Abstract
The ability of viruses to control and/or evade the host antiviral response is critical to the establishment of a productive infection. We have previously shown that West Nile virus NY (WNV-NY) delays activation of interferon regulatory factor 3 (IRF-3), a transcription factor critical to the initiation of the antiviral response. Here we demonstrate that the delayed activation of IRF-3 is essential for WNV-NY to achieve maximum virus production. Furthermore, WNV-NY utilizes a unique mechanism to control activation of IRF-3. In contrast to many other viruses that impose a nonspecific block to the IRF-3 pathway, WNV-NY eludes detection by the host cell at early times postinfection. To better understand this process, we assessed the role of the pathogen recognition receptor (PRR) retinoic acid-inducible gene I (RIG-I) in sensing WNV-NY infection. RIG-I null mouse embryo fibroblasts (MEFs) retained the ability to respond to WNV-NY infection; however, the onset of the host response was delayed compared to wild-type (WT) MEFs. This suggests that RIG-I is involved in initially sensing WNV-NY infection, while other PRRs sustain and/or amplify the host response later in infection. The delayed initiation of the host response correlated with an increase in WNV-NY replication in RIG-I null MEFs compared to WT MEFs. Our data suggest that activation of the host response by RIG-I early in infection is important for controlling replication of WNV-NY. Furthermore, pathogenic strains of WNV may have evolved to circumvent stimulation of the host response until after replication is well under way.
West Nile virus (WNV) is a member of the Flavivirus genus of the family Flaviviridae, which are enveloped single-strand positive-sense RNA viruses. In areas where WNV is endemic, such as the Middle East, Asia, and Africa, WNV is not considered a public health concern since infections are generally asymptomatic or associated with a mild febrile illness in children. In sharp contrast to this, recent outbreaks in Europe, Israel, and the United States have been associated with a marked increase in both the number of reported cases and the severity of disease among mammals and birds (31), suggesting that a more pathogenic strain has emerged. Since its introduction into the United States in 1999, outbreaks of WNV have become a yearly occurrence, and the virus has now been detected in nearly every state within the continental United States, as well as parts of Canada, Mexico, and the Caribbean (http://www.cdc.gov/ncidod/dvbid/westnile/index.htm) (7). The rapid spread and persistence of WNV indicates that it has firmly established itself in the Western hemisphere.
The first line of defense against an invading viral pathogen is the innate intracellular antiviral response, which is triggered when cellular pathogen recognition receptors (PRRs) detect the presence of pathogen-associated molecular patterns within products of viral replication (17, 42). Upon sensing the invading viral pathogen, the cell activates multiple distinct signaling pathways by inducing a number of latent transcription factors (36). One such transcription factor that is central to establishment of the host antiviral response is interferon regulatory factor 3 (IRF-3) (4). Two classes of PRRs have been determined to stimulate IRF-3 transcriptional activity in response to double-stranded RNA (dsRNA), a well-defined viral pathogen-associated molecular pattern. The first PRR recognized to stimulate IRF-3 in response to dsRNA was Toll-like receptor 3 (TLR3) (5). TLR3 is expressed on the cell surface or within endocytic vesicles in a cell-type-dependent manner (26, 29). Leucine-rich repeats located within the ectodomain of TLR3 are presumably responsible for detecting dsRNA, thus restricting TLR3 to the detection of either extracellular dsRNA or dsRNA within vesicles, including exogenous dsRNA that might enter the cell through endocytosis. Activation of TLR3 by dsRNA leads to the recruitment of the adaptor molecule TRIF (14, 30, 50). TRIF subsequently recruits two kinases, TBK1 and IKKɛ, that have been shown to phosphorylate IRF-3 (9, 38). Phosphorylated IRF-3 forms homodimers that are retained in the nucleus and interact with the CBP/p300 coactivator to induce the expression of multiple target genes, including beta interferon (IFN-β) (23, 34, 48, 53, 54).
Cells lacking TLR3 have been shown to induce interferon expression in response to viral infections or upon the introduction of dsRNA directly into the cytoplasm, which indicated the existence of TLR3-independent intracellular response (15, 49). A second dsRNA-responsive PRR, retinoic acid-inducible gene I (RIG-I), has recently been shown to mediate this intracellular response to dsRNA (41, 52). RIG-I consists of a C-terminal DExD/H box RNA helicase domain and two N-terminal caspase recruitment domains (CARD). Binding of dsRNA to the helicase domain of RIG-I is postulated to induce conformational changes that allow it to interact with downstream effector molecules via CARD. These interactions initiate a signaling cascade that results in the activation of IRF-3. TBK1 is also involved in RIG-I-dependent activation of IRF-3, suggesting that the TLR3 and RIG-I pathways converge at this point. Thus, stimulation of either TLR3 by extracellular dsRNA or RIG-I by intracellular dsRNA results in the activation of IRF-3 and the subsequent expression of IRF-3 target genes, such as IFN-stimulated gene 15 (ISG15), ISG54, ISG56, and IFN-β (12). It is the expression of these direct IRF-3 target genes that initiates the establishment of an antiviral state to block viral replication. Binding of secreted IFN-β to the type I IFN receptor amplifies the host antiviral response by triggering the activation of the Janus kinase and signal transducers and activators of transcription, JAK/STAT, signal transduction pathway. Activation of the JAK/STAT pathway leads to the induction of expression of a wide variety of ISGs, which are responsible for conferring the antiproliferative, antiviral, and proapoptotic actions of IFNs that serve to limit virus infection.
As eukaryotic antiviral programs evolved to combat invading pathogens, viruses evolved processes to escape the antiviral effects of these programs. The molecular mechanisms by which WNV overcomes the host cell antiviral response to establish a productive infection are beginning to be elucidated. Using microarray analysis we have recently demonstrated that the induction of ISGs in response to infection with WNV-NY is attenuated, which suggested that WNV-NY modulates the host antiviral response (11). Recently, several groups have shown that WNV is capable of attenuating signaling through the JAK/STAT pathway (13, 25). In addition, we found that WNV-NY delays activation of IRF-3 until approximately 12 to 16 h postinfection, with maximal activation occurring much later (11). This is in sharp contrast to a variety of other viruses that have been shown to induce IRF-3 activation within 3 to 10 h postinfection (8, 28, 32, 37, 40, 44, 54). The delayed activation of IRF-3 means that WNV-NY replicates virtually unchallenged by the host cell at early times postinfection. In this report we examine the importance of delaying activation of the host antiviral response to WNV-NY replication and the mechanism by which WNV-NY evades stimulation of IRF-3.
MATERIALS AND METHODS
Cells and viruses.
Vero, A549, 293, Huh7, and PH5CH8 cell lines (16) (kindly provided by Nobuyuki Kato and Stanley Lemon) and mouse embryo fibroblast (MEFs) (19) (kindly provided by Shizuo Akira) were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, antibiotic-antimycotic solution, and nonessential amino acids (complete DMEM). U-2 OS/NS3/4A cells were propagated in complete DMEM supplemented with 500 μg/ml G418, 1 μg/ml puromycin, and 1 μg/ml tetracycline. HEK293-pCDNA-TLR3-YFP cells (kindly provided by Kate Fitzgerald) were maintained in complete DMEM containing 400 μg/ml G418. Working stocks of WNV-NY were generated by passaging the virus recovered from the infectious clone pFLWNV (39) one time on 293 cells at a low multiplicity of infection (MOI), and aliquots were stored at −80°C. Titers of working viral stocks were determined for each of the cell lines listed above. The amount of virus added to cultures to achieve the indicated MOI was calculated using the titer of the viral stock on the respective cell line. Vesicular stomatitis virus encoding green fluorescent protein (VSV-GFP), a gift from Michael A. Whitt, and Sendai virus (SenV) Cantrell strain (Charles River) were amplified in baby hamster kidney cells and chicken embryos, respectively.
Huh7-WNV-2.
The generation of the Huh7-WNV-2 replicon cell line was previously described (46). Briefly, Huh7 cells were transfected with 10 μg of total RNA recovered from BHK cells harboring the WNV-Rluc/Neo subgenomic replicon (kindly provided by Pei-Yong Shi) (25a). Following transfection, cultures were incubated in the presence of 400 μg/ml Geneticin (G418) to select for cells harboring the neomycin-expressing replicon. Individual colonies of G418-resistant cells were isolated and expanded, and the levels of expression of Renilla luciferase and WNV proteins were examined. The established cell line used in this study, designated Huh7-WNV-2, was maintained in complete DMEM containing 200 μg/ml G418. G418 was removed from the culture medium prior to infection with SenV.
Plaque assays.
Monolayers of Vero cells in six-well plates were washed two times in serum-free DMEM followed by the addition of serial dilutions of viral samples. The cells were incubated in a 5% CO2 incubator for 1 h at 37°C with rocking, the inocula were removed, and a 0.9% agarose-complete DMEM overlay was added. Cell monolayers were incubated for 48 h, and a second overlay of agarose-complete DMEM containing 0.003% neutral red (MP Biomedicals) was added. The plates were incubated for an additional 48 h prior to counting plaques.
Virus growth curves.
Cultures of the indicated cell lines were infected with WNV-NY for 1 h at 37°C. The amount of virus added to cultures to achieve the indicated MOI was calculated using the titer of the viral stock on the respective cell line. The inoculum was removed, and complete DMEM was added. Culture supernatants were collected at the indicated time points. Cell debris was removed by low-speed centrifugation at 1,500 rpm for 5 min, and supernatants were transferred to new tubes and stored at −80°C until titers were determined by plaque assay on Vero cells.
UV inactivation of WNV.
Cell debris was removed from WNV-NY-infected 293 cell supernatants by low-speed centrifugation, and virions were recovered by ultracentrifugation (100,000 × g; 1.5 h) through a 20% sucrose cushion. The viral pellet was resuspended in phosphate-buffered saline (PBS), divided into aliquots, and stored at −80°C. UV inactivation of WNV-NY was carried out by exposing an aliquot of concentrated virus to UV (254 nm) for 25 min at room temperature in a Stratalinker model XL-1000 apparatus (Spectronics Corp.). Titers of UV-treated WNV-NY were below detectible levels on Vero cells, confirming complete inactivation of the virus stock. Titers of control untreated virus stocks were 2.8 × 1010 PFU/ml on Vero cells.
Northern blot analysis.
RNA was extracted from mock- or WNV-NY-infected A549 cells using TRIzol reagent as recommended by the manufacturer (Invitrogen Life Technologies, Inc.). Purified RNA was resuspended in water, quantified by spectrometry, and mixed with RNA loading buffer. After heating at 50°C for 10 min, 6 μg of RNA was separated through a 1% agarose gel containing 2.2 M formaldehyde, 20 mM morpholinepropanesulfonic acid (pH 7.0), 8 mM NaOAc, and 1 mM EDTA (pH 8.0). To process the gel for transfer of RNA, the gel was soaked in water for 1 h with gentle agitation followed by incubation in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min. RNA transfer onto a Nytran membrane was carried out using the Schleicher & Schuell Turboblotter downward transfer system as recommended by the manufacturer. DNA probes specific for ISG15, ISG56, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were generated using Klenow DNA polymerase and mixed nonomer random primers in a reaction mixture that contained [α-32P]dCTP. Hybridization reactions were carried out using the ULTRAhybe reagent (Ambion) and 106 cpm/ml of radiolabeled probe at 48°C for 16 h. Blots were rinsed twice for 5 min each with preheated 2× SSC-0.1% sodium dodecyl sulfate (SDS) wash buffer, followed by two 15-min washes with 0.1× SSC-0.1% SDS wash buffer. Blots were imaged using a Storm 820 PhosphorImager (Amersham).
Immunoblot analysis.
Cells were lysed in RIPA buffer (10 mM Tris, 150 mM NaCl, 0.02% Na-deoxycholate, 1% Triton X-100, 0.1% SDS) containing protease inhibitors (Sigma) and okadaic acid (1 mM; Sigma). Proteins (20 μg) were resolved on 10% polyacrylamide gels containing SDS. After electrophoresis, proteins were transferred to a NitroPure nitrocellulose transfer membrane (Micron Separations Inc.), and blots were blocked overnight at 4°C. The following monoclonal or polyclonal antibodies were used to probe the blots: rabbit anti-human IRF-3 serum (kindly provided by Michael David), rabbit anti-phospho serine 396 IRF-3 (kindly provided by John Hiscott), rabbit anti-human ISG56 (kindly provided by Ganes Sen), rabbit anti-mouse ISG54 (kindly provided by Ganes Sen), rabbit anti-RIG-I (kindly provided by Takashi Fujita), mouse anti-WNV (obtained from the Centers for Disease Control and Prevention), goat anti-actin (Santa Cruz), goat anti-GAPDH (Santa Cruz), mouse anti-GAPDH (Abcam), and peroxidase-conjugated secondary donkey anti-rabbit, donkey anti-mouse, or donkey anti-goat antibody (Jackson ImmunoResearch). Protein bands were visualized using the ECL Plus Western blotting detection reagents (Amersham Biosciences) followed by exposure of the blot to film. In some experiments bands were quantified using the Kodak 1D image analysis software.
Indirect immunofluorescence analysis (IFA).
The indicated cell lines were grown on tissue culture chamber slides and infected with either WNV-NY or Sendai virus (50 HA units). At the indicated times postinfection, slides were washed with PBS and fixed with 3% paraformaldehyde for 30 min at room temperature. Cell monolayers were permeabilized with a solution of PBS-0.2% Triton X-100 for 15 min, followed by 1 h of incubation in PBS containing 10% normal goat serum. After rinsing with PBS, cells were incubated for 1 h in the presence of a rabbit polyclonal anti-human IRF-3 antibody (1:500) and a mouse polyclonal anti-WNV antibody (1:750) in PBS-0.05% Tween 20-3% bovine serum albumin. Cells were washed three times with PBS-0.5% Tween-20 and incubated with goat anti-rabbit immunoglobulin G-Alexa 488 antibody conjugate (1:4,000; Molecular Probes), goat anti-mouse immunoglobulin G-rhodamine antibody conjugate (1:4,000; Jackson ImmunoResearch), and 4′,6′-diamidino-2-phenylindole stain for 1 h at room temperature. Cells were washed three times and allowed to dry, and the slides were overlaid with Vectashield solution (Vector Labs), after which coverslips were mounted and visualized with a Zeiss Axiovert fluorescence microscope equipped with a digital camera.
Treatment with pIC.
Stocks of poly(I):poly(C) (pIC; 1 mg/ml) were boiled for 10 min and allowed to cool to room temperature prior to being added to culture medium at a final concentration of 100 μg/ml.
Luciferase reporter assays.
Subconfluent cultures of Huh7 cells in a 48-well plate were mock infected or infected with WNV-NY (MOI, 5) and incubated for 3 h at 37°C. Cultures were subsequently transfected with 100 ng of pISG56-luc, 25 ng of pCMV-Renilla, and either 1 μg pIC or increasing concentrations of pEF-flagN-RIG (50 to 500 ng/well) using Lipofectamine 2000 (Invitrogen). The pISG56-luc plasmid (a kind gift from Ganes Sen) encodes the firefly luciferase gene under transcriptional control of the ISG56 promoter, the pCMV-Renilla (Promega) encodes the Renilla luciferase gene under the control of the constitutively active cytomegalovirus (CMV) early promoter, and pEF-flagN-RIG (a kind gift from Takashi Fujita) encodes the constitutively active N terminus of RIG-I (52). Cells were harvested at the indicated times, and the extracts were subjected to the dual luciferase assay as described by the reagent manufacturer (dual-luciferase reporter assay system; Promega). Luciferase activity was quantified with a Bio-Rad luminometer. Normalized luciferase levels were determined by dividing firefly luciferase levels by control Renilla luciferase levels. Determinations at all time points were performed in triplicate.
Quantitative real-time PCR.
RNA was extracted from mock- or WNV-infected WT and RIG-I null MEFs using TRIzol reagent as recommended by the manufacturer (Invitrogen Life Technologies, Inc.). Purified RNA was resuspended in water, quantified by spectrometry, and diluted to 5 ng/μl. Quantitative real-time PCR analyses were performed on an ABI 7500 real-time PCR system using SYBR Green RT-PCR reagents (ABI) with 25 ng of RNA per reaction mixture. All reactions were conducted in triplicate. The following primers were used to amplify murine ISG56 and GAPDH: mISG56 forward primer, 5′-TGGCCGTTTCCTACAGTTT-3′; mISG56 reverse primer, 5′ mGAPDH forward primer, 5′-CAACTACATGGTCTACATGTTC-3′; mGAPDH reverse primer, 5′-CTCGCTCCTGGAAGATG-3′.
RESULTS
The delayed activation of the host response is essential for efficient replication of WNV.
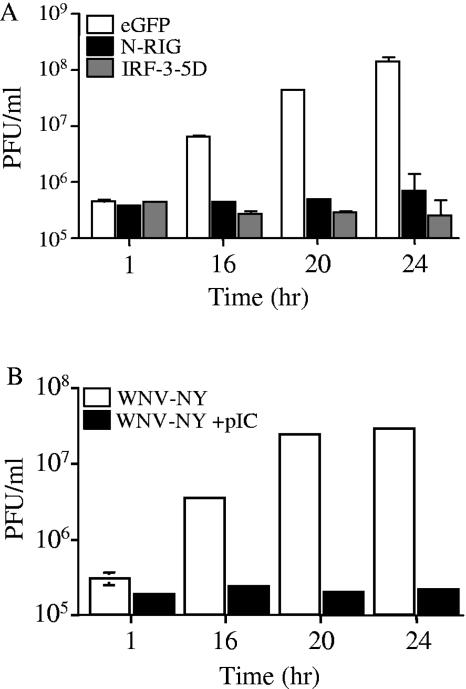
We hypothesized that the delayed activation of the host response to WNV-NY allows the virus to replicate to high levels at early times postinfection and that activation of the host response prior to infection would attenuate WNV-NY replication. To test this hypothesis, the ability of WNV-NY to replicate in the face of an activated host response was examined. The host response was artificially stimulated by transfecting cells with constitutively activate forms of IRF-3 (IRF-3-5D) (24) or RIG-I (N-RIG) (41, 52) (Fig. 1A) prior to infection with WNV-NY. Alternatively, the host antiviral response was stimulated through the TLR3 pathway by adding pIC, a synthetic dsRNA molecule, to the culture medium (Fig. 1B). Expression of constitutively active forms of either RIG-I or IRF-3 attenuated WNV-NY replication compared to control cultures transfected with an EGFP expression plasmid, as did pretreatment of cells with pIC. This suggests that WNV-NY is sensitive to the antiviral actions of the IRF-3 pathway, regardless of the route of stimulation. Therefore, the ability of WNV-NY to delay the activation of the host response is critical to achieving high titers early after infection.
FIG. 1.
Effect of IRF-3 activation on WNV-NY replication. (A) 293 cells transfected with either IRF-3-5D, N-RIG, or EGFP were incubated for 24 h and infected with WNV-NY (MOI, 0.5). (B) pIC (100 μg/ml) was added to supernatants of PH5CH8 cells, and cultures were incubated for 8 h at 37°C prior to infection with WNV-NY (MOI, 0.05). (A and B) Culture supernatants were recovered at the time points indicated, and infectious particle production was assessed by plaque assay on Vero cells.
Induction of the IRF-3 pathway requires establishment of a productive infection.
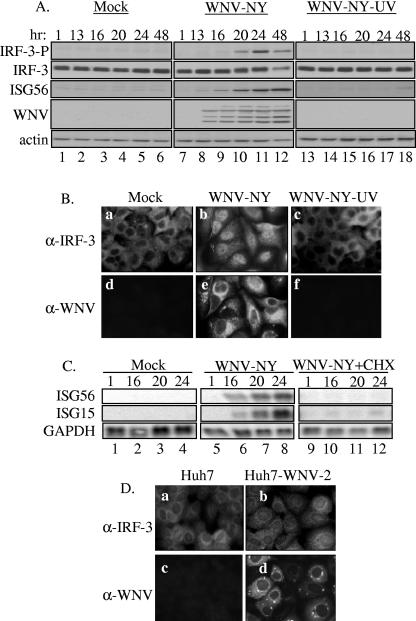
In order to gain insight into the mechanism by which WNV-NY delays the activation of IRF-3, it was necessary to first define the interactions between WNV-NY and IRF-3 that are required for activation. The role of WNV-NY replication in IRF-3 activation was assessed by exposing A549 cells to UV-inactivated virus at a concentration equivalent to an MOI of 5. Activation of IRF-3 was monitored by Western blot analysis of the phosphorylation state of IRF-3, as well as examination of the cellular localization of IRF-3 by IFA (Fig. 2A and B). As previously observed, IRF-3 phosphorylation (Fig. 2A, lanes 7 to 12) and nuclear retention (Fig. 2B, panel b) were detected in control cultures infected with untreated virus. In contrast, neither phosphorylation of IRF-3 (Fig. 2A, lanes 13 to 18) nor nuclear localization (Fig. 2B, panel c) was detected in cultures exposed to UV-inactivated virus, indicating that the IRF-3 pathway was not induced in the absence of viral replication.
FIG. 2.
WNV-NY replication is required for activation of IRF-3. (A and B) Examination of IRF-3 activation in response to UV-inactivated WNV-NY. A549 cells were mock infected, infected with WNV-NY (MOI, 5), or exposed to UV-inactivated virus at a concentration equivalent to an MOI of 5. (A) Cell lysates were recovered at the indicated times and subjected to immunoblot analysis. Phosphorylation of IRF-3 was detected using an antibody specific for the phosphoserine 396 isoform of IRF-3 (IRF-3-P). Steady-state protein levels of total IRF-3, ISG56, WNV, and actin were also examined. (B) IRF-3 localization in mock-infected (a and d), WNV-NY-infected (b and e), or UV-inactivated WNV-NY- (c and f) treated A549 cells was detected by IFA. IRF-3 was detected using an IRF-3 polyclonal antiserum and an Alexa 488-conjugated secondary antibody (a, b, and c). WNV protein expression (d, e, and f) was detected using a mouse polyclonal anti-WNV antibody and rhodamine-conjugated secondary antibody. (C) Effect of cycloheximide on WNV-NY-induced expression of IRF-3 target genes. A549 cells were infected with WNV-NY (MOI, 1) in the presence or absence of cycloheximide (50 μg/ml). Induction of ISG15 and ISG56 was assessed by Northern blot analysis of total RNA harvested at the indicated times postinfection. Levels of GAPDH expression were also assessed to control for loading. (D) Activation of IRF-3 in cells harboring the WNV replicon. Cellular localization of IRF-3 (a and b) and WNV protein expression (c and d) were examined in parental Huh7 (a and c) and Huh7-WNV-2 replicon (b and d) cell lines by IFA.
To confirm these results, cultures were infected with WNV-NY in the presence or absence of cycloheximide, which prevents viral replication by blocking translation of both cellular mRNA and incoming viral genomes. Since de novo synthesis of cellular proteins is not required for the activation of IRF-3 (32), cycloheximide will block WNV-NY replication without impeding activation of the IRF-3 pathway. Northern blot analysis was used to examine the level of expression of the IRF-3 target genes ISG15 and ISG56 in order to assess activation of the IRF-3 pathway (Fig. 2C). ISG15 and ISG56 expression was detected in control WNV-NY-infected cultures treated with dimethyl sulfoxide (Fig. 2C, lanes 5 to 8) but not in cultures exposed to cycloheximide (Fig. 2C, lanes 9 to 12). These data demonstrate that the synthesis of viral protein(s) and/or viral replication is required for activation of the IRF-3 pathway during WNV-NY infection.
The requirements for WNV-NY-induced activation of IRF-3 were further characterized using the WNV subgenomic replicon Huh7-WNV-2 (46). This replicon genome encodes the nonstructural genes of WNV-NY, which are sufficient for autonomous replication of the replicon within the host cell. Thus, cells harboring the WNV replicon are exposed to an actively replicating viral genome and the nonstructural proteins of WNV. Cellular localization of IRF-3 was examined by IFA in order to assess the activation of IRF-3 within the replicon-bearing cells (Fig. 2D). IRF-3 remained localized to the cytoplasm of cells harboring the WNV replicon (Fig. 2D, panel b), indicating that IRF-3 is inactive in these cultures. Therefore, either the presence of the actively replicating WNV replicon was not sufficient to induce activation of IRF-3 or Huh7-WNV-2 encodes a mechanism to block activation.
WNV-NY does not antagonize the activation of IRF-3.
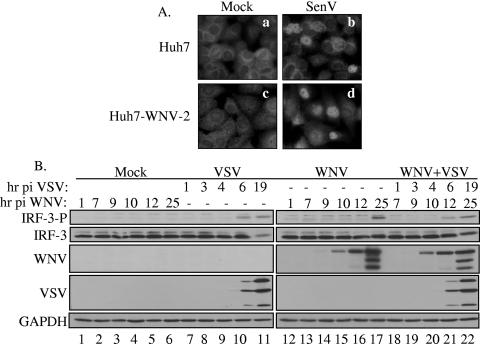
In order to determine whether WNV is capable of abrogating IRF-3 activation during viral replication, we assessed the ability of Huh7-WNV-2 replicon to respond to SenV, a potent activator of IRF-3. SenV infection triggered nuclear translocation of IRF-3 in both parental and Huh7-WNV-2 replicon cells (Fig. 3A, panels b and d), demonstrating that IRF-3 can be activated in these cell lines. These results indicate that the nonstructural proteins of WNV-NY do not impose a signaling blockade to the IRF-3 pathway. To confirm and extend these results, the ability of WNV-NY to impede IRF-3 activation in the context of a native infection was also assessed. WNV-NY-infected cultures were superinfected with VSV, a virus capable of inducing the activation of IRF-3 within the lag period prior to WNV-mediated activation (Fig. 3B). VSV infection rapidly induced the phosphorylation of IRF-3 in both the absence (Fig. 3B, lanes 7 to 11) and presence (Fig. 3B, lanes 18 to 22) of WNV infection. Therefore, WNV-NY infection does not prevent activation of IRF-3 by VSV. These results indicate that WNV-NY does not impose a blockade upon the virus-mediated activation of the IRF-3 pathway.
FIG. 3.
Effects of WNV-NY infection on IRF-3 induction by SenV and VSV. (A) Parental Huh7 cells (a and b) and Huh-WNV-2 replicon cells (c and d) were mock infected (a and c) or infected with SenV (b and d). IRF-3 localization was assessed by IFA. (B) Phosphorylation state of IRF-3 in mock- (lanes 1 to 6), VSV-GFP- (lanes 7 to 11), WNV-NY- (lanes 12 to 17), or VSV-GFP- and WNV-NY- (lanes 18 to 22) infected A549 cells. WNV-NY-infected cultures were incubated for 6 h prior to superinfection with VSV-GFP. Whole-cell lysates were recovered at the indicated times postinfection with WNV-NY, and Western blot analysis was performed with an antibody specific for the phosphoserine 396 isoform of IRF-3 (IRF-3-P). Blots were stripped and reprobed with antisera against total IRF-3, WNV, VSV, or GAPDH.
WNV-infected cultures remain responsive to stimulation of IRF-3 through both the TLR3- and RIG-I-dependent pathways.
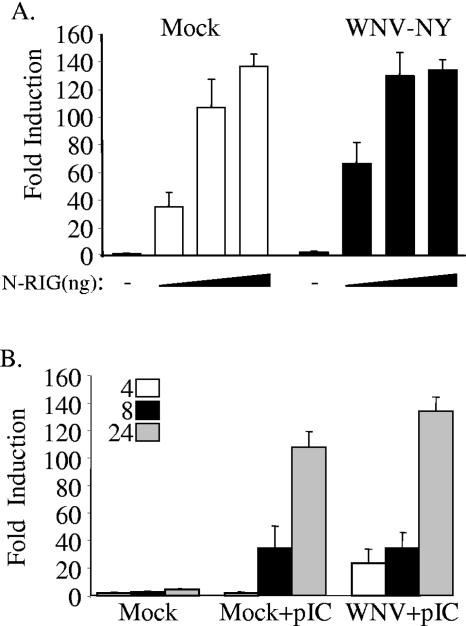
Virus activation of IRF-3 is triggered in response to stimulation of two distinct PRRs, TLR3 and RIG-I. Both SenV and VSV have been shown to activate IRF-3 in a RIG-I-dependent manner in most cell lines (19). Thus, these viruses would circumvent a possible WNV-NY-mediated block of TLR-3-dependent activation of IRF-3. To investigate this possibility, the effects of WNV-NY on the TLR-3 and RIG-I pathways were independently examined. The effect of WNV-NY on the RIG-I pathway was assessed by transfecting WNV-NY-infected Huh7 cells with an IRF-3-responsive reporter construct (pISG56-luc) and increasing concentrations of a plasmid encoding constitutively active N-RIG to stimulate IRF-3. The level of IRF-3-dependent luciferase expression at 8 h poststimulation with N-RIG was similar between control mock-infected and WNV-NY-infected cultures (Fig. 4A). The RIG-I pathway can also be stimulated by introducing pIC directly into the cytoplasm of the cell (41, 51). Therefore, the effect of WNV-NY on the intracellular response to dsRNA was also examined. Huh7 cells, which lack TLR3 expression and are refractory to extracellular pIC (20), were infected with WNV-NY for 3 h prior to cotransfection with ISG56-luc and pIC. pIC treatment consistently induced higher levels of IRF-3-dependent ISG56 promoter activity in WNV-NY-infected cultures compared to mock-infected cultures at 4 h posttreatment, suggesting that WNV-NY infection may potentiate the activation of IRF-3 early in infection. Nonetheless, similar levels of luciferase expression were observed in mock- and WNV-NY-infected cultures at all other time points (Fig. 4B). These data confirm that WNV-NY does not antagonize RIG-I-mediated signaling of IRF-3 activation.
FIG. 4.
Effect of WNV-NY infection on RIG-I-dependent induction of IRF-3. (A) Huh7 cells were infected in triplicate with WNV-NY (MOI, 5) for 3 h prior to transfection with pISG56-luc, pCMV-Renilla, and increasing concentrations of N-RIG (50, 250, and 500 ng). Cell extracts were recovered at 8 h posttransfection, and the level of luciferase expression was assessed using the Promega dual luciferase kit. A representative example from two independent experiments is shown. (B) Huh7 cells were infected with WNV-NY cells (MOI, 5) for 3 h and subsequently transfected with pISG56-luc, pCMV-Renilla, and pIC (1 μg). Cells were lysed at 4, 8, and 24 h posttransfection, and the level of luciferase expression was determined. A representative example from two independent experiments is shown.
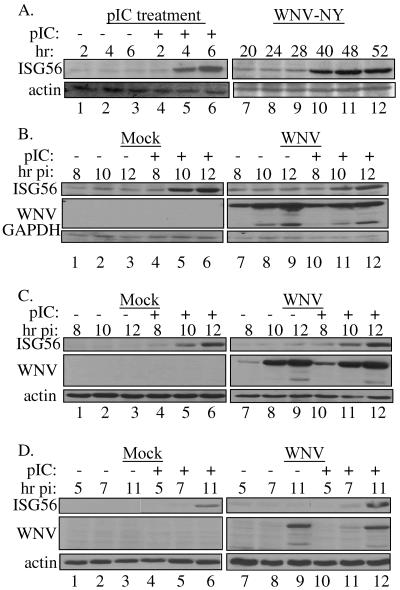
We next examined whether or not WNV-NY could regulate signaling of IRF-3 activation by TLR3 stimulation using 293T-pCDNA-TL3-YFP cells, which stably express a TLR3 transgene. The ability of 293T-pCDNA-TL3-YFP cells to activate IRF-3 in response to exogenous dsRNA or WNV-NY was monitored by Western blot analysis of ISG56 expression. Consistent with previous results, ISG56 expression in WNV-NY-infected cultures was only detected at late time points (11) (Fig. 5A, lanes 7 to 12). Thus, overexpression of TLR3 did not increase the rate of activation of IRF-3 in response to infection. In contrast, treatment of cells with exogenous pIC rapidly activated IRF-3, as demonstrated by the detection of expression of ISG56 at 4 h posttreatment (Fig. 5A, lanes 1 to7). To determine if WNV-NY could prevent TLR3 signaling of IRF-3 activation at early times postinfection, cultures were mock treated or infected with WNV-NY for 6 h prior to stimulation with exogenous pIC. IRF-3-dependent gene expression was then assessed at times points prior to WNV-NY-mediated activation of IRF-3. ISG56 expression was induced in mock- and WNV-NY-infected cells treated with pIC (Fig. 5B, lanes 4 to 6 and 10 to 12, respectively), indicating that the cells retained the ability to activate IRF-3 in response to stimulation of the TLR3 pathway. To ensure that the inability of WNV-NY to block activation of IRF-3 in these cells was not due to the overexpression of TLR3, two other cell lines, PH5CH8 and U-2 OS/NS3/4A, were examined. Both cell lines express TLR3 (data not shown) (21) and have been previously shown to respond to pIC in the culture medium (10, 21). pIC stimulation induced ISG56 expression in mock-infected PH5CH8 and U-2 OS/NS3/4A cells (lanes 4 to 6 of Fig. 5C and D, respectively), confirming that IRF-3 was activated by exogenous dsRNA. Likewise, pIC-induced ISG56 expression in both cell lines infected with WNV-NY (Fig. 5C and D, lanes 10 to 12). These results confirm that WNV-NY infection does not interfere with the activation of IRF-3 in response to TLR3-specific stimulation.
FIG. 5.
TLR3-mediated activation of IRF-3. (A) 293T-pCDNA-TL3-YFP cells were treated with pIC or infected with WNV-NY at an MOI of 5. Whole-cell lysates were recovered at the indicated times, and steady-state levels of ISG56 expression were examined by Western blotting. Blots were stripped and reprobed for actin to control for loading. (B) pIC was added to the culture supernatants of WNV-NY-infected (MOI, 3.6) 293T-pCDNA-TL3-YFP cells at 6 h postinfection. (C) U-2 OS/NS3/4A cells were infected with WNV-NY (MOI, 3) for 4 h prior to the addition of pIC to the culture medium. (D) PH5CH8 cells infected with WNV-NY (MOI, 1) were treated with pIC at 3 h postinfection. In panels B, C, and D, whole-cell lysates collected at the indicated times postinfection were analyzed for steady-state levels of ISG56 by immunoblotting. Blots were stripped and reprobed for WNV protein to assess viral replication and GAPDH or actin to control for loading.
RIG-I-dependent and -independent activation of IRF-3 by WNV-NY.
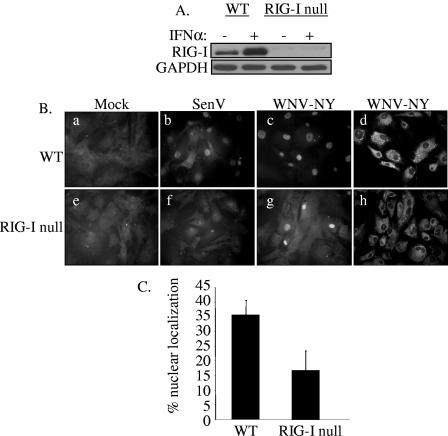
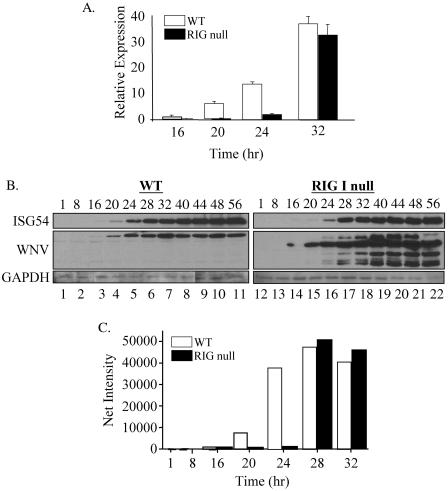
The role of RIG-I in WNV-NY-mediated activation of IRF-3 was examined using MEFs recovered from RIG-I null embryos (19). Western blot analysis of RIG-I null MEFs confirmed the absence of RIG-I protein expression in these cells (Fig. 6A). IRF-3 cellular localization in WT and RIG-I null MEFs infected with either SenV or WNV-NY was monitored by IFA. IRF-3 nuclear translocation was detected in both SenV- and WNV-NY-infected WT MEFs (Fig. 6B, panels b and c). As previously reported, SenV infection failed to induce activation of IRF-3 in RIG-I null MEFs (panel f), confirming ablation of RIG-I-dependent activation of IRF-3. In contrast, IRF-3 was detected in the nucleus of WNV-NY-infected RIG null MEFs (panel g), indicating that WNV-NY can induce activation of IRF-3 in the absence of RIG-I. However, at 24 h postinfection the percentage of cells with nuclear IRF-3 in cultures of WNV-NY-infected RIG-I null MEFS was consistently reduced compared to WT MEFs (Fig. 6C). To further assess this difference, the kinetics of induction of IRF-3 target genes in WNV-NY-infected WT and RIG-I null MEFs was examined. Since there are no available antibody reagents with which to detect the mouse homologue of ISG56, we used quantitative real-time PCR to examine induction of ISG56 mRNA expression. WNV-NY infection induced the expression of ISG56 mRNA in both WT and RIG-I null MEFs; however, the induction of ISG56 expression was delayed in RIG-I null MEFs compared to WT MEFs (Fig. 7A). To verify induction of IRF-3-dependent gene expression, Western blot analysis of a second and related IRF-3 target gene, ISG54 (45), was also examined. The induction profile of ISG54 confirmed that WNV-NY-mediated activation of IRF-3 was delayed in RIG-I null MEFs (Fig. 7B and C). This suggests that the RIG-I pathway mediates the initial activation of the host response to WNV; however, distinct secondary pathways are capable of mediating activation of IRF-3 in the absence of RIG-I.
FIG. 6.
IRF-3 localization in WNV-NY-infected WT and RIG-I null MEFs. (A) The RIG-I null genotype was confirmed by immunoblot analysis of lysates prepared from WT and IRF-3 null MEFs incubated in the presence or absence of 200 U/ml mouse IFN-α. Steady-state levels of RIG-I were assessed using a rabbit polyclonal antiserum to RIG-I. (B) Cellular localization of IRF-3 in WT (a to c) and RIG-I null (e to g) MEFs was examined. Mock- (a and e), SenV- (b and f), and WNV-NY- (c and g) infected cells were probed for IRF-3 using an IRF-3 polyclonal antiserum and an Alexa 488-conjugated secondary antibody (a through c and e through g). WNV protein expression (d and h) was detected using a mouse polyclonal anti-WNV antibody and rhodamine-conjugated secondary antibody. (C) Percent IRF-3 nuclear localization in WNV-NY-infected WT and RIG-I null MEFs. The number of cells with nuclear IRF-3 was divided by the total number of cells present in nine individual fields of WT and RIG-I null MEFs.
FIG. 7.
Kinetics of activation of the host antiviral response in WNV-NY-infected WT and RIG-I null MEFs. (A) Comparison of the kinetics of expression of ISG56 mRNA levels in WNV-NY-infected WT and RIG-I null MEFs. Total RNA was recovered from WNV-NY-infected WT and RIG-I MEFs at the indicated times postinfection. Quantitative real-time PCR was used to determine the levels of ISG56 and GAPDH mRNA present at each time point. Bars show the level of ISG56 mRNA relative to GAPDH in each sample. (B) Western blot analysis of ISG54 expression. Whole-cell lysates were collected at the indicated times postinfection, and steady-state levels of ISG54, WNV, and GAPDH were examined. (C) Quantitation of ISG54 expression in WNV-NY-infected WT and RIG-I null MEFs.
Contributions of the RIG-I and TLR3 pathways in controlling WNV infection.
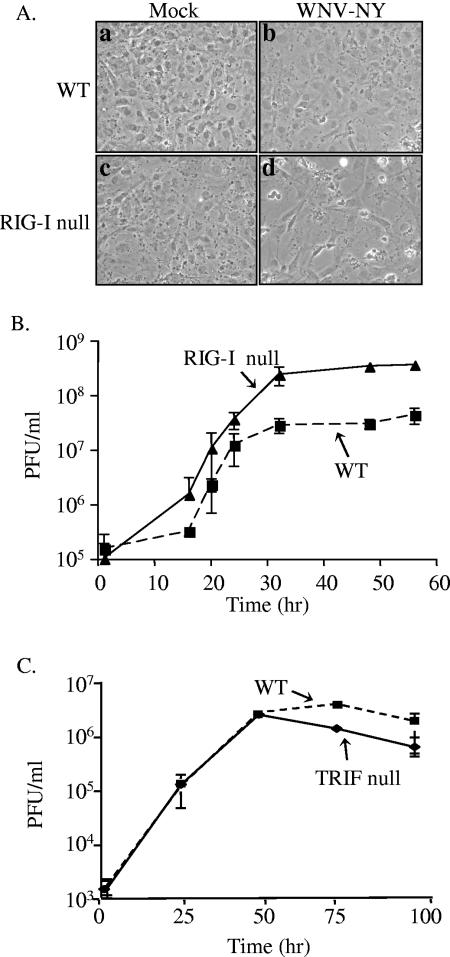
Despite the fact that WNV-NY induced a robust IRF-3 response in RIG-I null MEFs late in infection, WNV-NY protein expression was significantly enhanced in the absence of RIG-I (Fig. 6, compare panels d and h; Fig. 7B). In addition, extensive cytopathic effect (CPE) was observed in WNV-NY-infected RIG-I null but not WT MEFs (Fig. 8A). This suggests that activation of the host antiviral response through RIG-I contributes to the ability of cells to control WNV-NY replication and pathogenesis. The ability of RIG-I to repress WNV-NY replication was assessed by comparing the kinetics of viral replication within WT and RIG-I null MEFs (Fig. 8B). WNV-NY achieved higher peak viral titers in RIG-I null MEFs compared to WT MEFs, demonstrating that stimulation of the RIG-I pathway contributes to the ability of the host cell to control WNV-NY replication.
FIG. 8.
Replication of WNV-NY in WT, RIG-I null, and TRIF null MEFs. (A) Virus-induced CPE in WT and RIG-I null MEFs. Mock-infected (a and c) and WNV-NY-infected (b and d) cultures of WT (a and b) and RIG-I null (c and d) MEFs were visualized at 56 h postinfection using a Zeiss light microscope, and images were captured with a digital camera. (B) Infectious particle production by WNV-NY-infected WT and RIG-I null MEFs. Culture medium was removed from infected MEFs and cleared of cell debris by low-spin centrifugation. The presence of infectious virus particles was determined as PFU per milliliter by titrating supernatants on Vero cells in duplicate. The average of three independent experiments is shown. Solid line, RIG-I null; broken line, WT MEFs. (C) Infectious particle production by WNV-NY-infected TRIF null MEFs. Titers for supernatants removed from WNV-NY-infected WT and TRIF null MEFS were determined on Vero cells in duplicate. The average of three independent experiments is shown. Solid line, TRIF null; broken line, WT MEFs.
While WNV-NY can induce the IRF-3 pathway in the absence of TLR3, it is possible that activation of the host antiviral response through the TLR3 pathway also plays a role in controlling viral replication in WT MEFs. Therefore, MEFs deficient for TRIF, an adaptor molecule that is essential for TLR3 signaling (50), were used to assess the contribution of the TLR3 pathway in controlling WNV replication. Abrogation of the TLR3 pathway in TRIF null MEFs was confirmed by the inability of these cells to induce ISG54 expression in response to stimulation with pIC (data not shown). Titers of virus recovered from TRIF null MEFs were similar to titers recovered from WT MEFs (Fig. 8C), demonstrating that disruption of the TLR3 pathway had no effect on the replication of WNV-NY. Therefore, the TLR3 pathway is not essential for limiting viral replication in vitro.
DISCUSSION
The importance of IRF-3 in controlling viral infections is evident from the fact that many viruses have evolved to specifically block this pathway (1, 3, 6, 10, 18, 22, 27, 33, 43). Here we have demonstrated that, when stimulated prior to infection, the IRF-3 pathway initiates the establishment of an antiviral state that attenuates WNV-NY replication. Our data suggest that WNV-NY avoids triggering the IRF-3 pathway at points early in infection when it is most sensitive to host cell defenses. In order to begin to dissect the mechanism by which WNV-NY avoids activation of the host response early in infection, we first assessed the basic requirements for activation of IRF-3 by WNV. Both UV inactivation and cycloheximide treatment blocked WNV-NY-induced stimulation of IRF-3, demonstrating that establishment of a productive infection is necessary for activation. However, IRF-3 was not activated in cells harboring an actively replicating subgenomic replicon of WNV. This suggests that either (i) the expression of the nonstructural proteins of WNV and replication of the WNV RNA do not induce activation of IRF-3, (ii) the WNV replicon genome encodes a mechanism to block activation, or (iii) the process used to establish the replicon cell line selected for cells with a defective IRF-3 pathway. We assessed the last two possibilities by infecting Huh7-WNV-2 replicon cells with SenV. SenV infection induced activation of IRF-3, demonstrating that the IRF-3 pathway is intact in these cultures and that the replicon does not possess the ability to prevent activation of this pathway. Therefore, Huh7-WNV-2 cells lack the stimulus necessary for activation of the IRF-3 pathway. This suggests that the structural proteins of WNV-NY may be required for activation of IRF-3 during infection; however, we cannot rule out the possibility that the WNV agonist(s) of IRF-3 is simply expressed at levels too low in the replicon-bearing cells to stimulate activation.
The ability of WNV-NY to block IRF-3 activation in the context of a native viral infection was also evaluated. Cultures infected with WNV-NY remained responsive to dsRNA added directly to the culture medium or transfected into the cells. These results contradict recently published data that suggest that WNV is capable of blocking TLR3-mediated activation of IRF-3 in HeLa cells (35). Nonetheless, our data demonstrate that WNV-NY infection did not prevent stimulation of IRF-3 by dsRNA in a variety of responsive cell lines, including human embryonic kidney, osteosarcoma, and hepatocyte cell lines. This discrepancy is likely due to a cell line-specific effect in the ability of WNV to block stimulation of IRF-3 through the TLR3 pathway and suggests the possibility that IRF-3 activation is differentially regulated in distinct tissues in vivo. The ability of WNV-NY to modulate IRF-3 activation through the RIG-I pathway was also examined. Cultures infected with WNV-NY retained the ability to activate the IRF-3 pathway in response to both superinfection with VSV and introduction of dsRNA directly in the cytoplasm. Therefore, WNV-NY does not directly antagonize IRF-3 activation through either the TLR3 or RIG-I pathways. Our results demonstrated that the delayed activation of the IRF-3 pathway by WNV-NY is not due to a virus-directed block imposed on either the activation or the transcriptional activity of IRF-3. Instead WNV-NY appears to prevent the host cells from sensing viral replication at early times postinfection. The mechanism by which WNV-NY avoids detection by the host antiviral response early in infection remains to be determined. One possible explanation is that high levels of the WNV-NY agonist(s) are required for efficient activation of IRF-3, such that activation does not occur until sufficient levels of the viral agonist(s) have accumulated. The observation that WNV-NY infection enhanced the IRF-3 response at 4 h posttransfection with pIC (Fig. 4B) might indicate that replication is just below the threshold of activation of IRF-3. Alternatively, WNV-NY may have evolved to specifically mask the IRF-3 agonist(s) produced early in infection, thus blocking the accessibility of the viral agonist(s) to PRRs until the virus has established a productive infection.
Another important step towards elucidating the process by which WNV-NY evades the host response at early times postinfection is the identification of the host cell components involved in sensing WNV-NY infection. It has recently been demonstrated that in some cells the TLR system is dispensable for activation of IRF-3 in response to viral infection (19). In fibroblasts and conventional dendritic cells, activation of the host antiviral response by negative-strand RNA viruses was specifically mediated though RIG-I. Moreover, our observation that WNV-NY-mediated activation of IRF-3 occurs in 293 cells (11), a cell line that lacks TLR3 expression, suggests that WNV-NY can stimulate the host antiviral response in a TLR-3-independent manner. Characterization of the role of RIG-I in WNV-NY-induced activation of IRF-3 demonstrated that abrogation of the RIG-I pathway did not prevent WNV-NY-mediated activation of IRF-3 but resulted in a delayed induction of the host response. Thus, RIG-I is involved in triggering the initial antiviral response to WNV-NY; however, an additional PPR(s) functions to amplify and/or sustain the host response later in infection. One candidate PRR molecule for sensing WNV-NY infection is the RIG-I homologue MDA5, which has also been shown to be involved in activation of the host antiviral response (2, 51). Preliminary experiments indicate that disruption of signaling through both MDA5 and RIG-I completely abrogates the host response to WNV-NY, suggesting that MDA5 is responsible for the residual activation of the host response observed in RIG null cells (unpublished data). However, we cannot rule out the possibility that an as-yet-unidentified PPR is also involved in inducing the host response to WNV-NY.
We also examined the contribution of both the RIG-I and TLR3 pathways to controlling WNV-NY replication. Ablation of the RIG-I pathway resulted in an increase in WNV-NY titers and CPE in MEFs, demonstrating enhanced viral replication. Therefore, stimulation of the RIG-I pathway contributes to the establishment of an initial innate antiviral immune response that is capable of constraining WNV-NY replication. In contrast, disruption of the TLR3 pathway had no effect on WNV-NY replication in MEFs, suggesting that the TLR3 pathway does not function to control WNV-NY replication in vitro. However, the TLR3 pathway does appear to play a significant role in WNV-NY replication and pathogenesis in vivo. Wang et al. have previously demonstrated that viral loads are higher in the blood of WNV-infected TLR3 null mice compared to WT (47). Taken together with our data, this suggests that TLR3's inhibitory effect in vivo may be due to its involvement in the stimulation of professional immune cells, which act to constrain WNV-NY replication through an adaptive immune response, rather than triggering the host defense within the infected cell. Despite increased viral loads, WNV virulence was attenuated in TLR3 null mice. The enhanced virulence in WT mice appeared to be due to the increased permeability of the blood-brain barrier caused by induction of an inflammatory response by TLR3. This suggests that highly virulent strains of WNV-NY may have evolved to more efficiently stimulate the TLR3-mediated inflammatory response rather than to disrupt TLR3 signaling.
Our results demonstrate that, unlike many viruses, WNV-NY does not encode a general mechanism to shut down the IRF-3 pathway. Instead, WNV-NY specifically prevents activation of IRF-3 by its own replication at early times postinfection, which allows the virus to replicate to high titers before the host cells can mount an effective antiviral response. This idea is further supported by the observation that a prolonged lag period prior to induction of the host response, which resulted from abrogation of the RIG-I pathway, corresponded with increased viral titers. Our results suggest that rather than directly antagonizing the IRF-3 pathway, WNV-NY eludes detection by the host cell until a productive infection is established.
Acknowledgments
We thank S. Akira, T. Fujita, J. Hiscott, M. A. Whitt, G. Sen, M. David, K. A. Fitzgerald, S. M. Lemon, N. Kato, and P. Y. Shi for reagents. We thank Michael Diamond for helpful discussion and critical reading of the manuscript.
This work was supported by NIH grant AI057568 (M.G.). M.G. is the Nancy C. and Jeffery A. Marcus Scholar in Medical Research, in honor of Bill S. Vowell.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 5.Barton, G. M., and R. Medzhitov. 2003. Linking Toll-like receptors to IFN-alpha/beta expression. Nat. Immunol. 4:432-433. [DOI] [PubMed] [Google Scholar]
- 6.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dauphin, G., S. Zientara, H. Zeller, and B. Murgue. 2004. West Nile: worldwide current situation in animals and humans. Comp. Immunol. Microbiol. Infect. Dis. 27:343-355. [DOI] [PubMed] [Google Scholar]
- 8.Elco, C. P., J. M. Guenther, B. R. Williams, and G. C. Sen. 2005. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-κB, and interferon but not toll-like receptor 3. J. Virol. 79:3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 10.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 102:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredericksen, B. L., M. Smith, M. G. Katze, P. Y. Shi, and M. Gale, Jr. 2004. The host response to West Nile virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737-7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoebe, K., X. Du, P. Georgel, E. Janssen, K. Tabeta, S. O. Kim, J. Goode, P. Lin, N. Mann, S. Mudd, K. Crozat, S. Sovath, J. Han, and B. Beutler. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 424:743-748. [DOI] [PubMed] [Google Scholar]
- 15.Hoebe, K., E. M. Janssen, S. O. Kim, L. Alexopoulou, R. A. Flavell, J. Han, and B. Beutler. 2003. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat. Immunol. 4:1223-1229. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, M., K. Sugiyama, T. Mizutani, T. Tanaka, K. Tanaka, H. Sekihara, K. Shimotohno, and N. Kato. 1998. Human hepatocyte clonal cell lines that support persistent replication of hepatitis C virus. Virus Res. 56:157-167. [DOI] [PubMed] [Google Scholar]
- 17.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 18.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 20.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, R., P. Genin, Y. Mamane, M. Sgarbanti, A. Battistini, W. J. Harrington, Jr., G. N. Barber, and J. Hiscott. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800-811. [DOI] [PubMed] [Google Scholar]
- 23.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Lo, M. K., M. Tilgner, and P.-Y. Shi. 2003. Potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 77:12901-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto, M., K. Funami, M. Tanabe, H. Oshiumi, M. Shingai, Y. Seto, A. Yamamoto, and T. Seya. 2003. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J. Immunol. 171:3154-3162. [DOI] [PubMed] [Google Scholar]
- 27.Melroe, G. T., N. A. DeLuca, and D. M. Knipe. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiya, T., and A. L. DeFranco. 2004. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J. Biol. Chem. 279:19008-19017. [DOI] [PubMed] [Google Scholar]
- 30.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161-167. [DOI] [PubMed] [Google Scholar]
- 31.Petersen, L. R., and J. T. Roehrig. 2001. West Nile virus: a reemerging global pathogen. Emerg. Infect. Dis. 7:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato, M., N. Tanaka, N. Hata, E. Oda, and T. Taniguchi. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 425:112-116. [DOI] [PubMed] [Google Scholar]
- 35.Scholle, F., and P. W. Mason. 2005. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology 342:77-87. [DOI] [PubMed] [Google Scholar]
- 36.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 37.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 38.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 39.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Iκ B kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 41.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 43.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.tenOever, B. R., S. Sharma, W. Zou, Q. Sun, N. Grandvaux, I. Julkunen, H. Hemmi, M. Yamamoto, S. Akira, W. C. Yeh, R. Lin, and J. Hiscott. 2004. Activation of TBK1 and IKKɛ kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 78:10636-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terenzi, F., S. Pal, and G. C. Sen. 2005. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340:116-124. [DOI] [PubMed] [Google Scholar]
- 46.Wang, C., M. Gale, Jr., B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425-434. [DOI] [PubMed] [Google Scholar]
- 47.Wang, T., T. Town, L. Alexopoulou, J. F. Anderson, E. Fikrig, and R. A. Flavell. 2004. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med. 10:1366-1373. [DOI] [PubMed] [Google Scholar]
- 48.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640-643. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668-6672. [DOI] [PubMed] [Google Scholar]
- 51.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 52.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 53.Yoneyama, M., W. Suhara, and T. Fujita. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22:73-76. [DOI] [PubMed] [Google Scholar]
- 54.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]