Abstract
The gene for a bacterial enoyl-CoA hydratase (crotonase) homolog (HCHL) previously shown to convert 4-coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA to the corresponding hydroxybenzaldehydes in vitro provided an opportunity to subvert the plant phenylpropanoid pathway and channel carbon flux through 4-hydroxybenzaldehyde and the important flavor compound 4-hydroxy-3-methoxybenzaldehyde (vanillin). Expression of the Pseudomonas fluorescens AN103 HCHL gene in two generations of tobacco plants caused the development of phenotypic abnormalities, including stunting, interveinal chlorosis and senescence, curled leaf margins, low pollen production, and male sterility. In second generation progeny, the phenotype segregated with the transgene and transgenic siblings exhibited orange/red coloration of the vascular ring, distorted cells in the xylem and phloem bundles, and lignin modification/reduction. There was depletion of the principal phenolics concomitant with massive accumulation of novel metabolites, including the glucosides and glucose esters of 4-hydroxybenzoic acid and vanillic acid and the glucosides of 4-hydroxybenzyl alcohol and vanillyl alcohol. HCHL plants exhibited increased accumulation of transcripts for phenylalanine ammonia-lyase, cinnamate-4-hydroxylase, and 4-coumarate:CoA ligase, whereas β-1,3-glucanase was suppressed. This study, exploiting the ability of a bacterial gene to divert plant secondary metabolism, provides insight into how plants modify inappropriately accumulated metabolites and reveals the consequences of depleting the major phenolic pools.
INTRODUCTION
The plant phenylpropanoid pathway is responsible for the synthesis of a wide variety of secondary metabolic compounds, including lignins, salicylates, coumarins, hydroxycinnamic amides, flavonoid phytoalexins, pigments, UV light protectants, and antioxidants (Dixon and Paiva, 1995). In addition to their roles in the structure and protection of the plant, phenylpropanoids have an important effect on plant qualities such as texture, flavor, color, and processing characteristics. Molecular engineering provides a tool to determine the complex biochemical pathways involved in the synthesis and regulation of phenylpropanoids and to manipulate pathways to increase or initiate the production of economically desirable traits or compounds (Dixon et al., 1996). Expression of plant genes in heterologous plant systems can lead to improved or novel synthesis of valuable compounds (Yun et al., 1992) and improved disease resistance (Hain et al., 1993). The overexpression or downregulation of enzymes involved in phenylpropanoid and lignin biosynthesis has demonstrated that it is possible to alter the content and properties of lignin and its associated phenolics (reviewed in Boudet, 1998). This finding has important implications for the manipulation of plant quality with respect to pulping, forage digestibility, texture, and defense responses (Campbell and Sederoff, 1996). The expression of bacterial genes in transgenic plants also has proven to be effective, both in introducing new pathways to increase the accumulation of desired compounds (Fecker et al., 1993; Siebert et al., 1996) and in facilitating understanding of the plant defense response (Delaney et al., 1994).
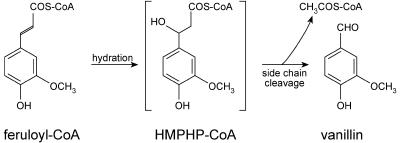
A novel bacterial enzyme was isolated previously from a Pseudomonas fluorescens strain (AN103) selected on the basis of its ability to metabolize ferulic acid via vanillin (Gasson et al., 1998; Narbad and Gasson, 1998). The bacterial enzyme, named 4-hydroxycinnamoyl-CoA hydratase/lyase (HCHL), is an enoyl-CoA hydratase with an aldolase function; in vitro, it can convert feruloyl-CoA into equimolar quantities of vanillin (4-hydroxy-3-methoxybenzaldehyde) and acetyl-CoA (Figure 1). Vanillin is the major flavor component of vanilla, one of the most important flavoring substances. The HCHL enzyme also converts 4-coumaroyl-CoA and caffeoyl-CoA in vitro to the corresponding hydroxybenzaldehydes (Mitra et al., 1999), one of which (4-hydroxybenzaldehyde) is a minor but important component of vanilla flavor. The enzyme does not act on sinapoyl-CoA. Expression of the Pseudomonas HCHL gene in plants has the potential to divert phenylpropanoid metabolism to produce novel metabolites based on hydroxybenzaldehyde derivatives. We have expressed the HCHL gene in two generations of tobacco and analyzed the phenotypic, biochemical, and molecular consequences.
Figure 1.
Conversion of Feruloyl-CoA to Vanillin by the HCHL Enzyme.
HCHL performs the hydration and cleavage of feruloyl-CoA to produce vanillin and acetyl-CoA in vitro. The enzyme also is able to reproduce feruloyl-CoA, in addition to vanillin and acetyl-CoA, from the artificial substrate and putative reaction intermediate 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl-CoA (HMPHP-CoA) (Gasson et al., 1998).
RESULTS
Generation of Tobacco Plants Expressing the Pseudomonas HCHL Gene
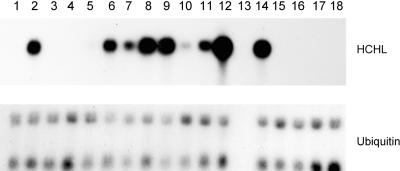
The wild-type form of the Pseudomonas HCHL gene and a modified form altered by one nucleotide to a more typical eukaryotic translation initiation context (atga to atgg; Kozak, 1986) were fused to the constitutive cauliflower mosaic virus (CaMV) 35S RNA promoter containing duplicated enhancer sequences. To express the bacterial gene in plants, we subcloned the expression cassettes into the Agrobacterium tumefaciens binary vector pBin19 and introduced them into tobacco by Agrobacterium-mediated leaf disc transformation. A total of 20 primary transformants were regenerated from leaf discs and eventually transferred to soil. The presence of the transgene in the regenerated plants was confirmed by genomic gel blot analysis (results not shown), and steady state mRNA accumulation was analyzed by hybridization of RNA gel blots with the HCHL gene fragment. Thirteen of the transformants expressed the transgene at detectable but varying levels (Figure 2); on longer exposures, all transformed plants expressed the transgene (results not shown). Three of these plants contained the wild-type form of the HCHL gene, whereas the rest contained the modified form.
Figure 2.
Steady State mRNA Levels of the HCHL Transcript in Independent Primary Transformants.
Total RNA from young leaves of primary transformants was hybridized to the HCHL gene fragment. Lanes 1 to 12, plants containing the pmHCHL construct: plants 3, 6, 7, 9, 11, 13, 21, 22, 27, 34, 37, and 39; lane 13, no sample; lanes 14 to 16, plants containing the pHCHL construct: plants 201, 210, and 224; lanes 17 and 18, control plants transformed with the empty pBin19 vector. The same blot was stripped and hybridized to a Datura stramonium ubiquitin probe.
Phenotypes of HCHL Primary Transformants and Their Progeny
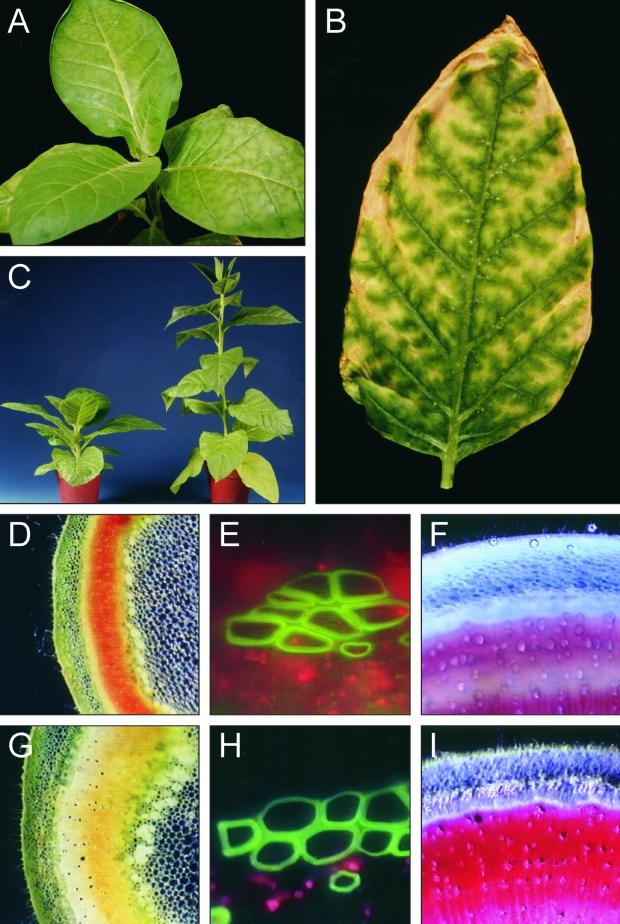
Of the 13 independent primary transformants expressing detectable levels of HCHL mRNA, three plants with the highest accumulation of the transcript (plants 201, 27, and 39) exhibited severely abnormal phenotypes from an early stage upon transfer to soil in the greenhouse. The abnormal phenotype was characterized by the development of interveinal leaf chlorosis and thin, curled leaf margins (Figure 3A). Leaf chlorosis began as paler green areas between veins that appeared relatively sunken and shiny compared with adjacent tissue. As the leaves aged, the lighter green areas became yellow; this was followed by a brown, senescent-like appearance, with the senescent-like tissue spreading out between the veins, which remained green (Figure 3B). The senescent-like areas of the transgenic leaves were paler than was the natural senescence of the old leaves of wild-type plants. During flowering, the three plants exhibiting leaf chlorosis produced little pollen and were male sterile. Three other plants (transformants 3, 6, and 22) did not develop interveinal leaf chlorosis but did produce first flowers with very reduced pollen content that failed to set seed after self-fertilization, although later flowers were male fertile. No obvious phenotypic abnormalities were detected in any of the other HCHL-expressing plants.
Figure 3.
Morphology and Microscopic Analysis of Transgenic Plants.
(A) Primary transformant 201 at 22 days after transfer to the greenhouse.
(B) Detached leaf of primary transformant 201 at 74 days after transfer (after flowering).
(C) to (I) Comparisons between transgenic and syngenic progeny of line 201. (C) shows sibling plants of line 201 at 90 days after sowing, demonstrating delayed elongation of stem internodes in transgenic plants (left). Unstained stem sections showing an orange/red coloration of the vascular ring in transgenic plants (D) compared with the yellow color observed in syngenic plants (G) and distortion of phloem fibers in the transgenic stem (E) compared with the syngenic plants (H) were viewed under UV light fluorescence. Phloroglucinol staining of vascular tissue in transgenic stems (F) was reduced compared with that of the syngenic samples (I).
Four of the primary transformants were selected for further study: three plants exhibiting the interveinal leaf chlorosis phenotype (plants 201, 39, and 27) and a fourth plant that expressed the HCHL transgene but did not exhibit phenotypic abnormalities (plant 13). Only plant 201 expressed the unmodified form of the transgene. To analyze segregating progeny of the second generation, we pollinated the three male-sterile transformants with wild-type pollen, whereas primary transformant 13 was self-fertilized. Resulting seed were sown directly onto soil, and the transgenic status of the progeny plants was assessed by gel blot analysis of leaf RNA (see below). Progeny from all four lines clearly segregated into transgenic and syngenic siblings, with a complete correlation between HCHL expression and abnormal phenotypes. It was not possible to easily categorize transgenic siblings into homozygous or heterozygous phenotypes. Transgenic progeny from all four lines exhibited the interveinal leaf chlorosis phenotype when young, which in the case of line 13 indicated penetration of the abnormal phenotype only in the second generation.
From an early stage, the transgenic progeny of line 201 developed thin, curled leaf margins and leaf tip chlorosis before the development of interveinal chlorosis. Transgenic progeny of this line were markedly stunted compared with the syngenic siblings (Figure 3C). Eventually, the internodes of the transgenic line 201 progeny elongated, so that flowering occurred at approximately the same height as in the syngenic siblings but with a delay of 2 to 3 weeks. Initial flowers of the transgenic line 201 plants were male sterile, but later flowers were male fertile. None of the transgenic progeny from the other three lines exhibited stunting or flowering delay, and the interveinal leaf chlorosis was much less severe than in line 201 plants. Flowers of these other lines became male fertile soon after the beginning of flowering, with only a few flowers failing to develop seed capsules after self-pollination. In all four lines, the petals of the transgenic plants were noticeably paler that those of the syngenic siblings.
Examination of detached leaves of line 201 segregating progeny under UV light (365 nm) revealed a distinct difference in fluorescence between transgenic and syngenic siblings (results not shown). In syngenic leaves, the red-orange autofluorescence emitted by chloroplasts in response to excitation by UV light was shielded by phenolic compounds, so that the leaves appeared dark green/blue (Chapple et al., 1992; Tamagnone et al., 1998). In contrast, transgenic leaves appeared red-orange under UV light, suggesting depletion of the phenolic content.
Microscopic Examination of Second Generation Segregating Progeny
Hand-cut sections of stem taken from nodes 10 and 12 of transgenic and syngenic plants of line 201 during late flowering were examined unstained using light microscopy. The vascular ring in stems of transgenic plants had developed a red-orange coloring compared with the yellow of the syngenic siblings (Figures 3D and 3G). At higher magnification, the xylem vessels in transgenic and syngenic plant stems appeared to have a similar cell wall thickness, but some distortion and buckling was apparent in both xylem vessels and phloem bundle cells of transgenic plants (Figures 3E and 3H). The effect of HCHL expression on lignin content was examined by microscopic analysis of phloroglucinol-stained stem sections. Deep cherry-red coloring of syngenic plant stem sections was reduced in intensity and extent in the transgenic siblings (Figures 3F and 3I), indicating a change in the quantity and/or quality of lignin.
Effect of the HCHL Transgene on Expression of Genes of the Phenylpropanoid Pathway
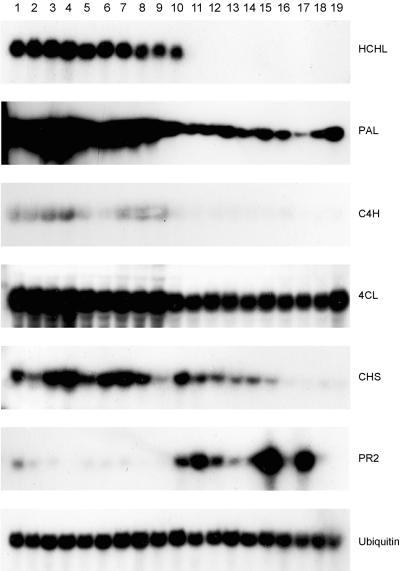
A total of 79 second generation progeny plants of lines 201, 39, 27, and 13 were examined by RNA gel blot analysis, confirming 100% linkage of HCHL expression with the development of phenotypic abnormalities. To assess the effect of transgene expression on the mRNA accumulation of the phenylpropanoid pathway biosynthetic enzymes, probes for phenylalanine ammonia-lyase (PAL), cinnamate-4-hydroxylase (C4H), and 4-coumarate:CoA ligase (4CL) as well as chalcone synthase (CHS) and the pathogenesis-related protein β-1,3-glucanase (PR2) were used to hybridize to total leaf RNA from segregating progeny of line 201 (Figure 4). Leaves were analyzed before the development of the chlorosis phenotype. Steady state mRNA levels for the phenylpropanoid pathway enzymes PAL, C4H, and 4CL were clearly increased, and CHS was generally increased in the transgenic progeny, whereas the stress-associated PR2 gene was generally suppressed. We do not know whether these changes were a direct consequence of changes in phenylpropanoid metabolite levels or an indirect response to the cellular perturbation occurring in the transgenic leaves.
Figure 4.
Steady State mRNA Levels of the HCHL Message and Endogenous Plant Genes in Progeny of Line 201.
Lanes 1 to 10, transgenic; lanes 11 to 19, syngenic.
HCHL Enzyme Synthesis and Activity in Transgenic Progeny
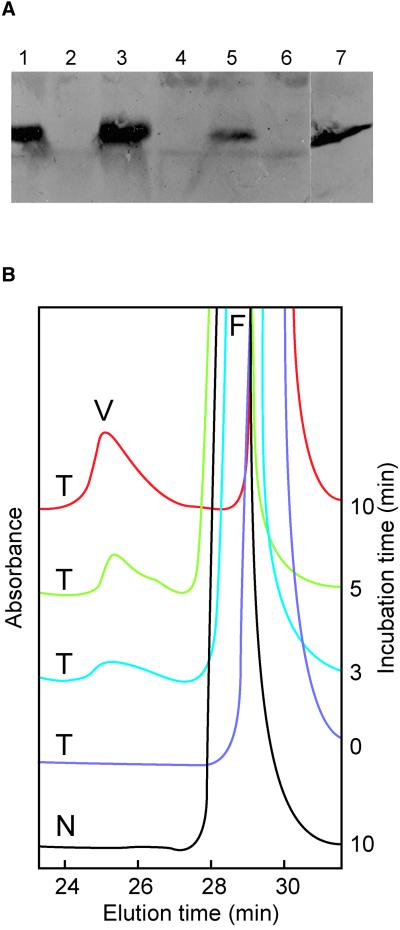
Synthesis of the HCHL protein in transgenic leaves was confirmed by protein gel blotting and detection with a polyclonal antibody (Figure 5A). The antibody did not recognize any proteins in the syngenic leaf samples. One protein band of ∼31 kD was detected in extracts from transgenic plants and was similar in size to the protein found in the original Pseudomonas strain that was the source of the HCHL gene (Gasson et al., 1998).
Figure 5.
HCHL Enzyme Synthesis in Young Leaves of Transgenic Progeny and Activity in Vitro.
(A) Protein gel blot analysis of total protein extracts from lines 201 and 27. Lane 1, transgenic, line 201, set 1; lane 2, syngenic, line 201, set 1; lane 3, transgenic, line 201, set 2; lane 4, syngenic, line 201, set 2; lane 5, transgenic, line 27; lane 6, syngenic, line 27; lane 7, total protein extract from Pseudomonas strain AN103. Line 201 was sown on two separate occasions (sets 1 and 2).
(B) In vitro production of vanillin (V) by the incubation of 10 μL of cell-free extracts from a transgenic offspring (T) of line 201 with feruloyl-CoA (F) at 30°C for 0, 3, 5, and 10 min. Incubation with extracts from a syngenic plant (N) showed no accumulation of vanillin at any time. HPLC analysis was viewed at 310 nm.
HCHL enzyme activity was measured using feruloyl-CoA as a substrate with subsequent detection of vanillin production by HPLC (Figure 5B, Table 1). The identification of vanillin was confirmed by gas chromatography–mass spectrometry (results not shown). Extracts from young leaves of second generation plants were made from the same liquid nitrogen–frozen ground powder used for RNA extraction and biochemical analysis. In all cases, in vitro vanillin production and hence HCHL activity were absent in syngenic plant extracts, whereas all transgenic plant extracts produced vanillin in varying quantities. Both the control and transgenic plant extracts contained unidentified activity that depleted feruloyl-CoA to some extent, producing an unidentified peak running at 11 min, so the incubation times for the HCHL assays were reduced to 10 min. Under these conditions, exhaustion of the substrate was prevented and the enzyme assay was linear. The HCHL enzyme activities correlated well with the level of HCHL mRNA, with line 13 progeny having more than twofold less activity than lines 201 and 39. Line 27 was an exception in that its HCHL activity was lower than predicted from the relative HCHL mRNA accumulation in the parent plant.
Table 1.
In Vitro HCHL Enzyme Activities in Young Leaves from Transgenic Progeny
| Plant Linea | No. Plants Sampled (n) |
Mean Enzyme Activity ±se (pmol vanillin per sec/μg protein) |
|---|---|---|
| 201, set 1, T | 4 | 0.152 ± 0.019 |
| 201, set 1, N | 4 | NDb |
| 201, set 2, T | 10 | 0.144 ± 0.019 |
| 201, set 2, N | 9 | ND |
| 27, T | 12 | 0.070 ± 0.009 |
| 27, N | 7 | ND |
| 39, T | 10 | 0.138 ± 0.010 |
| 39, N | 1 | ND |
| 13, T | 5 | 0.061 ± 0.014 |
| 13, N | 5 | ND |
| 201, set 1, F, T | 5 | 0.221 ± 0.051 |
| 201, set 1, F, N | 3 | ND |
a F, taken during flowering; T, transgenic; N, syngenic. Line 201 was sown on two separate occasions (set 1 and set 2).
b ND, not detected.
Biochemical Changes in HCHL Second Generation Plants
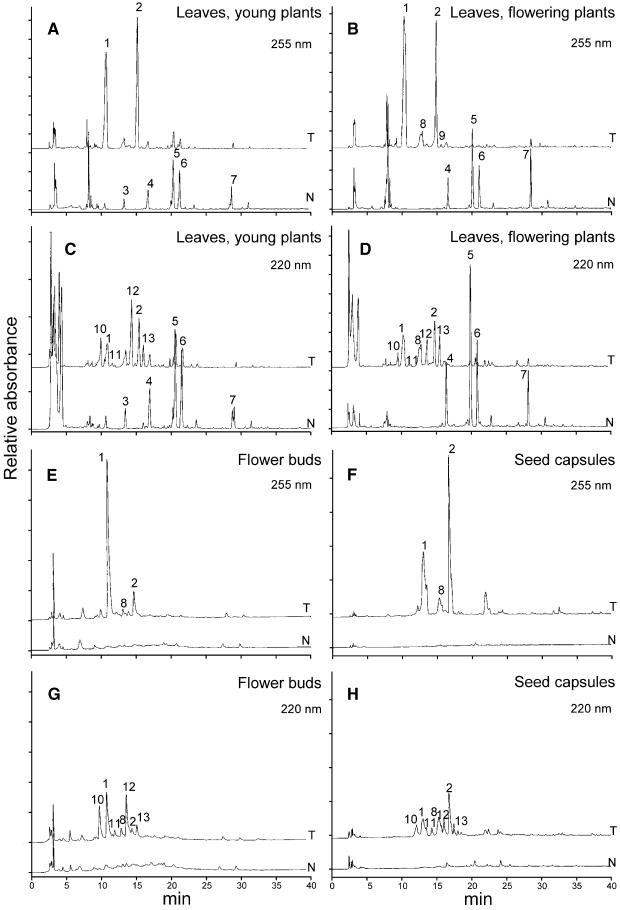
To determine changes in the level of soluble phenolics in HCHL-expressing transgenic progeny, we used HPLC with UV light detection at 220 and 255 nm (Figure 6) to analyze methanol extracts of leaves, unopened flower buds, and seed capsules. Marked differences in phenolic content were observed in young leaves of transgenic and syngenic plants before flowering. In young leaves of syngenic plants, the major phenolic peaks corresponded to chlorogenic acid (5-caffeoylquinic acid), its isomers (4-caffeoylquinic acid and 3-caffeoylquinic acid), and the major flavonoid rutin (quercetin 3-β-d-rutinoside). In young leaves of transgenic plants, these phenolics were depleted severely, although the results for rutin were less consistent. The relative depletion of these phenolics was more pronounced in the late leaves of flowering plants of line 201, with an average 93% reduction in chlorogenic acid in the transgenic progeny (Table 2). All of the major phenolics in syngenic plants were present at higher levels in the later leaves of flowering plants compared with its presence in young leaves before flowering. Depletion of these major phenolics was less pronounced in lines 27 and 13, which also exhibited lower HCHL enzyme activities.
Figure 6.
HPLC Analysis of Soluble Phenolics in Young Leaves, Unopened Flower Buds, and Seed Capsules of the Progeny of Line 201.
Phenolic profiles of transgenic (T) and syngenic (N) progeny of line 201 were compared at 255 and 220 nm in young leaves from young plants ([A] and [C]) and flowering plants ([B] and [D]), flower buds ([E] and [G]), and seed capsules ([F] and [H]). Key to peak identity: 1, 4-hydroxybenzoic acid glucoside; 2, 4-hydroxybenzoic acid glucose ester; 3, caffeoyl putrescine; 4, 3-caffeoylquinic acid; 5, chlorogenic acid; 6, 4-caffeoylquinic acid; 7, rutin; 8, vanillic acid glucoside; 9, vanillic acid glucose ester; 10, 4-hydroxybenzyl alcohol 4-O-glucoside; 11 and 13, vanillyl alcohol glucosides; 12, 4-hydroxybenzyl alcohol glucoside. Peaks were confirmed by mass spectrometry (see Figure 7).
Table 2.
Accumulation of Major Soluble Phenolics in Young Leaves of HCHL Transgenic and Syngenic Progeny
| Mean ±se (μg/g−1 fresh weight)b
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Plant Linea | n | 3-CG | % | CGA | % | 4-CG | % | Rutin | % |
| 201, set 1, T | 4 | 79 ± 12 | 40 | 200 ± 41 | 86 | 160 ± 30 | 76 | 9 ± 2 | 82 |
| 201, set 1, N | 4 | 132 ± 16 | 1424 ± 45 | 641 ± 14 | 49 ± 11 | ||||
| 201, set 2, T | 10 | 99 ± 8 | 63 | 238 ± 25 | 81 | 184 ± 16 | 70 | 25 ± 5 | 72 |
| 201, set 2, N | 9 | 271 ± 27 | 1258 ± 180 | 604 ± 60 | 90 ± 8 | ||||
| 27, T | 13 | 132 ± 8 | 14 | 500 ± 39 | 48 | 277 ± 16 | 33 | 32 ± 6 | 60 |
| 27, N | 7 | 153 ± 4 | 970 ± 139 | 412 ± 41 | 20 ± 7 | ||||
| 39, T | 12 | 114 ± 12 | 11 | 370 ± 50 | 76 | 209 ± 25 | 64 | 50 ± 10 | 49 |
| 39, N | 1 | 128 | 1540 | 579 | 99 | ||||
| 13, T | 5 | 83 ± 6 | 33 | 542 ± 35 | 48 | 240 ± 12 | 30 | 55 ± 15 | 26 |
| 13, N | 5 | 124 ± 16 | 1033 ± 116 | 341 ± 23 | 74 ± 9 | ||||
| 201, set 1, TF | 5 | 155 ± 12 | 68 | 124 ± 8 | 93 | 105 ± 4 | 86 | 63 ± 7 | 66 |
| 201, set 1, NF | 3 | 493 ± 101 | 1715 ± 637 | 776 ± 147 | 186 ± 64 | ||||
a Line 201 was sown on two separate occasions (set 1 and set 2). T, transgenic; N, syngenic; F, taken during flowering.
b 3-CG, 3-caffeoylquinic acid; CGA, chlorogenic acid; 4-CG, 4-caffeoylquinic acid. Percentage change between syngenic and transgenic mean values.
The rerouting of the phenylpropanoid pathway away from the major phenolics became evident with the concomitant appearance of two major peaks at 255 nm (Figure 6) in extracts from young leaves of young plants. Comparison with known spectra and standards suggested that the peaks represented 4-hydroxybenzoic acid glucoside (4-HBAG) and 4-hydroxybenzoic acid glucose ester (4-HBAGE). This identification was confirmed by positive and negative ion mass spectrometry (Figure 7A). In the late leaves from older flowering plants of line 201, these peaks were more prominent, and the average fresh weight content of 4-HBAG was >0.2% (Table 3). Mass spectrometry also demonstrated the presence of vanillic acid glucoside and vanillic acid glucose ester (Figure 7B). These peaks were noticeable in HPLC traces from late leaves of transgenic flowering plants, unopened flower buds, and brown seed capsules (Figure 6). Flower buds and seed capsules of line 201 transgenic plants accumulated high levels of 4-HBAG and 4-HBAGE, with the average level of 4-HBAG reaching ∼0.5% of fresh weight in seed capsules (Table 3).
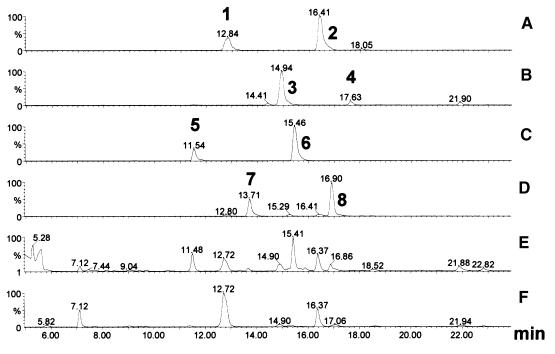
Figure 7.
Liquid Chromatography–Mass Spectrometry Analysis of Benzaldehyde Derivatives.
Positive ion mass chromatograms of the sodium ([M + Na]+) adducts of the molecules of mass 323 (peak 1, 4-hydroxybenzoic acid glucoside; peak 2, 4-hydroxybenzoic acid glucose ester) (A); 353 (peak 3, vanillic acid glucoside; peak 4, vanillic acid glucose ester) (B); 309 (peaks 5 and 6, 4-hydroxybenzyl alcohol glucosides) (C); 339 (peaks 7 and 8, vanillyl alcohol glucosides) (D); and the corresponding UV light chromatograms at 220 nm (E) and 254 to 256 nm (F). The identities of these molecules were confirmed by their characteristic full scan spectra combined with the retention times of known standards. After subtraction of background peaks, the following characteristic mass spectra were obtained (only peaks with intensities >20% of base are reported): peak 1, 323 (100), 139 (67), 121 (22), 95 (70); peak 2, 323 (36), 139 (96), 121 (52), 95 (100); peak 3, 353 (100), 169 (69), 134 (30), 125 (72), 111 (35); peak 4, 353 (100), 125 (27); peak 5, 309 (14), 120 (22), 107 (44), 105 (100); peak 6, 309 (35), 107 (100); peak 7, 339 (70), 137 (100); peak 8, 339 (40), 137 (100).
Table 3.
Quantitative Analysis of Soluble Phenolics in HCHL Transgenic Progenya
| Mean ±se (mg/g−1 fresh weight)
|
||||||
|---|---|---|---|---|---|---|
| Organ | Plant Lineb | n | 4-HBAG | 4-HBAGE | 4-HBOHG | VAGc |
| Leaves | 201, set 1 | 5 | 0.54 ± 0.11 | 0.18 ± 0.03 | 0.06 ± 0.02 | ND |
| 201, set 2 | 10 | 0.98 ± 0.05 | 0.36 ± 0.04 | 0.23 ± 0.01 | ND | |
| 27 | 12 | 0.56 ± 0.05 | 0.17 ± 0.01 | 0.08 ± 0.01 | ND | |
| 39 | 10 | 0.60 ± 0.05 | 0.25 ± 0.02 | 0.14 ± 0.01 | ND | |
| 13 | 5 | 0.32 ± 0.04 | 0.11 ± 0.02 | 0.07 ± 0.02 | ND | |
| 201, set 1, F | 5 | 2.33 ± 0.20 | 0.59 ± 0.04 | 0.20 ± 0.00 | 0.37 ± 0.04 | |
| Buds | 201, set 1 | 5 | 1.65 ± 0.10 | 0.15 ± 0.01 | 0.47 ± 0.02 | 0.12 ± 0.02 |
| Capsules | 201, set 1 | 3 | 4.58 ± 0.22 | 3.13 ± 0.36 | 1.03 ± 0.16 | 1.77 ± 0.20 |
a Mean accumulation of hydroxybenzaldehyde derivatives in young leaves, unopened flower buds, and mature seed capsules of transgenic HCHL progeny.
b Line 201 was sown on two separate occasions (set 1 and set 2). F, taken during flowering.
c VAG, vanillic acid glucoside; ND, not determined.
At 220 nm, four new peaks were identified in the leaves of transgenic plants. These were identified by mass spectrometry as vanillyl alcohol derivatives (Figure 7C) and 4-hydroxybenzyl alcohol glucosides (Figure 7D), and one (Figure 6, peak 10) had a retention time identical to that of a 4-hydroxybenzyl alcohol 4-O-glucoside (4-HBOHG) standard. The highest levels of 4-HBOHG were found in brown seed capsules and unopened flower buds, and the highest levels of all of the 4-hydroxybenzoic acid, 4-hydroxybenzyl alcohol, and vanillic acid derivatives were found in seed capsules of line 201 transgenic plants. The ratio of vanillic acid glucoside to 4-HBAG was higher in seed capsules (0.386) than in leaves (0.159) or green buds (0.07). Mass spectrometry confirmed that there was no detectable accumulation of protocatechuic aldehyde derivatives.
Another consequence of this production of new phenolic compounds in flowers was a decrease in petal anthocyanin levels. Line 201 transgenic plants, which exhibited the strongest reduction in chlorogenic acid, also presented the greatest reduction in anthocyanin. The mean level of anthocyanins in petals of line 201 syngenic plants was 75.4 ± 6.1 μmol/g fresh weight, whereas the line 201 transgenic plants contained 32.2 ± 0.9 μmol/g fresh weight. Together, these results suggest that expression of the HCHL gene caused a major diversion from the phenylpropanoid pathway to the production of novel 4-hydroxybenzaldehyde and vanillin derivatives.
DISCUSSION
The activity of the HCHL transgene in tobacco had two principal biochemical consequences: depletion of the major phenolic pools and accumulation of the novel chain-shortened C6C1 derivatives. Development of the abnormal morphology of the transgenic plants most likely was caused by the biochemical perturbation. There was a good correlation between the level of HCHL enzyme activity and the severity of the phenotype. The altered morphological phenotype of the HCHL transgenic plants was very similar to that of plants with reduced PAL activity. Cosuppression of PAL in transgenic tobacco by overexpression of a bean PAL construct led to stunted plants with curled leaves that developed localized lesions that were fluorescent in UV light and that were identical to the interveinal chlorosis seen with the HCHL plants (Elkind et al., 1990). In addition, flowers of one PAL-suppressed plant produced anthers with less pollen and decreased pollen viability (Elkind et al., 1990).
In the second generation of severely PAL-suppressed plants, the pattern of soluble phenolics in leaves was similar to that in control plants, but there was substantial depletion of chlorogenic acid and rutin (98 and 91%, respectively). Similarly, the severely affected HCHL line 201 showed up to 93 and 82% depletion of chlorogenic acid and rutin, respectively. Because chlorogenic acid represents 60% and rutin represents 10% of the total phenolic pool in tobacco (Snook et al., 1986), expression of HCHL activity and PAL suppression both cause major depletion of total original phenolics in tobacco. Because an 88% depletion of chlorogenic acid caused by antisense inhibition of C4H did not result in interveinal chlorosis (Blount et al., 2000), this phenotypic abnormality seen with PAL suppression and expression of HCHL activity cannot be attributed solely to depletion of the main phenolic pool. Sense cosuppression of 4CL in tobacco (Kajita et al., 1996) and antisense suppression in Arabidopsis (Lee et al., 1997) did not lead to a leaf chlorosis phenotype. It is possible that some differences exist between the various types of transgenic plants, leading to subtly different spatial expression of the transgene, which may account for the varying morphological phenotypes with similar levels of phenolic pool depletion.
The reduction in pollen amount and viability seen in HCHL-expressing primary transformants and PAL-suppressed plants may be attributable to a reduction in the general flavonoid pathway and, more specifically, in flavonols. We did not quantitate flavonol content in the HCHL plants, but the major flavonoid rutin was depleted by up to 82% in leaves of line 201. The reduced fertility of the HCHL transgenic pollen in relation to the severe depletion of the flavonoid pool agrees with the proposed role of flavonoids in pollen development (Mo et al., 1992; van der Meer et al., 1992; Ylstra et al., 1992). In parallel with reduced pollen viability in HCHL-expressing plants, there was a severe reduction in anthocyanin content in petals of transgenic progeny (>70% in line 201). Some petals in PAL-suppressed tobacco were white, again suggesting extreme depletion of anthocyanins (Elkind et al., 1990). Similar floral phenotypes in tobacco were seen when the grapevine stilbene synthase was expressed from a CaMV 35S promoter (Fischer et al., 1997). Stilbene synthase uses malonyl-CoA and 4-coumaroyl-CoA as substrates to form the trihydroxystilbene resveratrol, and this rerouting of 4-coumaroyl-CoA into resveratrol resulted in flowers with inviable pollen and pale pink, almost white petals (Fischer et al., 1997).
Morphological and biochemical phenotypes similar to those caused by expression of the HCHL gene also were observed in other transgenic tobacco plants. A single tobacco transformant with reduced cinnamoyl-CoA reductase (CCR) activity exhibited a morphological phenotype that was very similar to the HCHL phenotype, with stunting, small leaves with curled margins, and interveinal discoloration or chlorosis (Piquemal et al., 1998). MYB transcription factors seem to repress the phenylpropanoid pathway after the cinnamic acid stage, and a series of transgenic tobacco plants were produced overexpressing the Antirrhinum majus Myb308 and Myb330 genes (Tamagnone et al., 1998). Myb308 plants were stunted, with reduced elongation of stem internodes during development and a striking leaf phenotype consisting of reduced leaf expansion and interveinal leaf necrosis. The leaf phenotype differed from that of HCHL plants in that the interveinal necrosis produced areas of white, dead tissue rather than the brown, senescent-like tissue seen with the HCHL plants. Flowers of Myb308 plants also had petals with reduced pigmentation, but the pollen was fertile. Leaf extracts of the Myb308 plants were deficient in all detectable soluble phenolics, with chlorogenic acid being reduced on average by 80% and flavonoid compounds being reduced to a lesser extent.
Lignin is a major component of secondary cell walls and is made by the polymerization of cinnamyl alcohols. Reduction of carbon flux through the phenylpropanoid pathway evident in the HCHL plants might be expected to have profound effects on lignin formation. Coloration of the xylem ring was described previously in tobacco plants with reduced 4CL activity (Kajita et al., 1996), reduced CCR activity (Piquemal et al., 1998), and reduced cinnamyl alcohol dehydrogenase (CAD) activity (Halpin et al., 1994; Hibino et al., 1995; Yahiaoui et al., 1998). Xylem coloration also was observed in a loblolly pine mutant carrying a null CAD allele (MacKay et al., 1995), in poplar with downregulated CAD (Baucher et al., 1996), in a maize mutant of caffeic acid O-methyltransferase (COMT) (Vignols et al., 1995), and in transgenic trees with reduced COMT (van Doorsselaere et al., 1995; Tsai et al., 1998). A similar downregulation of COMT in tobacco did not lead to colored xylem (Atanassova et al., 1995), nor was it seen in the xylem ring of Myb308 and Myb330 plants (Tamagnone et al., 1998). The coloration of lignin may be attributable to the incorporation of nonlignin phenolics, with different phenolics imparting different colors (Higuchi et al., 1994; Piquemal et al., 1998; Yahiaoui et al., 1998).
The results we obtained with phloroglucinol staining indicate a quantitative and/or qualitative change in the lignin composition of HCHL plants. Similar decreases in phloroglucinol staining were observed in planta with modified phenylpropanoid and lignin metabolism, for example, in PAL- and CCR-suppressed tobacco plants (Bate et al., 1994; Piquemal et al., 1998). The most severely CCR-suppressed transformant exhibited collapsed vessels similar to those seen in HCHL plants (Piquemal et al., 1998), whereas Elkind et al. (1990) observed thinning of xylem cell walls in PAL-suppressed plants.
The singular aspect of using the Pseudomonas HCHL gene in tobacco was the extensive rerouting of carbon flow from phenylpropanoids into the synthesis of novel metabolites, consisting of the glucosides and glucose esters of the acid and alcohol derivatives of 4-hydroxybenzaldehyde and vanillin but with a complete absence of the protocatechuic forms. The extent of this rerouting was massive: the 4-HBAG in seed capsules of line 201 plants accumulated to approximately 4.5 mg/g fresh weight (i.e., 0.45% of the total fresh weight content). It is not surprising that the HCHL products are glucosylated, because complete glucosylation of the artificial metabolite 4-hydroxybenzoate produced in tobacco cell cultures by expression of the bacterial ubiC gene encoding chorismate pyruvate lyase was observed previously (Siebert et al., 1996). Two constitutive glucosyltransferase activities producing the glucoside and glucose ester forms of 4-hydroxybenzoic acid were able to glucosylate 4-hydroxybenzoate to produce glucosylated derivatives representing 0.52% of the dry weight of the suspension cultures (Siebert et al., 1996) without a concomitant increase of the glucosyltransferase activities (Li et al., 1997). Less predictable in the HCHL plants was a complete absence of the aldehyde forms of the chain-shortened phenylpropanoids, with the acid form predominating.
The decreased accumulation of the vanillin-derived compounds compared with the 4-hydroxybenzaldehyde–derived compounds may be attributable to the kinetics of the metabolic diversion, because 4-coumaroyl-CoA is the precursor of caffeoyl-CoA and feruloyl-CoA. The vanillic acid derivatives accumulated to high levels in seed capsules, and the ratio between the vanillic acid forms and 4-hydroxybenzoate also was higher in seed capsules. Complete lack of the protocatechuic acid forms (i.e., the forms derived from caffeoyl-CoA) may result from metabolic channeling causing less access to the HCHL enzyme of the caffeoyl-CoA than of the feruloyl-CoA. Channeling was proposed previously for the phenylpropanoid and flavonoid pathways (Rasmussen and Dixon, 1999; Winkel-Shirley, 1999). Similar rapid conversion of 4-hydroxybenzaldehyde and vanillin in the HCHL plants to the acid and alcohol forms before glucosylation could explain the lack of glucosylated aldehydes. No benzyl alcohol glucoside derivatives were reported in tobacco cell lines or plants or in Lithospermum erythrorhizon hairy root cultures expressing the Escherichia coli ubiC gene (Siebert et al., 1996; Li et al., 1997; Sommer and Heide, 1998; Sommer et al., 1999). However, the HPLC analysis used absorbance at 254 nm, whereas the benzyl alcohol derivatives are detected optimally at 220 nm. Thus, we do not know whether 4-HBOG also was converted to the alcohol form in these systems.
Site-directed mutagenesis of the HCHL open reading frame to render the translation initiation context more typically eukaryotic (Kozak, 1986) did not produce a clear indication of whether the modification increased HCHL activity. Our sample size of the unmodified HCHL gene plants probably was too small to evaluate the effect of the modification confidently. Sommer and Heide (1998) performed a similar modification to the E. coli ubiC gene but did not determine the effect of this modification in planta.
The molecular response of the HCHL plants of line 201 was intriguing. Transcripts of PAL, C4H, and 4CL clearly were upregulated, CHS also was upregulated in general, but the PR2 gene was downregulated markedly. It is not possible to determine from our data whether the upregulation of the phenylpropanoid pathway and CHS was caused by feedback control by metabolite levels or was an indirect response to generalized cellular stress present in the HCHL plants. Stress induction of PAL, 4CL, and CHS transcripts has been reported in a number of systems (reviewed in Dixon and Paiva, 1995). However, the repression of the stress-inducible PR2 gene (Ward et al., 1991) suggests that the upregulation of the phenylpropanoid pathway and the CHS gene was attributable not to stress but to metabolic feedback. Transcript levels of PAL, C4H, 4CL, and CAD were repressed in Myb308 plants (Tamagnone et al., 1998), even though phenolic levels were depleted severely in these plants. The contrasting behavior of the HCHL and Myb308 phenylpropanoid pathway genes in the presence of a depleted phenolic pool may be explained by the proposed role of the Myb308 protein as a competitive inhibitor of other activating factors (Tamagnone et al., 1998), thereby suppressing transcription of the phenylpropanoid pathway genes.
In conclusion, expression of the Pseudomonas HCHL gene in tobacco caused massive rerouting of carbon flux from the phenylpropanoid pathway to novel C6C1 derivatives. The aldehyde products of HCHL activity were not detected, suggesting rapid conversion to the acid and alcohol forms. It is possible that glucosylation occurred after the dehydrogenation step, because no glucosylated aldehyde ester was detected. The ratio of vanillic acid to 4-hydroxybenzoic acid derivatives was higher in seed capsules, indicating where future refinement of the metabolic engineering for flavor compounds should be targeted.
METHODS
Vector Construction
Molecular experiments were performed using standard protocols (Sambrook et al., 1989) or according to manufacturers' instructions. Restriction enzymes were obtained from Promega (Southampton, UK); general biochemicals were obtained from Sigma (Poole, Dorset, UK) unless stated otherwise.
Cosmid pFI1039 (Gasson et al., 1998) was restricted with EcoRI and BamHI to excise an 864-bp fragment encoding the 4-hydroxycinnamoyl-CoA hydratase/lyase (HCHL) enzyme, extending from −29 upstream of the ATG to 6 bp downstream of the stop codon. The fragment was ligated into the corresponding sites in pBluescript SK+ (Stratagene) and then excised with BamHI and HindIII and subcloned into the BamHI and HindIII sites of pJIT60, placing the sequence in a sense orientation between a cauliflower mosaic virus (CaMV) 35S RNA promoter with duplicated enhancer sequences and the CaMV termination sequence (Guerineau et al., 1992). The plasmid was restricted first with SacI and then partially with EcoRV, and the 2322-bp cassette was ligated into the SacI and SmaI sites of pBin19 (Bevan, 1984) to produce the construct pHCHL.
A second construct, pmHCHL, was produced based on a polymerase chain reaction (PCR)–amplified copy of the HCHL coding sequence incorporating a 1-bp change after the start codon (ATGAGC to ATGGGC) to improve the translational initiation context for plant expression (Kozak, 1986). Oligonucleotide primers (sense, 5′-ATCGCCATGGGCACATACGAAGGTC-3′; antisense, 5′-TCCTTCAGCGTTTATACGC-3′) to produce a 1-bp modification were synthesized in an ABI synthesizer (Applied Biosystems, Cheshire, UK). An 843-bp sequence (extending from −6 upstream of the ATG to 6 bp downstream of the stop codon) was amplified from pFI1039 with Pfu DNA polymerase (Stratagene) according to the manufacturer's instructions. The PCR product was ligated into the EcoRV site of pBluescript SK+, excised with EcoRI and SalI, and subcloned into the corresponding sites of pJIT60. The 2332-bp cassette was excised with EcoRV and KpnI and subcloned into the SmaI-KpnI sites of pBin19. pHCHL and pmHCHL constructs and an empty pBin19 vector were transferred into Agrobacterium tumefaciens strain LBA4404 by triparental mating as described by Bevan (1984).
Plant Material and Transformation
Leaf discs of tobacco (Nicotiana tabacum cv Xanthi XHFD8) were transformed with A. tumefaciens (Horsch et al., 1985). Discs were incubated for 2 days on 1 × Murashige and Skoog (1962) medium (Melford Laboratories, Ipswich, UK) and then transferred to selective medium (1 × Murashige and Skoog medium incorporating 300 mg/L carbenicillin disodium [Melford Laboratories], 100 mg/L kanamycin sulfate, 10 mg/L dimethylaminopurine, and 1 mg/L indole acetic acid). After 3 to 5 weeks, plantlets were transferred to rooting medium (1 × Murashige and Skoog medium incorporating 300 mg/L carbenicillin and 50 mg/L kanamycin). Rooted plants were transferred to pots in a greenhouse (day temperature, 20°C; night temperature, 18°C; 16-hr day lit by high pressure sodium lamps). Plants were fed weekly with Solufeed high potash fertilizer (Kings Horticulture, Essex, UK).
Molecular Analysis of Plants
For RNA, DNA, and biochemical analyses, young leaves were taken from plants before flowering at nodes 9 and 10. Leaf samples also were taken during the early stages of flowering (approximately node 20). Total RNA and genomic DNA were extracted together using the method of Verwoerd et al. (1989). DNA and RNA gel blot analysis were performed as described (Sambrook et al., 1989) using Hybond-C extra nitrocellulose membrane (Amersham Pharmacia Biotech). RNA molecular weight markers were from Promega. The 843-bp PCR product excised from pBluescript SK+ was used as a probe to detect expression of the transgene. Expression of genes associated with phenylpropanoid and flavonoid biosynthesis was assessed using probes generously provided by Dr. Cathie Martin (John Innes Centre, Norwich, UK). A Datura stramonium ubiquitin sequence was generated by PCR from genomic DNA using primers derived from the tomato sequence (sense, 5′-ATGCAGATCTTCGTGAAAAC-3′; antisense, 5′-CTTAACCTTCTTCTTCTTCTGCTT-3′ [Hoffman et al., 1991]). Tobacco ubiquitin and β-1,3-glucanase (PR2) coding sequences were amplified by PCR from tobacco genomic DNA using oligonucleotide primers derived from published sequences (ubiquitin sense, 5′-TGGCTCAGGATGAACGCTGGC-3′; antisense, 5′-CATCTT-TGAGACCTCAGTAGAC-3′ [Karrer et al., 1998]; PR2 sense, 5′-CATGCAAACAATTTACCATCAGACC-3′; antisense, 5′-CCAGGTTTCTTT-GGAGTTCCTGCC-3′ [Ward et al., 1991]). DNA probes were gel purified (Qiaex II; Qiagen, Crawley, West Sussex, UK) and radiolabeled to high specific activity by random priming with High Prime (Roche Diagnostics, East Sussex, UK) using 32P-dCTP (Amersham Pharmacia Biotech). Radiolabeled probe was separated from unincorporated nucleotides using a NICK column (Amersham Pharmacia Biotech). Filters were hybridized overnight in QuikHyb (Strategene) at 65°C, rinsed twice in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) at room temperature, and then washed twice for 30 min in 0.1 × SSC and 0.1% SDS at 65°C. Radioactivity was detected on x-ray film (RX; Fuji, Tokyo, Japan) using a Fuji BAS 1500 phosphorimager.
Assay of HCHL Activity and Protein Gel Blotting
The same ground frozen leaf powder used for RNA analysis was used to examine the HCHL enzyme activity and the soluble phenolic content. Enzyme activity in cell-free extracts was assessed by the method of Mitra (1999). Cell-free extracts were made at 4°C from 200 mg of frozen powder mixed with 20 mg of polyvinylpolypyrrolidone and 0.9 mL of extraction buffer (EB; 100 mM Tris-HCl, pH 8.5, 20 mM DTT, and 10 mM Na2EDTA). Extracts were spun for 30 min at 13,000g and then applied to a PD10 column (Amersham Pharmacia Biotech) preequilibrated with 25 mL of EB. The column was washed with 1.6 mL of EB, and the sample was eluted in 1.26 mL of EB. Ten microliters of the eluate was incubated for 10 min at 30°C with 150 μM feruloyl-CoA in 100 mM Tris-HCl, pH 8.5, in a total volume of 100 μL. Total amounts of protein per reaction varied from 2 to 14 μg (usually 2 to 5 μg). The reaction was stopped with an equal volume of acidified methanol (12% glacial acetic acid and 88% methanol) and stored at −70°C.
Production of vanillin by HCHL activity in vitro was measured by gradient HPLC (apparatus from Spectra Physics, San Jose, CA) using a Lichrosorb 10-μm C18 column (25 cm × 4.6 mm; Phase Sep, Deeside, Clwyd, UK). Solvent A was 20 mM NaOAc, pH 6.0, solvent B was methanol, and the gradient ran for 50 min at 1.2 mL/min with the following concentrations: 0 min: 100% A, 0% B; 15 min: 90% A, 10% B; 40 min: 50% A, 50% B; 45 min: 30% A, 70% B; and 50 min: 100% A, 0% B. The column eluant was monitored using a Spectra Focus scanning detector (Spectra Physics). A vanillin standard was used to calculate the response factor in this system.
Protein concentration of the PD10 samples was determined by the method of Bradford (1976) with the Bio-Rad protein assay dye reagent (Bio-Rad Laboratories, Hemel Hempstead, Hertfordshire, UK) using BSA as a standard. Expression of the HCHL protein in transgenic plants was confirmed by protein gel blotting using a polyclonal antibody (Mitra, 1999). Total protein was extracted from frozen, powdered leaf material by shaking overnight at 4°C in 0.1 M Tris-HCl, pH 6.8, 10% glycerol, 1% SDS, and 0.001% bromphenol blue at 40 mg (fresh weight)/mL. DTT was added to 0.5%, and samples were heated for 5 min at 95°C and spun to pellet debris before electrophoresis through a 12% polyacrylamide gel using reagents supplied by National Diagnostics (from Flowgen, Lichfield, Staffordshire, UK). Protein gel blotting was performed as described by Carter et al. (1997) using ProBlot membrane (Applied Biosystems) and the 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium liquid substrate system (Sigma). A duplicate gel was stained to visualize the proteins and the size marker (Gibco BRL, Paisley, UK) using silver stain reagents (Sigma).
HPLC and Liquid Chromatography–Mass Spectrometry Analysis of Soluble Phenolics
Soluble phenolics were extracted by vortexing 30 mg of frozen, powdered leaf material briefly in 300 μL of 70% methanol. After the addition of 100 μL of distilled water, the extract was vortexed again and then spun for 10 sec at 13,000g to pellet the debris. The extract was analyzed by HPLC using a Columbus 5-μm C18 column (25 cm × 4.6 mm; Phenomenex, Macclesfield, Cheshire, UK). Solvent A was 1 mM trifluoroacetic acid, solvent B was acetonitrile, and the gradient was run for 65 min at a flow rate of 1 mL/min with the following concentrations: 0 min: 100% A, 0% B; 50 min: 60% A, 40% B; 60 min: 30% A, 70% B; and 65 min: 100% A, 0% B.
Combined liquid chromatography–mass spectrometry was performed using a Hewlett-Packard 1050 HPLC system (Agilent Technologies, Stockport, UK) and a Quattro II mass spectrometer (Micromass, Manchester, UK) equipped with a Z-spray electrospray ion source. The HPLC conditions were as described for the analysis of soluble phenolic compounds. The flow exiting the HPLC column (1 mL/min) was split 8:1 between the diode array detector and the mass spectrometer ion source. In positive ion mode, the electrospray capillary voltage was set to 3.5 kV and the cone voltage was set to 29 V; the corresponding voltages in negative ion mode were 2.87 kV and 29 V, respectively. The source temperature was set to 140°C, and the desolvation temperature was set to 350°C. Mass spectra were scanned, at unit mass resolution, from mass-to-charge ratio of 50 to 1100 at a rate of 2 sec per scan, with an interscan time of 0.1 sec. Diode array UV light spectra were recorded between 190 and 400 nm, with a step interval of 2 nm and scan interval of 0.1 min. All data were processed and displayed using MassLynx software, version 3.4 (Micromass).
Standards for 4-hydroxybenzoic acid glucoside (4-HBAG) and 4-hydroxybenzoic acid glucose ester (4-HBAGE) were identified by comparison with standards synthesized by Dr. Shigeo Tawaka (Kyoto University, Japan) and kindly provided by Prof. Dr. Lutz Heide (University of Tübingen, Tübingen, Germany). The response factors of these standards in this system were determined by alkaline hydrolysis (4-HBAGE) or β-glucosidase digestion (4-HBAG and vanillic acid glucoside) of known volumes of standards to liberate free 4-hydroxybenzoic acid or vanillic acid, which were then quantified. Hydrolysis by the method of Cooper-Driver et al. (1972) was used to remove glucose from the 4-hydroxybenzyl alcohol glucosides, and the liberated alcohol was compared with a 4-hydroxybenzyl alcohol standard. A vanillic acid glucoside standard was kindly provided by the Takasago International Corporation (Tokyo, Japan).
Gas Chromatography–Mass Spectrometry
Gas chromatography–mass spectrometry was performed on a Fisons Instruments (Altrincham, UK) Trio 1-S mass spectrometer equipped with a Hewlett-Packard 5890B Series II gas chromatograph. Injections of 1 μL of sample were made in splitless mode with an injector temperature of 250°C and with the purge valve on after 1 min. The column used was from J&W Scientific (Folsom, CA; DB5-MS, 15 m, 0.32-mm i.d., 0.25-μm film thickness). The program was 60°C for 1 min, 10°C/min until 250°C was reached, and then hold for 10 min. Samples were ionized by electron ionization at 70 eV and analyzed by cyclic continuous scanning from mass-to-charge ratio of 35 to 600 at a rate of 0.9 sec per scan, with an interscan time of 0.1 sec.
Measurement of Anthocyanin
Anthocyanin was extracted by the method of Martin et al. (1985). Petals from tobacco flowers were weighed, cut into small pieces, and then extracted overnight at 4°C in sealed bottles containing 10 mL of 97% methanol and 3% HCl. The peak absorbance at 530 nm was measured in a spectrophotometer at 530 nm, and the anthocyanin content was calculated given that 1 OD = 33 mM anthocyanin (Martin et al., 1985).
Microscopic Examination of Tobacco Stems
Hand-cut, unstained sections of tobacco stems above leaf node 12 were taken from second generation transgenic and syngenic plants of line 201 during mid flowering. The vascular ring was observed under a dark field (Wild Heerbrugg stereomicroscope [Wild Leitz, Heerbrugg, Switzerland]). Cellular structure was examined by light microscopy (Leitz Ortholux II microscope [Leitz, Wetzlar, Germany] with a HBO 50-W mercury arc lamp and an exciter and barrier filter combination with transmission of 450 to 490 nm and >515 nm, respectively). Phloroglucinol staining was performed using the method of Speer (1987), and sections were viewed by light microscopy.
Acknowledgments
We thank Dr. John Payne for oligonucleotide synthesis and sequencing services, Dr. Cathie Martin for the gift of cDNA clones, Dr. Fran Mulholland for helpful advice, John Eagles for gas chromatography–mass spectrometry services, Lionel Perkins for maintaining the plants, and IFR Communications for photography and graphics. We thank Julie Hofer for critically reading the manuscript. M.J.M. was supported by Biotechnology and Biological Science Research Council Realising Our Potential Award Grant C&M07233.
References
- Atanassova, R., Favet, N., Martz, F., Chabbert, B., Tollier, M.-T., Monties, B., Fritig, B., and Legrand, M. (1995). Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 8, 465–477. [Google Scholar]
- Bate, N.J., Orr, J., Ni, W., Meromi, A., Nadler-Hassar, T., Doerner, P.W., Dixon, R.A., Lamb, C.J., and Elkind, Y. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-limiting step in natural product synthesis. Proc. Natl. Acad. Sci. USA 91, 7608–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher, M., Chabbert, B., Pilate, G., Van Doorsselaere, J., Tollier, M.-T., Petit-Conil, M., Cornu, D., Monties, B., Van Montagu, M., Inze, D., Jouanin, L., and Boerjan, W. (1996). Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 112, 1479–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount, J.W., Korth, K.L., Masoud, S.A., Rasmussen, S., Lamb, C., and Dixon, R.A. (2000). Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol. 122, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet, A.-M. (1998). A new view of lignification. Trends Plant Sci. 3, 67–71. [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Campbell, M.M., and Sederoff, R.R. (1996). Variation in lignin content and composition: Mechanisms of control and implications for the genetic improvement of plants. Plant Physiol. 110, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, A.J., Beiche, F., Hove-Jensen, B., Narbad, A., Barker, P.J., Schweizer, L.M., and Schweizer, M. (1997). PRS1 is a key member of the gene family encoding phosphoribosylpyrophosphate synthase in Saccharomyces cerevisiae. Mol. Gen. Genet. 254, 148–156. [DOI] [PubMed] [Google Scholar]
- Chapple, C.C.S., Vogt, T., Ellis, B.E., and Somerville, C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Driver, G., Corner-Zamodits, J.J., and Swain, T. (1972). The metabolic fate of hydroxybenzoic acids in plants. Z. Naturforsch. 27, 943. [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A., Lamb, C.J., Masoud, S., Sewalt, V.J.H., and Paiva, N.L. (1996). Metabolic engineering: Prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses. A review. Gene 179, 61–71. [DOI] [PubMed] [Google Scholar]
- Elkind, Y., Edwards, R., Mavandad, M., Hedrick, S.A., Ribak, O., Dixon R.A., and Lamb, C.J. (1990). Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc. Natl. Acad. Sci. USA 87, 9057–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecker, L.F., Rugenhagen, C., and Berlin, J. (1993). Increased production of cadaverine and anabasine in hairy root cultures of Nicotiana tabacum expressing a bacterial lysine decarboxylase gene. Plant Mol. Biol. 23, 11–21. [DOI] [PubMed] [Google Scholar]
- Fischer, R., Budde, I., and Hain, R. (1997). Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant J. 11, 489–498. [Google Scholar]
- Gasson, M.J., Kitamura, Y., McLauchlan, W.R., Narbad, A., Parr, A.J., Parsons, E.L.H., Payne, J., Rhodes, M.J.C., and Walton, N.J. (1998). Metabolism of ferulic acid to vanillin: A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J. Biol. Chem. 273, 4163–4170. [DOI] [PubMed] [Google Scholar]
- Guerineau, F., Lucy, A., and Mullineaux, P. (1992). Effect of two consensus sequences preceding the translation initiator codon on gene expression in plant protoplasts. Plant Mol. Biol. 18, 815–818. [DOI] [PubMed] [Google Scholar]
- Hain, R., Reif, H.J., Krause, E., Langebartels, R., Kindl, H., Vornam, B., Wiese, W., Schmelzer, E., Schreier, P.H., Stocker, R.H., and Stenzel, K. (1993). Disease resistance results from foreign phytoalexin expression in a novel plant. Nature 361, 153–156. [DOI] [PubMed] [Google Scholar]
- Halpin, C., Knight, M.E., Foxon, G.A., Campbell, M.M., Boudet, A.M., Boon, J.J., Chabbert, B., Tollier, M.-T., and Schuch, W. (1994). Manipulation of lignin quality by downregulation of cinnamyl alcohol dehydrogenase. Plant J. 6, 339–350. [Google Scholar]
- Hibino, T., Takabe, K., Kawazu, T., Shibata, D., and Higuchi, T. (1995). Increase of cinnamaldehyde groups in lignin of transgenic tobacco plants carrying an antisense gene for cinnamyl alcohol dehydrogenase. Biosci. Biotechnol. Biochem. 59, 929–931. [Google Scholar]
- Higuchi, T., Ito, T., Umezawa, T., Hibino, T., and Shibata, D. (1994). Red-brown color of lignified tissues of transgenic plants with antisense CAD gene: Wine-red lignin from coniferyl aldehyde. J. Biotechnol. 37, 151–158. [Google Scholar]
- Hoffman, N.E., Ko, K., Milkowski, D., and Pichersky, E. (1991). Isolation and characterization of tomato cDNA and genomic clones encoding the ubiquitin gene ubi3. Plant Mol. Biol. 17, 1189–1201. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B., Fry, J.E., Hoffman, N.L., Eichholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Kajita, S., Katayama, Y., and Omori, S. (1996). Alterations in the biosynthesis of lignin in transgenic plants with chimeric genes for 4-coumarate:coenzyme A ligase. Plant Cell Physiol. 37, 957–965. [DOI] [PubMed] [Google Scholar]
- Karrer, E.E., Beachy, R.N., and Holt, C.A. (1998). Cloning of tobacco genes that elicit the hypersensitive response. Plant Mol. Biol. 36, 681–690. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Lee, D., Meyer, K., Chapple, C., and Douglas, C.J. (1997). Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9, 1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.-M., Wang, Z.-X., Wemakor, E., and Heide, L. (1997). Metabolization of the artificial secondary metabolite 4-hydroxybenzoate in ubiC-transformed tobacco. Plant Cell Physiol. 38, 844–850. [Google Scholar]
- MacKay, J.J., Liu, W., Whetten, R., Sederoff, R.R., and O'Malley, D.M. (1995). Genetic analysis of cinnamyl alcohol dehydrogenase in loblolly pine: Single gene inheritance, molecular characterization and evolution. Mol. Gen. Genet. 247, 537–545. [DOI] [PubMed] [Google Scholar]
- Martin, C., Carpenter, R., Sommer, H., Saedler, H., and Coen, E.S. (1985). Molecular analysis of instability in flower pigmentation of Antirrhinum majus following isolation of the pallida locus by transposon tagging. EMBO J. 4, 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A. (1999). 4-Hydroxycinnamoyl-CoA Hydratase/Lyase from Pseudomonas fluorescens AN103: Characterisation and Effects of Expression in Transformed Root Cultures of Datura stramonium. PhD Dissertation (University of East Anglia, Norwich, UK).
- Mitra, A., Kitamura, Y., Gasson, M.J., Narbad, A., Parr, A.J., Payne, J., Rhodes, M.J.C., Sewter, C., and Walton, N.J. (1999). 4-Hydroxycinnamoyl-CoA hydratase/lyase (HCHL): An enzyme of phenylpropanoid chain cleavage from Pseudomonas. Arch. Biochem. Biophys. 365, 10–16. [DOI] [PubMed] [Google Scholar]
- Mo, Y., Nagel, C., and Taylor, L.P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 89, 7213–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Narbad, A., and Gasson, M.J. (1998). Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly-isolated strain of Pseudomonas fluorescens. Microbiology 144, 1397–1405. [DOI] [PubMed] [Google Scholar]
- Piquemal, J., Lapierre, C., Myton, K., O'Connell, A., Schuch, W., Grima-Pettenati, J., and Boudet, A.-M. (1998). Down-regulation of cinnamoyl-CoA reductase induces significant changes in lignin profiles in transgenic tobacco plants. Plant J. 13, 71–83. [Google Scholar]
- Rasmussen, S., and Dixon, R.A. (1999). Transgene-mediated and elicitor-induced perturbation of metabolic channelling at the entry point into the phenylpropanoid pathway. Plant Cell 11, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Siebert, M., Sommer, S., Li, S.-m., Wang, Z.-w., Severin, K., and Heide, L. (1996). Genetic engineering of plant secondary metabolism: Accumulation of 4-hydroxybenzoate glucosides as a result of the expression of the bacterial ubiC gene in tobacco. Plant Physiol. 112, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook, M.E., Mason, P.F., and Sisson, V.A. (1986). Polyphenols in the Nicotiana species. Tob. Sci. 30, 43–49. [Google Scholar]
- Sommer, S., and Heide, L. (1998). Expression of the bacterial chorismate pyruvate-lyase in tobacco: Evidence for the presence of chorismate in the plant cytosol. Plant Cell Physiol. 39, 1240–1244. [DOI] [PubMed] [Google Scholar]
- Sommer, S., Köhle, A., Yazaki, K., Shimomura, K., Bechthold, A., and Heide, L. (1999). Genetic engineering of shikonin biosynthesis hairy root cultures of Lithospermum erythrorhizon transformed with the bacterial ubiC gene. Plant Mol. Biol. 39, 683–693. [DOI] [PubMed] [Google Scholar]
- Speer, O.E. (1987). A method of retaining phloroglucinol proof of lignin. Stain Technol. 62, 279–280. [DOI] [PubMed] [Google Scholar]
- Tamagnone, L., Merida, A., Parr, A., MacKay, S., Culianez-Macia, F.A., Roberts, K., and Martin, C. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10, 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.-J., Popko, J.L., Mielke, M.R., Hu, W.-J., Podila, G.K., and Chiang, V.L. (1998). Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol. 117, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, I.M., Stam, M.E., van Tunen, A.J., Mol, J.N., and Stuitje, A.R. (1992). Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorsselaere, J., et al. (1995). A novel lignin in poplar trees with a reduced caffeic acid/5-hydroxyferulic O-methyltransferase activity. Plant J. 8, 855–864. [Google Scholar]
- Verwoerd, T.C., Dekker, B.M.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNA. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignols, F., Rigau, J., Torres, M.A., Capellades, M., and Puigdomenech, P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.R., Payne, G.B., Moyer, M.B., Williams, S.C., Dincher, S.S., Sharkey, K.C., Beck, J.J., Taylor, H.T., Ahl-Goy, P., Meins, F., Jr., and Ryals, J.A. (1991). Differential regulation of β-1,3-glucanase messenger RNAs in response to pathogen infection. Plant Physiol. 96, 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (1999). Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107, 142–149. [Google Scholar]
- Yahiaoui, N., Marque, C., Myton, K.E., Negrel, J., and Boudet, A.M. (1998). Impact of different levels of cinnamyl alcohol dehydrogenase down-regulation on lignins of transgenic tobacco plants. Planta 204, 8–15. [DOI] [PubMed] [Google Scholar]
- Ylstra, B., Touraev, A., Moreno, R.M.B., Stöger, E., Van Tunen, A.J., Vicente, O., Mol, J.N.M., and Heberle-Bors, E. (1992). Flavonols stimulate development, germination and tube growth of tobacco pollen. Plant Physiol. 100, 902–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, D.-J., Hashimoto, T., and Yamada, Y. (1992). Metabolic engineering of medicinal plants: Transgenic Atropa belladonna with an improved alkaloid composition. Proc. Natl. Acad. Sci. USA 89, 11799–11803. [DOI] [PMC free article] [PubMed] [Google Scholar]