. Wain-Hobson, and J. P. Vartanian. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turelli, P., and D. Trono. 2005. Editing at the crossroad of innate and adaptive immunity. Science 307:1061-1065. [DOI] [PubMed] [Google Scholar]
- 38.Vodicka, M. A., W. C. Goh, L. I. Wu, M. E. Rogel, S. R. Bartz, V. L. Schweickart, C. J. Raport, and M. Emerman. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193-198. [DOI] [PubMed] [Google Scholar]
- 39.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 42.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 43.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 44.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]