Abstract
Nuclear domain 10 (ND10s), or promyelocytic leukemia protein (PML) nuclear bodies, are spherical nuclear structures that require PML proteins for their formation. Many viruses target these structures during infection. The E4 Orf3 protein of adenovirus 5 (Ad5) rearranges ND10s, causing PML to colocalize with Orf3 in nuclear tracks or fibers. There are six different PML isoforms (I to VI) present at ND10s, all sharing a common N terminus but with structural differences at their C termini. In this study, PML II was the only one of these six isoforms that was found to interact directly and specifically with Ad5 E4 Orf3 in vitro and in vivo; these results define a new Orf3 activity. Three of a series of 18 mutant Orf3 proteins were unable to interact with PML II; these were also unable to cause ND10 rearrangement. Moreover, in PML-null cells that contained neoformed ND10s comprising a single PML isoform, only ND10s formed of PML II were rearranged by Orf3. These data show that the interaction between Orf3 and PML II is necessary for ND10 rearrangement to occur. Finally, Orf3 was shown to self-associate in vitro. This activity was absent in mutant Orf3 proteins that were unable to form tracks and to bind PML II. Thus, Orf3 oligomerization may mediate the formation of nuclear tracks in vivo and may also be important for PML II binding.
The infection of a cell by adenovirus type 5 (Ad5) results in the virus genome entering the nucleus, where it becomes active for transcription and, after the production of necessary viral proteins, DNA replication. During this colonization process, the virus alters the cell environment in various ways so as to facilitate replication and also to counteract host responses to the infection that would otherwise interfere in these events. One of the viral effectors of change within the cell nucleus during this early phase of infection is the E4 gene open reading frame 3 protein product (Orf3). Ad5 Orf3 has a number of functions ascribed to it. First, it influences differential splicing in the viral major late transcription unit (34). Second, it prevents the activation and recruitment of components of the host double-strand DNA break repair pathway to viral replication centers and so prevents the concatenation of linear viral genomes (4, 37). Third, it is necessary and sufficient for the disruption of nuclear structures termed variously nuclear domain 10 (ND10s) or promyelocytic leukemia protein (PML) bodies (8, 9). Last, it is necessary for another viral protein, E1b 55K, to localize to ND10s (28, 29) and for blocking the E1b 55K effect on p53 activation (26).
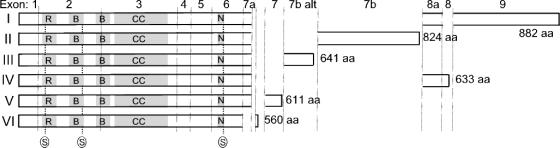
ND10s are dense granular nuclear bodies that are visualized by immunofluorescence as discrete dots numbering 10 to 20 per nucleus (11, 25, 40). Multiple proteins localize to ND10s (32), but PML protein is the key component as other ND10 proteins depend on PML for their localization to these structures (22, 42). A number of PML isoforms arise by translation from differentially spliced mRNA (17). Six of these isoforms (PML I to VI) have a common N-terminal domain of some 550 residues linked to distinct C termini of up to 330 amino acids (Fig. 1); other isoforms lack parts of the common domain necessary for nuclear localization and so are cytoplasmic (23). PML protein diversity is further increased by posttranslational coupling to SUMO-1, a ubiquitin-homology family member (10), and by mitosis-specific phosphorylation (16). There is growing evidence that this structural diversity in PML protein is reflected in its functional diversity at ND10 (2, 41).
FIG. 1.
Sequence relationship between PML isoforms I to VI. The proteins are represented as horizontal bars with lengths indicated in amino acid residues (aa). Vertical alignment of sequences indicates identity. The exons encoding each piece of sequence are indicated at the top of the figure, with boundaries denoted by vertical lines. Exon 7b alt indicates translation of exon 7b in an alternative reading frame through use of an alternate splice acceptor. Three sites of potential posttranslational modification with SUMO-1 are indicated (S). The ring (R), B-boxes (B), and coiled-coil motifs (CC) important for ND10 localization and PML multimerization and the nuclear localization signal (N) are shaded.
The organization of ND10s is sensitive to various stresses (7, 24, 30), and they are disrupted in disease states such as acute promyelocytic leukemia. Many viruses also affect ND10 composition and/or organization, though the specific effects are different in each case (reviewed in references 15, 27, and 35); during Ad5 infection, Orf3 disrupts ND10s, causing PML protein to form track-like structures (8, 9). Also, the incoming genomes of the nucleus-replicating DNA viruses, including Ad5, localize adjacent to ND10s early in infection (21). The generality of these virus interactions with ND10s suggests that there might be a common purpose underlying them. ND10s and/or PML protein have been implicated in a broad range of key cellular processes, including senescence, apoptosis, DNA damage responses, the innate immune response, and control of gene expression (reviewed in reference 3). Viruses might therefore alter ND10s either to disrupt or to harness for their benefit one or more of these functions.
Given the complexity of ND10 composition, the disruption of ND10s by Ad5 E4 Orf3 might be mediated via interaction with or effects on many different ND10 components. However, PML proteins are unique among known ND10 components in being required for the organization of other ND10 components, and, since Orf3 causes a gross rearrangement of PML and other ND10 proteins, PML proteins represent a likely molecular target for Orf3. In this report, we show for the first time that Ad5 E4 Orf3 does interact with PML protein. The interaction is directly with one specific isoform of PML protein, PML II, and this interaction is required for ND10 disruption.
MATERIALS AND METHODS
Cell culture, transfection, and antibodies.
HEp-2 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, whereas U2OS cells were maintained in similarly supplemented McCoy's 5A medium. Primary mouse embryo fibroblasts (MEFs) were generated from day 13 embryos of PML−/− sv129S2 mice (39); grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin; and used at passage 3. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. Anti-FLAG antibodies were M2 mouse monoclonal antibody or rabbit monospecific antiserum (Sigma). Orf3 was detected with rat monoclonal antibody 6A11 (33), kindly provided by T. Dobner. Other primary antibodies were anti-PML mouse monoclonal antibody PG-M3 and anti-nibrin goat polyclonal antibody C-19 (both Santa Cruz). Secondary antibodies were, for Western blotting, horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Santa Cruz) or goat anti-rat IgG (Chemicon) and, for immunofluorescence, Alexa488 goat anti-mouse IgG, Alexa594 rabbit anti-goat IgG, Alexa546 goat anti-rat IgG, and Alexa488 goat anti-rat IgG (Molecular Probes).
DNA manipulation.
To create a eukaryotic expression vector for Ad5 Orf3, the coding sequence (genome positions 34704 to 34288) was amplified by PCR from viral genomic DNA and cloned as a BamHI-EcoRI fragment in pcDNA3.1 using restriction sites incorporated into the primers. Variants of this plasmid encoding proteins with specific amino acid substitutions or deletions were constructed by a two-step PCR protocol. Primers were designed corresponding to both DNA strands at the target site for mutation and containing the intended sequence changes, as outlined at the protein level in Fig. 3. Each primer was used in a PCR with either the 5′ or 3′ flanking primer used to create the parental clone, to give a pair of overlapping DNA fragments. This pair of fragments was then used as template in a second PCR with both 5′ and 3′ flanking primers to give the full-length altered Orf3 gene. These PCR products were subsequently digested with BamHI and EcoRI and cloned into the pcDNA3.1 vector, and the changes were verified by DNA sequencing. FLAG-tagged PML isoforms were expressed from individual cDNA clones in vector pCINeo as described previously (1).
FIG. 3.
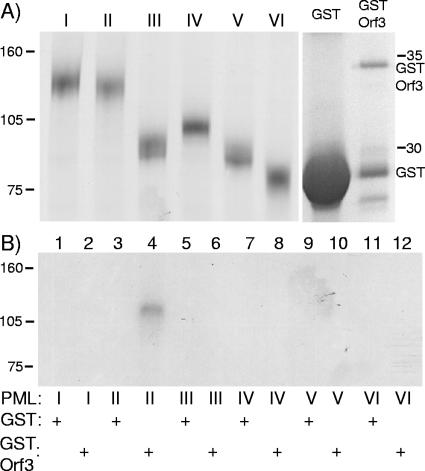
Interaction of Ad5 Orf3 with PML II in vitro. 35S-labeled, FLAG-tagged PML isoforms synthesized in vitro were incubated with crude bacterial cell lysates containing either GST or GST-Orf3 fusion protein. Affinity-purified GST and associated proteins were then analyzed by SDS-PAGE, and radiolabeled proteins were detected by autoradiography. (A) (Left panel) Labeled PML protein inputs to the assay. (Right panel) Affinity-purified GST proteins at the conclusion of the assay, detected by Coomassie blue staining. (B) Postbinding, glutathione affinity-purified labeled proteins from samples containing PML isoforms and either GST or GST-Orf3 as indicated below each lane. The positions to which marker proteins of known size migrated are indicated to the left and right of the figure.
Coimmunoprecipitation analysis.
Experiments were performed essentially according to Guccione et al. (19). All steps were carried out at 4°C or on ice, using precooled solutions. Transfected cells (7.5 × 106) were washed twice with 2 ml phosphate-buffered saline; lysed in a mixture of 0.5 ml 25 mM HEPES, pH 7, 0.1% NP-40, 0.5 M NaCl, 1 mM sodium butyrate, 1% protease inhibitor cocktail (Sigma, P8340), and 1% phosphatase inhibitor cocktail (Sigma, P5726); and scraped from the dish. Extracts were allowed to stand for 20 min, sonicated for 10s with a microtip sonicator, clarified by centrifugation (12,000 × g, 20 min) and then diluted with an equal volume of lysis buffer containing reduced salt (50 mM NaCl). For FLAG-PML precipitation, 20 μl of M2 antibody-coupled agarose beads (50% slurry; Sigma, A2220), prewashed twice with wash buffer (50 mM Tris HCl, 150 mM NaCl, pH 7.4, containing 0.01% protease and phosphatase inhibitor cocktails), once with 0.2 ml of 0.1 M glycine-HCl, pH 3.5, and then three times with wash buffer, was incubated with extract for 3 h. Agarose beads were then washed six times with wash buffer. Bound proteins were eluted at room temperature from bead pellets by adding 40 μl of 2× sodium dodecyl sulfate (SDS) gel sample buffer lacking reducing agent and boiling for 3 min. Samples were separated from the beads by centrifugation before adding 10 μl of 1 M dithiothreitol. Following separation by SDS-polyacrylamide gel electrophoresis (PAGE), proteins were transferred to Hybond-C filters (Amersham). Filters were blocked and probed using antibodies as described above followed by detection with the ECL Advance enhanced chemiluminescence reagent (Amersham) according to the manufacturer's protocol.
GST pull-down analysis.
Radiolabeled PML or Orf3 proteins were expressed by in vitro translation with [35S]methionine, using the TNT T7 coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. The Orf3 coding sequence was cloned into pGEX-2T (Amersham) to produce a glutathione S-transferase (GST) fusion. GST-tagged Orf3 protein and GST alone were each expressed in Escherichia coli cells grown in Luria-Bertani (LB) broth either for 4 h postinduction with 1 mM isopropylthiogalactoside (GST) or in LB broth supplemented with 0.5 M sorbitol and 2.5 mM betaine, for 20 h postinduction (GST-tagged Orf3). Bacterial cells were resuspended in phosphate-buffered saline containing protease inhibitor cocktail (Roche) and lysed by French press, and the crude lysates, clarified by centrifugation at 27,200 × g, were mixed with radiolabeled, in vitro-translated proteins and glutathione-agarose beads (Sigma). Binding was allowed to occur at 4°C for 3 h. After washing, proteins were eluted from the beads with SDS gel sample buffer and resolved by SDS-PAGE, and radioactive proteins were detected by exposure to Fuji RX film.
Immunofluorescence analysis.
HEp-2 cells and MEFs were grown on coverslips, transfected, fixed, and stained for specific antigens as described previously (28). Images were generated using a Leica SP2 confocal system, employing sequential channel scanning for multiply stained samples to eliminate fluorescence crossover between channels, and all images shown are maximal projections through a z-series. To define the nuclei of cells visualized by immunofluorescence, DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). Images were captured as full-color snapshots within the Leica confocal software and assembled for display without subsequent adjustment.
RESULTS
E4 Orf3 interacts specifically and directly with PML II.
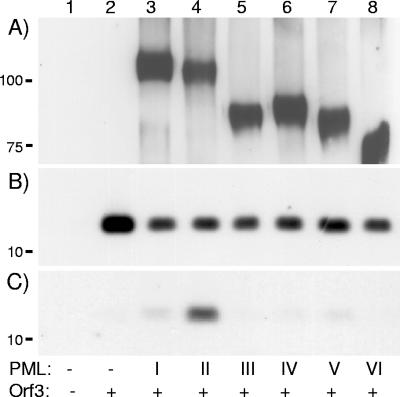
The E4 Orf3 proteins of different Ad serotypes have been shown to colocalize with ND10s and to cause these spherically shaped structures to assume a fibrous or track-like appearance (8, 13, 38). Since PML proteins are the only component of ND10s known to be required for their formation, we tested whether Orf3 might interact with one or more PML protein isoforms. U2OS cells were transiently transfected with expression plasmids for Ad5 E4 Orf3 and FLAG-tagged PML isoforms I to VI, and the cell extracts were analyzed by coimmunoprecipitation with anti-FLAG antibody. Each FLAG-PML isoform was equivalently expressed and immunoprecipitated from the relevant transfection (Fig. 2A). Each culture cotransfected with Orf3 and PML expressed Orf3 to similar levels (Fig. 2B). However, when FLAG-PML immunoprecipitates from these cells were examined for the presence of E4 Orf3, only PML II was found to coprecipitate this protein (Fig. 2C, lane 4). Identical results were obtained when Orf3 was provided by a superinfection of PML-transfected cells with wild-type Ad5 (data not shown).
FIG. 2.
Interaction of Ad5 Orf3 with PML II by coimmunoprecipitation. Cultures containing 7.5 × 106 U2OS cells were transfected with 6.4 μg of each PML expression plasmid and/or with 8.8 μg Orf3 expression plasmid as indicated below each lane, and lysates were prepared 24 h posttransfection. A fraction of the extract was retained for direct Western blot analysis, and the remainder was immunoprecipitated with anti-FLAG antibody immobilized on agarose beads. (A) Anti-FLAG immunoprecipitates analyzed by SDS-PAGE and Western blotting with anti-FLAG antibodies, detecting individual exogenously expressed PML isoforms. (B) Cell extract analyzed as in panel A, but with anti-Orf3 antibodies, detecting total expressed Orf3. (C) Anti-FLAG immunoprecipitates analyzed as in panel A, but with anti-Orf3 antibodies, detecting coimmunoprecipitated Orf3. The positions to which marker proteins of known size migrated are indicated to the left of the figure.
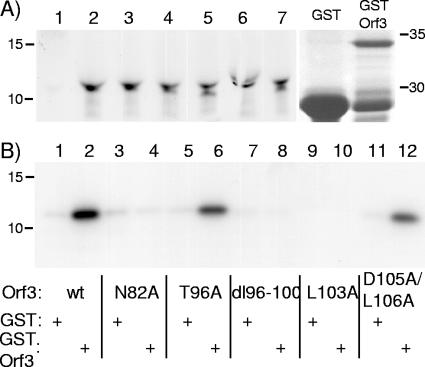
ND10s are multiprotein assemblies, and PML is known to participate in multiple protein-protein interactions (32). Thus, the specific coprecipitation of Orf3 via PML II might reflect either a direct interaction between the proteins or an association mediated via other components. To determine whether the interaction between E4 Orf3 and PML II was direct, a GST pull-down assay was performed. Prior quantitation of the GST-Orf3 protein for these assays was not possible because a significant proportion was cleaved during purification and/or storage. Thus, experiments were done with proteins immediately postextraction from bacteria, and the amounts present were quantified postanalysis. The amount of GST present in the control assays was routinely greater than the amount of GST-Orf3 (Fig. 3A), so the potential for nonspecific sticking of radiolabeled target protein during the assays was, if anything, greater for the control GST reactions than for the experimental samples. Each of the FLAG-PML isoforms was equivalently expressed in vitro as labeled protein (Fig. 3A). When these proteins were individually incubated with control GST protein, there was minimal association detected in each case (Fig. 3B). However, when incubated with GST-Orf3, labeled PML II was specifically bound (Fig. 3B, lane 4), confirming the coimmunoprecipitation results. None of the other PML isoforms interacted with GST-Orf3 in this assay. The fact that PML II bound to Orf3 in the absence of any other eukaryotic cell protein indicates that the interaction between these proteins is direct.
PML II-Orf3 interaction requires the C-terminal part of Orf3.
To investigate the sequence requirements in Orf3 for its interaction with PML II, a series of specific mutations were constructed in an Orf3 expression plasmid. Mutagenesis targets were initially selected from an alignment of four Orf3 sequences (Ad5, Ad9, Ad12, and Ad40) on the basis that residues important for this interaction would be conserved between Ad serotypes. This alignment was subsequently expanded, and further targets for mutation were selected; additional mutations were also made based on other published studies of Orf3 function (14, 37). In total, 2 deletions, (positions 38 to 42 [dl38-42] and 96 to 100 [dl96-100]), 14 single-amino-acid alanine substitutions, and 2 double-amino-acid substitutions were constructed (Fig. 4).
FIG. 4.
Alignment of the amino acid sequences of E4 Orf3 proteins from human Ad serotypes 5, 9, 12, and 40, representing subgroups C, D, A, and F, respectively, showing the position of single (single arrows) and double (underlined double arrows) alanine substitution mutations and in-frame deletion mutations (boxed) constructed in Ad5 Orf3 expression plasmids in this study. Sequences were taken from GenBank database entries AP_000230 (Ad5), AAB37506 (Ad9), AP_000139 (Ad12), and AAC13983 (Ad40).
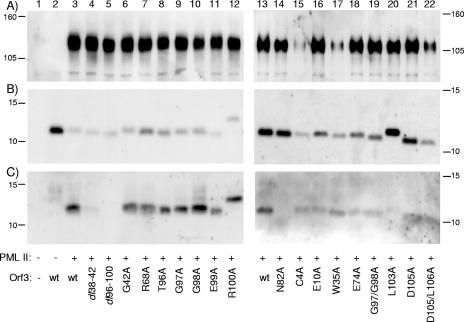
Each member of the panel of Orf3 mutants was tested for interaction with PML II by coimmunoprecipitation analysis from cotransfected cell extracts (Fig. 5). Similar amounts of PML II were expressed in all but two samples (Fig. 5A). The reduced amount of PML II in these two samples (Fig. 5A, lanes 15 and 17) was nevertheless sufficient to show a positive interaction with the coexpressed mutant Orf3 protein. Each of the mutant Orf3 proteins was expressed, although not all were expressed to the same level (Fig. 5B). Orf3 R100A and L103A reproducibly migrated more slowly than wild-type Orf3 on protein gels (Fig. 5B, lanes 12 and 20). Most of the mutated Orf3 proteins were coimmunoprecipitated with FLAG-PML II (Fig. 5C), although not all with the same efficiency; dl38-42 in particular showed reduced binding (Fig. 5C, lane 4). However, three mutant proteins, dl96-100, N82A, and L103A, were unable to interact with PML II in repeated experiments (Fig. 5C, lanes 5, 14, and 20). These data suggest that the C-terminal part of the 116-residue Orf3 protein is important for its interaction with PML II.
FIG. 5.
Mapping the interaction site of PML II on Orf3. Wild-type (wt) or mutant Orf3 was coexpressed with FLAG-PML II in U2OS cells, as indicated below each lane, and interactions were assessed as in Fig. 2. (A) Western blot of cell extracts probed with anti-FLAG antibodies, detecting exogenously expressed PML II. (B) Cell extract analyzed as in panel A, but with anti-Orf3 antibodies, detecting total expressed Orf3. (C) Anti-FLAG immunoprecipitates analyzed as in panel A, but with anti-Orf3 antibodies, detecting coimmunoprecipitated Orf3. The positions to which marker proteins of known size migrated are indicated to the left and right of the figure.
The Orf3 function in ND10 rearrangement requires interaction with PML II.
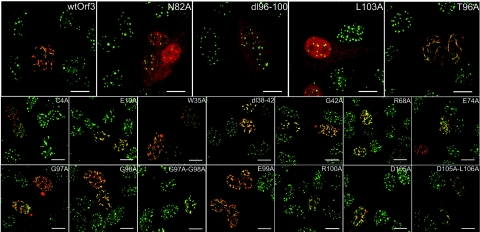
One of the well-documented functions of Orf3 is its ability to colocalize with ND10s in the cell nucleus and to rearrange PML and other ND10 proteins into a large number of tracks. To investigate which regions of the Orf3 protein were important for these functions, cells transfected with the mutated Orf3 expression plasmids were analyzed by double-label immunofluorescence for Orf3 and PML. HEp-2 cells were used for this analysis because their endogenous PML is readily detectable by this technique. Most of the mutated Orf3 proteins were able to colocalize with PML and to form tracks in the nucleus similar to wild-type Orf3 (Fig. 6). However, three mutants, dl96-100, N82A, and L103A (Fig. 6, top row), each showed a diffuse staining throughout the nucleus and cytoplasm and the ND10s in cells expressing these proteins, as revealed by PML staining, were not rearranged. The inability of N82A and L103A to rearrange ND10s and the aberrant localization of the latter mutant protein are in agreement with previously published results (14, 37). Each of the three mutant proteins that failed to rearrange ND10s was also unable to bind PML II (Fig. 5), suggesting that this interaction is necessary for ND10 rearrangement.
FIG. 6.
Mapping the region of Orf3 required for ND10 rearrangement. HEp2 cells grown on coverslips were transfected with expression plasmids for wild-type (wt) Ad5 Orf3 or mutant Orf3 as indicated, fixed and stained for Orf3 (red) and for endogenous PML proteins (green), and visualized by confocal microscopy. Yellow indicates colocalization of PML and Orf3 proteins. The images shown are overlays of red and green channels collected sequentially to avoid cross talk between the fluors. Scale bar, 12 μm.
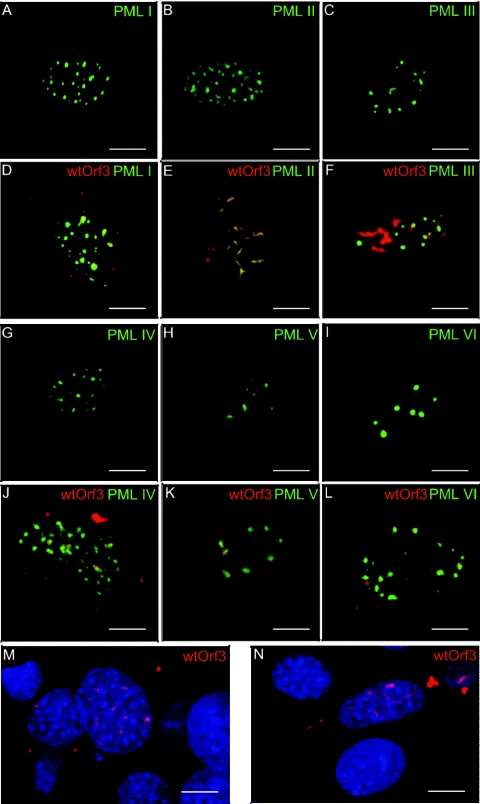
To further investigate this question, the ability of Orf3 to interact with and disrupt ND10 structures containing only one form of PML was tested. Neoformed ND10s were generated in PML-null primary MEFs by transfecting the six FLAG-PML expression plasmids individually. The effect of wild-type Orf3 on these ND10s was then examined by double-label immunofluorescence and confocal microscopy. In the absence of Orf3, each of the six PML isoforms formed nuclear bodies reminiscent of ND10s (Fig. 7A to C and G to I). The number of bodies formed by PML VI was fewer than for the other isoforms, as has been previously observed for PML VI overexpression in the presence of endogenous PML (1). When Orf3 was coexpressed with FLAG-PML, there was a clear distinction between PML II and the other isoforms. While FLAG-PML II bodies were disrupted into filamentous structures that colocalized with Orf3 (Fig. 7E), bodies formed from any of the other five isoforms remained essentially unaltered (Fig. 7D, F, and J to L). Although Orf3 did localize close to some of the PML I, III, IV, V, and VI bodies in a subset of the cells examined, as shown in Fig. 7, disruption of these PML bodies was never seen. Thus, the interaction of Orf3 with PML II is necessary for ND10 rearrangement.
FIG. 7.
PML II is required in ND10s for rearrangement by Orf3. PML−/− mouse embryo fibroblasts grown on coverslips were transfected with expression plasmids for wild-type Ad5 Orf3 and/or individual FLAG-PML isoforms as indicated. Cells were fixed and stained for Orf3 (red) and for FLAG-PML (green) and visualized by confocal microscopy. Yellow indicates colocalization of PML and Orf3 proteins. Nuclei were visualized by staining DNA (DAPI; blue). The images shown are overlays of channels collected sequentially to avoid cross talk between the fluors. Scale bar, 8 μm.
Orf3 track formation correlates with Orf3-Orf3 interaction.
Orf3 is a small protein of around 11 kDa that would be expected to diffuse freely between nucleus and cytoplasm. However, its actual localization pattern, to tracks in the nucleus, and the fact that it is very hard to solubilize from cells (36) suggest that it is anchored via interactions with other components. In contrast to wild-type Orf3, three mutant proteins were diffusely distributed across both the nucleus and the cytoplasm of the cell, suggesting that these mutations had abolished the ability of Orf3 to anchor in the nucleus. These mutant proteins were also unable to bind to PML II (Fig. 5), raising the possibility that Orf3 might localize to nuclear tracks through its interaction with PML II. However, wild-type Orf3 expressed in PML-null MEFs still adopted the characteristic track-like nuclear localization, albeit with an increased frequency of cytoplasmic aggregates (Fig. 7M and N), indicating that PML II is not required for Orf3 to form tracks in the nucleus.
Both Orf3 track formation and its resistance to solubilization from cells might be explained by oligomerization in the nucleus. To test this possibility, wild-type Orf3 and selected mutant Orf3 proteins were tested for binding to wild-type Orf3 in a GST pull-down assay. As shown in Fig. 8A, equivalent amounts of each labeled Orf3 protein were used. The amount of GST used as a control was greater than that of GST-Orf3 (Fig. 8A), as explained for Fig. 3. Wild-type Orf3 bound efficiently to GST-Orf3, but not to the GST control (Fig. 8B, lanes 1, 2), indicating that Orf3 has the ability to self-associate. However, each of the three mutants N82A, dl96-100, and L103A that lacked track formation, ND10 rearrangement, and PML II binding activities was unable to bind significantly to GST-Orf3 (lanes 3, 4, and 7 to 10). This finding suggests that Orf3 self-association is required for these activities.
FIG. 8.
Self-interaction of Ad5 Orf3 with mutant Orf3 proteins in vitro. 35S-labeled wild-type (wt) or mutant Orf3 synthesized in vitro was incubated with crude bacterial cell lysates containing either GST or GST-Orf3 fusion protein. Affinity-purified GST and associated proteins were then analyzed by SDS-PAGE, and radiolabeled proteins were detected by autoradiography. (A) (Left panel) Labeled Orf3 protein used in the assays. Lanes: 1, control translation; 2, wtOrf3; 3, N82A; 4, T96A; 5, dl96-100; 6, L103A; and 7, D105A/L106A. (Right panel) Affinity-purified GST proteins at the conclusion of the assay, detected by Coomassie blue staining. (B) Postbinding, glutathione affinity-purified labeled proteins from samples containing labeled Orf3 and GST or GST-Orf3 as indicated below each lane. The positions to which marker proteins of known size migrated are indicated to the left and right of the figure.
Mutants N82A and L103A were shown previously to be unable to target Mre11-Rad50-Nbs1 (MRN) complexes in the DNA double-strand break repair pathway (14, 37). We confirmed this result, using anti-Nbs1 staining, and found that the third mutant that lacked the Orf3 self-association function, dl96-100, behaved in the same way (data not shown), raising the possibility that failure to self-associate might also be linked with this MRN complex reorganization. However, mutant D105A/L106A, which also fails to target the MRN complex but forms tracks normally and reorganizes ND10s like wild-type Orf3 (14), retained Orf3 self-interaction ability similar to the wild-type protein (Fig. 8, lanes 11 and 12). Moreover, T96A, a second mutant that did not colocalize normally with Nbs1, although some rearrangement of Nbs1 into foci was apparent in cells transfected with this mutant (data not shown), was also still able to interact normally with wild-type Orf3 (Fig. 8, lanes 5 and 6). All other mutants analyzed in this study behaved like wild-type Orf3 with respect to the MRN complex (data not shown). Thus, Orf3 self-association ability does not correlate with the MRN complex targeting activity of the protein, but does correlate with its ability to form tracks, bind PML II, and reorganize ND10s.
DISCUSSION
The Ad5 E4 Orf3 protein is necessary and sufficient for the rearrangement of ND10s from their normal spherical appearance into a larger number of track-like structures (8, 9). The data presented here show that the molecular basis of this effect is an interaction of Orf3 with PML protein, which is a key ND10 component (22, 42). Orf3 was shown to bind specifically and directly with PML II, one of the six PML protein isoforms that is located in the nucleus at ND10s (1). Since no other PML isoforms could bind Orf3, sequences encoded by exon 7b, the only region unique to PML II (Fig. 1), must be required for binding to occur, though they may not be sufficient. Among a series of Orf3 mutants, each of the three mutants unable to bind PML II was also unable to rearrange ND10s, while the remainder retained both activities. Moreover, in cells containing artificial ND10s formed of single PML isoforms, only those ND10s that were formed of PML II were rearranged by wild-type Orf3. Thus, we conclude that the PML II-Orf3 interaction is necessary for ND10 rearrangement.
Two previous studies have failed to detect an interaction between Orf3 and PML proteins in coimmunoprecipitation experiments. Doucas et al. (9) used only one isoform, PML VI, in their work, and thus their negative result agrees with the present study. Evans and Hearing (13) reported that no Orf3 could be detected in Western blots of samples from A549 cells expressing epitope-tagged Orf3 and immunoprecipitated for endogenous PML. There are two possible explanations for this result in the light of the findings reported here. First, it is not known what fraction of endogenous PML protein is comprised of PML II. If the amount of this isoform is low, the assay might not be sufficiently sensitive to detect the interaction with Orf3, although there is clearly a sufficient amount of this isoform present in natural ND10s to allow their rearrangement by Orf3. Second, the conditions needed to solubilize endogenous PML for the assay might have been too harsh to preserve the interaction with Orf3. By using overexpression in the present study, much of the PML II present would be expected to be in a nonsumoylated, more readily extractable, form. In the experiments described here, mild extraction conditions produced an extract containing significant amounts of PML protein, typically greater than 50% of the total exogenous PML protein isoform present in the cells.
Wild-type Orf3 localizes to nuclear tracks. In contrast, three Orf3 mutant proteins showed aberrant localization, being diffusely distributed throughout the nucleus and cytoplasm. These mutants also lacked PML II binding activity, suggesting this might be involved in Orf3 localization. However, PML II was not required for Orf3 to localize to nuclear tracks, suggesting that some other activity of Orf3 was necessary for this localization and that this had also been compromised by these mutations. One novel property of Orf3, which our data showed to be specifically abrogated in mutant Orf3s that were unable to form tracks, was an ability to self-associate in vitro, suggesting that Orf3 may have the ability to form oligomers in vivo. Such activity could account for the adoption by Orf3 of its characteristic subnuclear localization pattern, when a protein of its size would otherwise be small enough to be freely diffusible. However, if this is the case, it is not clear yet what prevents such oligomers from also forming in the cytoplasm. Since each of the mutant Orf3 proteins that lacked PML II binding was also unable to bind wild-type Orf3 in vitro, Orf3 self-association may be necessary for PML II binding.
The three Orf3 mutant proteins that lacked PML II binding, ND10 rearrangement, and self-association activities all accumulated in cells to levels similar to that of wild-type Orf3. These mutations are therefore unlikely to be causing a gross disruption of protein folding. However, it is possible that these proteins lack interactions with cellular partners other than PML II and that these are relevant for track formation. A role in this process for an interaction with the MRN complex of the DNA double-strand break repair pathway could be excluded based on the properties of specific Orf3 mutant proteins (14, 37; this study). Also, the localization pattern of other known Orf3-interacting cellular partners CBP, p300, and DNA-PK (4, 13), is not at all similar to that of Orf3, and thus, these cannot be serving as a prelocalized nuclear anchor for Orf3. However, it is possible that other interactions of Orf3 that might be important for track formation remain to be discovered.
The specific distribution pattern of Orf3 within the nucleus, particularly in cells showing strong overexpression following plasmid transfection (data not shown), is reminiscent of that formed by the intermediate filament protein vimentin, when artificially directed to the nucleus by a heterologous nuclear localization signal (6, 18). Tracks or fibers of nuclear vimentin were shown to form in interchromatin spaces that surrounded chromosome territories; ND10s were also located in these spaces. It is possible that wild-type Orf3 normally behaves similarly to the experimental vimentin construct. This would explain the occasional colocalization of Orf3 with neoformed ND10s containing PML protein isoforms other than PML II in our experiments, since they would both be confined within the same region of the nucleus.
The purpose of ND10 reorganization by Orf3 in the context of virus infection remains uncertain. Mutant Ad5 unable to express Orf3, but having all other functions intact, is essentially wild type for growth on standard cell lines in culture (5, 20). However, a growth effect of the Orf3 mutation is manifest in a genetic background which is deficient in E1b 55K or another E4 protein, Orf6, indicating that there is some redundancy of function between these proteins. The Orf3 function that is responsible for this growth effect in cell culture has been shown to be the blocking of DNA double-strand break repair (14). However, although Orf3 is conserved among all human adenoviruses, only Ad5 Orf3 has the ability to target MRN; this is thought to be a property exclusive to Orf3 from group (species) C adenoviruses (38). Thus, some other Orf3 function has driven the evolution of this protein and its retention within human adenoviruses. This is likely to be related to the ND10 disruption function of Orf3, which is conserved in Orf3 from all serotypes tested (38).
The fact that Orf3 targets ND10s via PML II suggests that this PML protein isoform may have specific functions that are relevant to Ad5 infection. However, in contrast to some other PML protein isoforms, there is no functional information available about PML II that might indicate why the virus targets this isoform specifically. ND10 interaction and/or disruption is a common feature among many viruses (reviewed in reference 27), and for herpes simplex virus at least, there is good evidence that the interaction is important in counteracting the innate immune response (12, 31). Further experiments are needed to determine whether the E4 Orf3 effect on ND10s has a similar role during adenovirus infection.
Acknowledgments
The authors gratefully acknowledge the gifts of cDNA clones used to create PML expression plasmids from E. Solomon (Guys Hospital, London), P. G. Pelicci (European Institute of Oncology, Milan), A. Dejean (Institute Pasteur, Paris), and R. Everett (Institute of Virology, Glasgow); Orf3 antibody from T. Dobner (University of Regensberg, Regensberg, Germany); and U2OS cells from P. Jat (Ludwig Institute for Cancer Research, UCL, London). PML-null mice were generously provided by P. Salomoni (MRC Toxicology Unit, University of Leicester, Leicester, United Kingdom) with kind permission from P. P. Pandolfi (Memorial Sloane-Kettering Cancer Center, New York, N.Y.).
S.J.B. and J.D. were supported by research studentships from the Biotechnology and Biological Sciences Research Council, U.K., and Medical Research Council, U.K., respectively.
REFERENCES
- 1.Beech, S. J., K. J. Lethbridge, N. Killick, N. McGlincy, and K. N. Leppard. 2005. Isoforms of the promyelocytic leukemia protein differ in their effects on ND10 organization. Exp. Cell Res. 307:109-117. [DOI] [PubMed] [Google Scholar]
- 2.Bischof, O., O. Kirsh, M. Pearson, K. Itahana, P. G. Pelicci, and A. Dejean. 2002. Deconstructing PML-induced premature senescence. EMBO J. 21:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden, K. L. B. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 5.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridger, J. M., H. Herrmann, C. Munkel, and P. Lichter. 1998. Identification of an interchromosomal compartment by polymerization of nuclear-targeted vimentin. J. Cell Sci. 111:1241-1253. [DOI] [PubMed] [Google Scholar]
- 7.Carbone, R., M. Pearson, S. Minucci, and P. G. Pelicci. 2002. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21:1633-1640. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmofonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 10.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 11.Dyck, J. A., G. G. Maul, W. J. Miller, J. D. Chen, A. Kakizuka, and R. M. Evans. 1994. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76:333-343. [DOI] [PubMed] [Google Scholar]
- 12.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, J. D., and P. Hearing. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D., P. Lamonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112:4581-4588. [DOI] [PubMed] [Google Scholar]
- 17.Fagioli, M., M. Alcalay, P. P. Pandolfi, L. Venturini, A. Mencarelli, A. Simeone, D. Acampora, F. Grignani, and P. G. Pelicci. 1992. Alternative splicing of PML transcripts predicts expression of several carboxyterminally different protein isoforms. Oncogene 7:1083-1091. [PubMed] [Google Scholar]
- 18.Gorisch, S., P. Lichter, and K. Rippe. 2005. Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments. Histochem. Cell Biol. 123:217-228. [DOI] [PubMed] [Google Scholar]
- 19.Guccione, E., K. J. Lethbridge, N. Killick, K. N. Leppard, and L. Banks. 2004. HPV E6 proteins interact with specific PML isoforms and allow distinctions to be made between different POD structures. Oncogene 23:4662-4672. [DOI] [PubMed] [Google Scholar]
- 20.Huang, M.-M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. H. Yeh, J. F. Strauss, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 24.Kamei, H. 1996. Reversible large-body formation from nuclear bodies upon amino acid(s) starvation in T24 cells. Exp. Cell Res. 224:302-311. [DOI] [PubMed] [Google Scholar]
- 25.Koken, M. H. M., F. Puvion-Dutilleul, M. C. Guillemin, A. Viron, G. Linarescruz, N. Stuurman, L. Dejong, C. Szostecki, F. Calvo, C. Chomienne, L. Degos, E. Puvion, and H. de The. 1994. The t(1517) translocation alters a nuclear-body in a retinoic acid-reversible fashion. EMBO J. 13:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konig, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J. Virol. 73:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leppard, K. N., and J. Dimmock. Virus interactions with PML nuclear bodies. In J. Hiscox (ed.), Viruses and the nucleus, in press. John Wiley, New York, N.Y.
- 28.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 29.Lethbridge, K. J., G. E. Scott, and K. N. Leppard. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84:259-268. [DOI] [PubMed] [Google Scholar]
- 30.Maul, G. G., E. Yu, A. M. Ishov, and A. L. Epstein. 1995. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell Biochem. 59:498-513. [DOI] [PubMed] [Google Scholar]
- 31.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 33.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 73:1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nordqvist, K., K. Öhman, and G. Akusjärvi. 1994. Human adenovirus encodes 2 proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 36.Sarnow, P., P. Hearing, C. W. Anderson, N. Reich, and A. J. Levine. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J. Mol. Biol. 162:565-583. [DOI] [PubMed] [Google Scholar]
- 37.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 38.Stracker, T. H., D. V. Lee, C. T. Carson, F. D. Araujo, D. A. Ornelles, and M. D. Weitzman. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 40.Weis, K., S. Rambaud, C. Lavau, J. Jansen, T. Carvalho, M. Carmo-Fonseca, A. Lamond, and A. Dejean. 1994. Retinoic acid regulates aberrant nuclear-localization of PML-RAR-alpha in acute promyelocytic leukemia cells. Cell 76:345-356. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Z. X., W. X. Zou, P. Lin, and K. S. Chang. 2005. A role for PML3 in centrosome duplication and genome stability. Mol. Cell 17:721-732. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2753. [PubMed] [Google Scholar]