Abstract
A gene, HDC1, related to the Saccharomyces cerevisiae histone deacetylase (HDAC) gene HOS2, was isolated from the filamentous fungus Cochliobolus carbonum, a pathogen of maize that makes the HDAC inhibitor HC-toxin. Engineered mutants of HDC1 had smaller and less septate conidia and exhibited an ∼50% reduction in total HDAC activity. Mutants were strongly reduced in virulence as a result of reduced penetration efficiency. Growth of hdc1 mutants in vitro was normal on glucose, slightly decreased on sucrose, and reduced by 30 to 73% on other simple and complex carbohydrates. Extracellular depolymerase activities and expression of the corresponding genes were downregulated in hdc1 mutant strains. Except for altered conidial morphology, the phenotypes of hdc1 mutants were similar to those of C. carbonum strains mutated in ccSNF1 encoding a protein kinase necessary for expression of glucose-repressed genes. These results show that HDC1 has multiple functions in a filamentous fungus and is required for full virulence of C. carbonum on maize.
INTRODUCTION
Reversible histone acetylation is a major mechanism by which chromatin structure and gene regulation are integrated. The level of acetylation of histones in chromatin is a product of the balance between the activities of two enzymes, histone acetyltransferase (HAT) and histone deacety-lase (HDAC), and both enzymes have been demonstrated to be necessary for the correct expression of a number of genes (Struhl, 1998).
Transcriptionally active chromatin is hyperacetylated compared with inactive chromatin. Consistent with this, HATs are present in many transcriptional activation complexes, whereas HDACs are present in many repressor complexes (Struhl, 1998; Ng and Bird, 2000). In many cases, HATs and HDACs have been shown to bind to DNA binding proteins either directly or via coactivators and corepressors, respectively (Doetzlhofer et al., 1999; Knoepfler and Eisenman, 1999; Miska et al., 1999). Although a number of genes have been shown to be sensitive to the acetylation state of their associated histones, with acetylation promoting expression and deacetylation promoting repression, the expression of other genes appears to be independent of the acetylation state of their associated histones (Van Lint et al., 1996a, 1996b; Luo et al., 1998; Rundlett et al., 1998; Knoepfler and Eisenman, 1999; Bernstein et al., 2000). A further complication in the analysis of HDAC function in yeast and other organisms has been genetic redundancy. For example, in yeast, the mutation of individual HDAC genes often gives a subtle phenotype or no phenotype, yet phenotypes including lethality are seen when multiple HDAC genes are mutated simultaneously (Watson et al., 2000).
Because histone deacetylation is typically associated with gene repression, inhibition of HDAC activity or mutation of HDAC genes is predicted to cause enhanced gene expression, as has been found in many specific cases (Schlake et al., 1994; Chen et al., 1997; Rundlett et al., 1998; Selker, 1998). However, there are some examples in which suppression of HDAC activity (through mutation or inhibition) causes downregulation of gene expression. For example, c-Myc mRNA levels decrease after treatment of mammalian cells with trichostatin A (Van Lint et al., 1996a), and deletion of HDACs in yeast and Drosophila melanogaster increase silencing at telomeres and other heterochromatic regions (De Rubertis et al., 1996; Rundlett et al., 1996; Kim et al., 1999; Sun and Hampsey, 1999). In addition, the yeast HDAC gene RPD3 is required for the full induction of yeast PHO5 (Vidal and Gaber, 1991; Rundlett et al., 1996). By microarray analysis, a number of yeast genes also were shown to be downregulated when RPD3 was mutated or cells were treated with trichostatin A (Bernstein et al., 2000). However, in contrast to our understanding of HDACs as corepressors, in no case has the mechanism by which HDACs contribute to gene expression been elucidated.
The yeast Saccharomyces cerevisiae has five related HDAC genes (HDA1, RPD3, HOS1, HOS2, and HOS3) and a sixth unrelated HDAC, SIR2, that encodes an NAD+-dependent HDAC. The products of all of these except HOS1 and HOS2 have been shown to have intrinsic HDAC activity (Carmen et al., 1996, 1999; Rundlett et al., 1996; Imai et al., 2000; Landry et al., 2000). Humans have at least eight HDACs, all of which are related to RPD3 and HDA1 (Grozinger et al., 1999; Kao et al., 2000). Plants have at least three distinct classes of HDAC. One class is related to Rpd3p (Rossi et al., 1998; Lechner et al., 2000); another, HD2, appears to be unique to plants (Lusser et al., 1997; Wu et al., 2000); the third HDAC class, also from maize, has been purified but not yet characterized genetically (Brosch et al., 1996).
To understand the function of HDACs in filamentous fungi in general and in Cochliobolus carbonum in particular, especially in relation to the fact that this fungus produces a potent HDAC inhibitor, we investigated the structure and function of the HDAC genes of C. carbonum. Here we report on the cloning and mutational analysis of a C. carbonum gene, HDC1, which shows closest homology with HOS2 of yeast. We demonstrate that it is required for full virulence on maize, for growth on substrates other than glucose or sucrose, and for the inducible expression of several glucose-repressed genes.
RESULTS
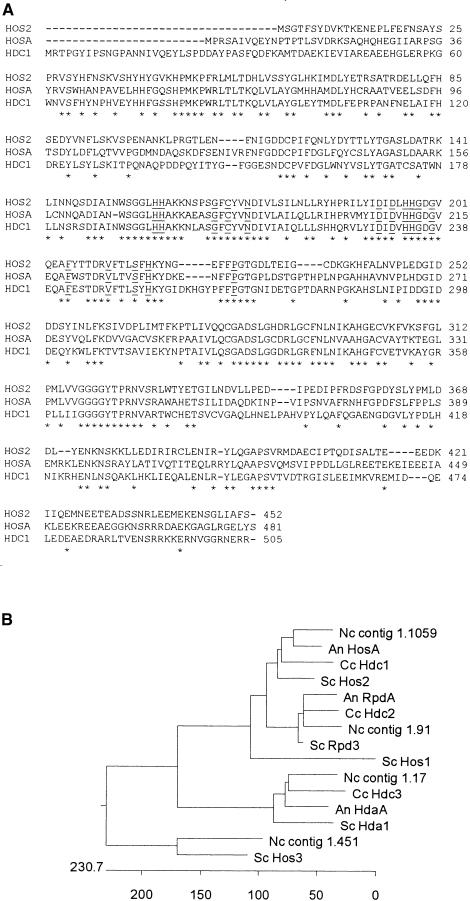
HDC1 was isolated using polymerase chain reaction (PCR) primers based on amino acid sequences that are conserved in known HDAC genes from other organisms. The gene contains no introns based on comparison of its genomic and cDNA sequences (data not shown). The closest matches of Hdc1p to proteins in the public databases were the products of HOSA, a putative HDAC from Aspergillus nidulans (Graessle et al., 2000), followed by HOS2 of yeast (Rundlett et al., 1996). The predicted molecular masses and pI values of the products of HDC1, HOSA, and HOS2 are similar: 56.9 kD and 5.7, 53.4 kD and 5.9, and 51.4 kD and 5.1, respectively. Hdc1p has an overall amino acid identity of 46% to the product of HOSA from A. nidulans, 44% to yeast HOS2, 40% to hda1+ (phd1+) of Schizosaccharomyces pombe, 38% to yeast RPD3, and <20% to yeast HOS1, HDA1, and HOS3. Sixteen of the 17 amino acid residues that are conserved in all known HDACs are present in Hdc1p (Hassig et al., 1998), with the exception of position 239, which also differs in HosAp (Figure 1A). Hdc1p also contains all seven of the motifs characteristic of class I HDACs (which includes Rpd3p and human HDAC1, HDAC2, and HDAC3) but lacks the characteristic features of class II HDACs (which includes yeast Hda1p and human HDAC4, HDAC5, and HDAC6) (Grozinger et al., 1999). Hdc1p and the other yeast HDACs, except HOS1, also show strong similarity to predicted proteins in the currently available genomic sequence of Neurospora crassa (www.genome.wi.mit.edu/annotation/fungi/neurospora/). On the basis of the clustering pattern, filamentous fungi contain sequences related to at least four of the five yeast HDAC genes related to RPD3, namely, RPD3, HDA1, HOS2, and HOS3 (Figure 1B).
Figure 1.
Alignment of the Predicted Amino Acid Sequences of HDC1 and Related Genes.
(A) Proteins shown are the products of HOS2 of S. cerevisiae (Rundlett et al., 1996; SWISS-PROT accession number P53096), HOSA of A. nidulans (Graessle et al., 2000; GenBank accession number AF164342), and HDC1 of C. carbonum (GenBank accession number AF306507) using CLUSTAL W (Thompson et al., 1994). Amino acids that are identical in all three proteins are indicated by asterisks. Sixteen of the 17 amino acids that are highly conserved in all RPD3-like HDAs and related bacterial proteins are underlined (Hassig et al., 1998).
(B) Unrooted cladogram showing relatedness of HDAC proteins. HosA and RpdA are from A. nidulans (An; Graessle et al., 2000). Hda1, Rpd3, Hos1, Hos2, and Hos3 are from S. cerevisiae (SC; Vidal and Gaber, 1991; Rundlett et al., 1996). Hdc1, Hdc2, and Hdc3 are from C. carbonum (Cc; this article; D. Baidyaroy and J.D. Walton, unpublished data; E-M. Brandtner, S. Graessle, and G. Brosch, unpublished data). Contigs 1.1059, 1.91, 1.17, and 1.451 represent predicted proteins from the eponymous genomic DNA sequences of N. crassa (Nc; http://www.genome.wi.mit.edu/annotation/fungi/neurospora/). The units on the scale at bottom are numbers of substitution events.
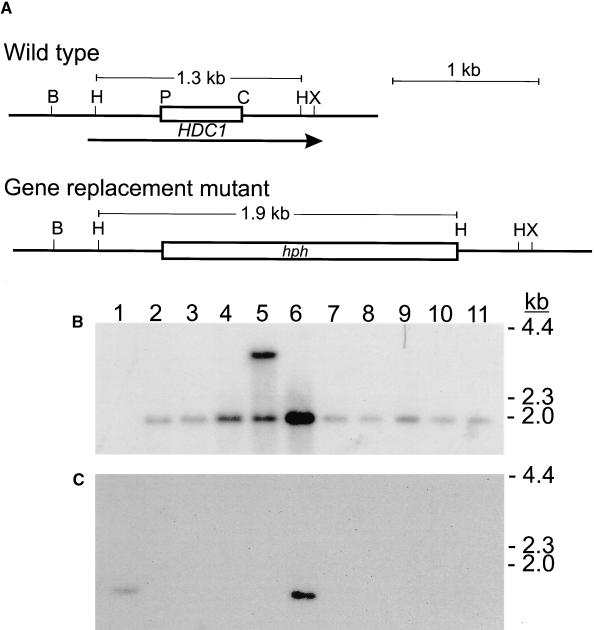
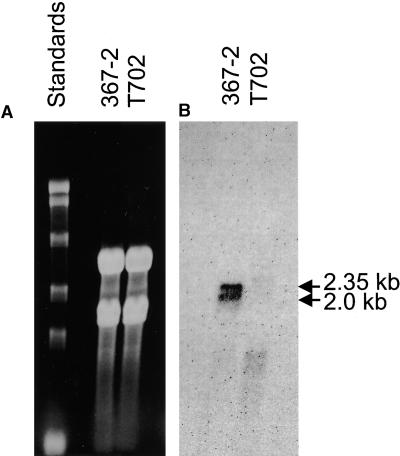
DNA blotting and hybridization indicated that HDC1 was a single-copy gene in C. carbonum (data not shown). Strains of C. carbonum that were specifically mutated in HDC1 were constructed by targeted gene replacement. Eight of 10 independent transformants had the expected pattern of DNA hybridization for a simple gene replacement (Figure 2). This rate of homologous integration of transforming DNA is consistent with the rate found in disruption experiments with other genes in C. carbonum (Pitkin et al., 1996). Mutant hdc1 strains grew more slowly than did wild-type strains on standard protoplast regeneration medium and on V8 agar plates. Consistent with HDC1 having been functionally deleted, no HDC1 mRNA could be detected by RNA blotting in any of the mutants (Figure 3 and data not shown). HDC1 was found to produce two sizes of mRNA transcripts that differed by ∼0.35 kb (Figure 3). Because the gene contains no introns, these two transcripts probably are attributable to the use of different transcriptional start sites or to different polyadenylation sites.
Figure 2.
Analysis of Engineered hdc1 Mutants
(A) Restriction map of the genomic region of wild-type HDC1 and predicted map of the gene replacement mutant. The open box in the wild-type map indicates the fragment of HDC1 DNA replaced by the hph gene encoding hygromycin phosphotransferase in the mutant. The arrow indicates the location of the HDC1 coding region. B, BamHI; H, HindIII; P, PstI; C, ClaI; X, XhoI.
(B) DNA gel blot probed with hph. Lane 1, wild-type strain 367-2. Lanes 2 to 11, 10 independent transformants: lane 2, T702.1; lane 3, T702.2; lane 4, T702.3; lane 7, T702.4; lane 8, T702.5.
(C) The same blot probed with the deleted segment of HDC1. On the basis of the hybridization pattern, all of the transformants except those shown in lanes 5 and 6 have undergone gene replacement by simple double crossover homologous integration. Sizes of DNA markers in kilobases are shown at right.
Figure 3.
RNA Gel Blot Analysis of HDC1.
(A) Gel stained with ethidium bromide. RNA marker sizes, from top to bottom, are 9.5, 7.5, 4.4, 2.4, and 1.4 kb. Fifteen micrograms of total RNA was loaded in each lane.
(B) The same gel blotted and probed with HDC1.
367-2 is the wild type and T702 is the hdc1 mutant T702.1.
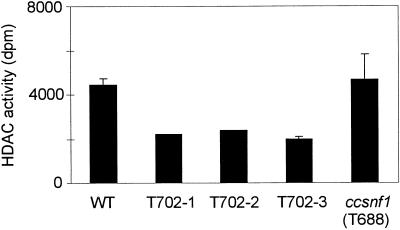
HDAC activity in crude extracts of the hdc1 mutants was reduced consistently by ∼50% (Figure 4). This finding strongly suggests that HDC1 encodes a functional HDAC. However, because HDACs associate with each other in mammals and yeast, an indirect effect of disrupting HDC1 on other HDAC activities cannot be excluded (Grozinger et al., 1999; Wu et al., 2001). As further assurance that the HDAC activity phenotype was attributable specifically to the mutation of HDC1, HDAC activity was measured in a ccsnf1 mutant. This was done because hdc1 mutants and ccsnf1 mutants share many of the same phenotypes, including slower growth on certain substrates (see below; Tonukari et al., 2000). HDAC activity in the ccsnf1 mutant T688 was not significantly different from that in the wild type (Figure 4). This result indicates that the reduced HDAC activity in the hdc1 mutant was not a side effect of any of the phenotypic abnormalities shared by the two mutants (see below).
Figure 4.
HDAC Activity.
HDAC activity in wild-type 367-2 (WT), three hdc1 mutants (T702.1, T702.2, and T702.3), and the ccsnf1 mutant T688 (Tonukari et al., 2000).
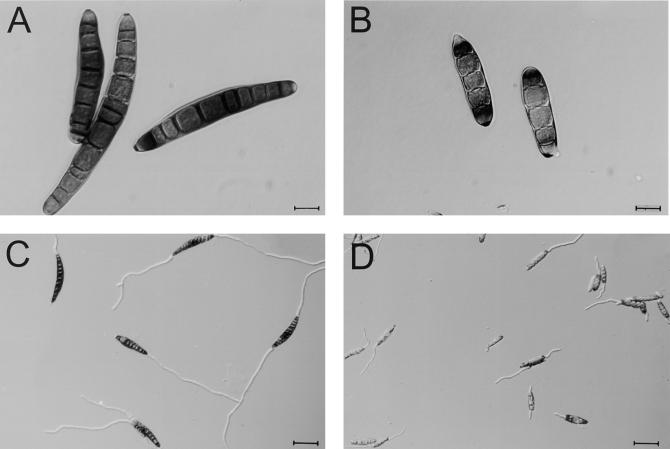
A striking developmental effect of mutating HDC1 was a reduction in conidial size and septum number. However, hdc1 conidia germinated at wild-type rates in vitro (Figure 5).
Figure 5.
Conidial Morphology of the Wild Type and the hdc1 Mutant T702.1.
(A) Ungerminated conidia of the wild type.
(B) Ungerminated conidia of mutant T702.1.
(C) Conidia of the wild type after germination on glass slides for 6 hr.
(D) Conidia of the mutant T702.1 after germination on glass slides for 6 hr.
Strains T702.2, T702.3, T702.4, and T702.5 (all hdc1/Tox2+) and five Tox2− hdc1 mutant strains (T717.1 through T717.5) showed a conidial morphology similar to that of T702.1 (data not shown). Bar in (A) and (B) = 12.5 μm; bar in (C) and (D) = 50 μm.
The virulence of the hdc1 mutants on maize was reduced greatly because of a reduction in the number of lesions formed (Figure 6A). Lesions that developed had similar morphology and rates of expansion as did those lesions caused by the wild-type fungus. Even at high inoculation densities (105 conidia/mL) and extended periods of disease development (>14 days), hdc1 mutants never killed plants, unlike the wild type, which eventually colonized and killed seedlings (Figure 6B). Conidia of hdc1 mutants could be seen microscopically to adhere efficiently to maize leaves, but whereas most lesions caused by the wild type could be attributed to single conidia, all lesions formed by the hdc1 mutants were associated with clumps of conidia (Figure 7). These results suggest that HDC1 may control a virulence factor or factors that are important for successful penetration of the maize epidermis, and they suggest that many hdc1 conidia in close proximity can compensate for the virulence factor or factors in which they are defective.
Figure 6.
Virulence of hdc1 Mutants.
Conidia (104/mL in 0.1% Tween 20) were sprayed on 3-week-old maize plants of genotype hm1/hm1 grown in a greenhouse, and the plants were covered overnight with plastic bags.
(A) Infected leaves 4 days after inoculation. From left to right, the leaves were inoculated with wild type (367-2), T702.1, T702.2, T702.3, T702.4, or T702.5, or uninoculated.
(B) Infected plants 2 weeks after inoculation. From left to right, the plants were inoculated with wild type (367-2), T702.1, T702.2, T702.3, T702.4, or T702.5. All of the plants are still alive except the plant sprayed with the wild-type conidia.
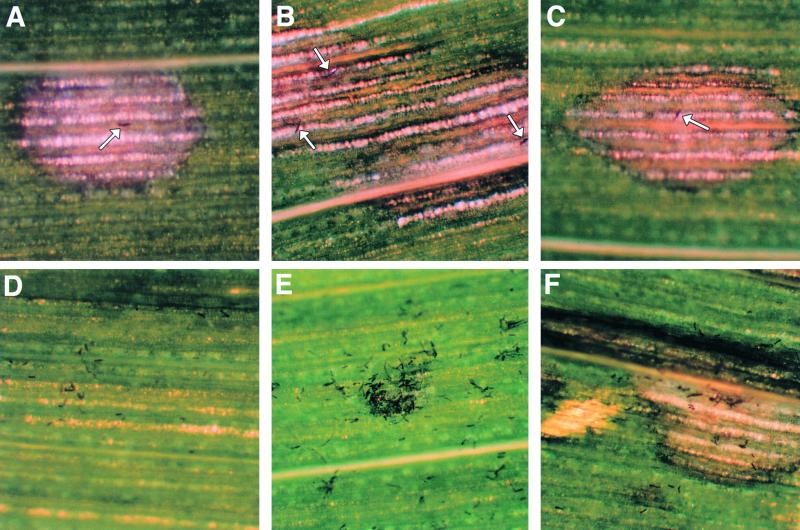
Figure 7.
Infection by hdc1 Mutant.
Low magnification (×40) microscopy of maize leaves infected with wild type (367-2) or hdc1 mutant T702.1. Photographs were taken 3 days after inoculation. Conidia (104/mL in 0.1% Tween 20) were sprayed on 3-week-old maize plants of genotype hm1/hm1 grown in a greenhouse, and the plants were covered overnight with plastic bags.
(A) to (C) Wild type. Arrows indicate individual conidia that established infection.
(D) to (F) hdc1 mutant. In (D) and (E), conidia failed to cause infection, whereas (F) shows a rare successful penetration by the hdc1 mutant.
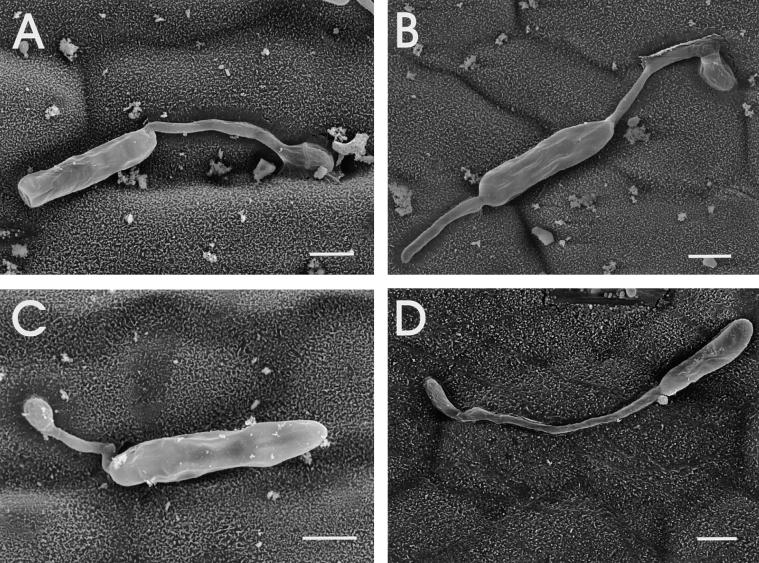
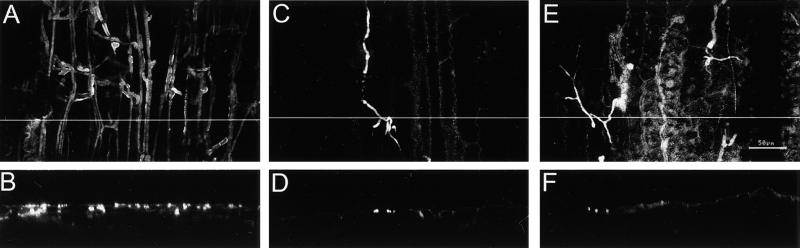
To determine whether the reduced virulence of the hdc1 mutant was caused by a reduced efficiency of germination or appressorium formation on leaves, inoculated leaves were examined by scanning electron microscopy. No difference was found in the germination rates of hdc1 and wild-type conidia on leaves (data not shown). The hdc1 mutants formed appressoria of normal morphology preferentially at the junctions between leaf epidermal cells, as has been reported for wild-type C. carbonum and other species of Cochliobolus (Figure 8) (Jennings and Ullstrup, 1957; Murray and Maxwell, 1975). This finding indicated that the defect in virulence of the hdc1 mutants was at a stage after germination and appressorium formation. Confocal microscopy with reconstructed cross-sectional views also indicated that although conidia of the hdc1 mutants germinated and grew along the surface of the maize leaf, they did not penetrate efficiently (Figure 9).
Figure 8.
Behavior of hdc1 Conidia on Maize Leaves.
Scanning electron micrographs were taken 24 hr after inoculation. All four panels show conidia of mutant T702.1. Formation of appressoria at the junctions between leaf epidermal cells is visible in (A), (B), and (D) (Jennings and Ullstrup, 1957; Murray and Maxwell, 1975). Bars = 10 μm.
Figure 9.
Fluorescence Confocal Microscopy of Infection of Maize Leaves by C. carbonum Wild Type and hdc1 Mutant.
(A) and (B) Wild type 367-2.
(C) to (F) hdc1 mutant T702.1.
Leaves were harvested 48 hr after inoculation. Fungal mycelia were stained with fluorescein isothiocyanate–conjugated wheat germ agglutinin. Images in (A), (C), and (E) were made looking down on the adaxial surface of the leaf. Images in (B), (D), and (F) show reconstructed cross-sectional views of the same leaves through the plane indicated by the white lines in (A), (C), and (E). The numerous bright fluorescent strands in (A) represent fungal mycelium ramifying through the leaf. The thin bright strands in (C) and (E) are fungal hyphae. The bright specks of fluorescence in (B) represent cross-sections of fungal mycelium on the surface and within the leaf, whereas the specks of fluorescence in (D) and (F) show the mutant restricted to the leaf surface. Some background fluorescence can be seen from the plant cell walls, particularly in (C) and (E). Bar in (E) = 50 μm for (A) to (F).
HC-toxin is an essential virulence determinant for C. carbonum (Walton, 1996). In vitro, the hdc1 mutants produced HC-toxin at concentrations similar to those in the wild type (data not shown). Therefore, their reduction in virulence could not be attributed to a decrease in HC-toxin production, and HDC1 does not regulate HC-toxin biosynthesis.
Mutation of the HDAC gene hda1+ (phd1+) in S. pombe results in increased sensitivity to trichostatin A (Kim et al., 1998; Olsson et al., 1998). Therefore, it is possible that the phenotype of the hdc1 mutants, made in a Tox2+ background, was attributable to self-inhibition of its remaining HDACs by HC-toxin. To test this notion, engineered hdc1 mutants were constructed in strain 164R1, which is toxin nonproducing (Tox2−) because it naturally lacks the genes for HC-toxin biosynthesis (Panaccione et al., 1992). Five independent hdc1/Tox2− mutants were recovered and analyzed (strains T717.1 through T717.5). All had the same small conidium phenotype as the hdc1/Tox2+ mutants had, and one transformant analyzed in detail showed the same growth phenotype on alternative carbon sources (Table 1; see below). Therefore, the phenotype of the T702 strains was attributable specifically to the mutation of HDC1 and was not a secondary effect caused by the self-inhibition of other HDACs by HC-toxin.
Table 1.
Growth of hdc1 Mutants on Various Carbon Sources and at Increased Temperature
| Growthb
|
|||
|---|---|---|---|
| Carbon Sourcea | Wild Type (mm/day) | Mutant (mm/day) | (Mutant/Wild Type) × 100 |
| Experiment 1 (367-2 vs. T702.1) | |||
| Glucose | 4.1 ± 0.3 | 4.1 ± 0.6 | 100 |
| Sucrose | 4.3 ± 0.2 | 4.1 ± 0.2 | 95 |
| Arabinose | 3.0 ± 0.3 | 0.8 ± 0.1 | 27 |
| Xylose | 7.7 ± 0.5 | 4.8 ± 0.2 | 62 |
| Xylan | 7.9 ± 0.4 | 5.5 ± 0.1 | 70 |
| Pectin | 5.7 ± 0.2 | 2.7 ± 0.4 | 47 |
| Maize cell walls | 8.2 ± 0.7 | 4.4 ± 0.6 | 54 |
| V8 juice (25°C) | 7.4 ± 0.3 | 4.8 ± 0.3 | 65 |
| V8 juice (30°C) | 6.1 ± 0.1 | 4.2 ± 0.4 | 69 |
| Experiment 2 (164R1 vs. T717.1) | |||
| Glucose | 3.8 ± 0.6 | 3.6 ± 0.2 | 95 |
| Xylan | 7.3 ± 0.1 | 4.7 ± 0.1 | 65 |
| Pectin | 6.2 ± 0.2 | 3.1 ± 0.1 | 50 |
| Maize cell walls | 6.7 ± 0.2 | 3.3 ± 0.4 | 49 |
367-2 is wild type Tox2+, and 164R1 is wild type Tox2−. T702.1 is an hdc1 mutant of 367-2, and T717.1 is an hdc1 mutant of 164R1.
b Growth on solid (agar) medium supplemented with the indicated sole carbon source was measured every day for 5 days. Data are average radial growth (n = 3) per day ±1 sd of the mean. Experiments were performed at 25°C unless indicated otherwise.
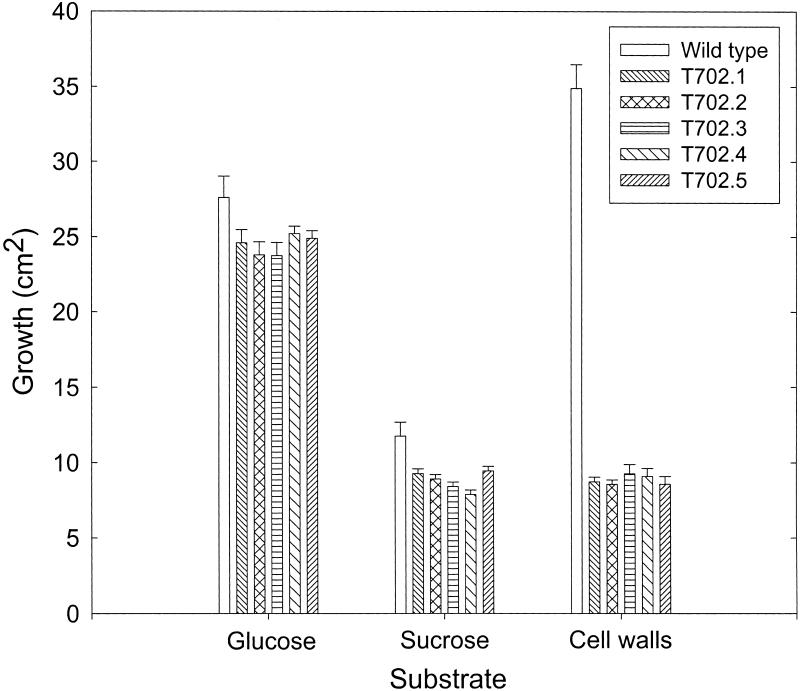
The reduced virulence phenotype of the hdc1 mutants was similar to that of strains of C. carbonum mutated in ccSNF1, a homolog of yeast SNF1 (Tonukari et al., 2000). SNF1 encodes a protein kinase that is required for derepression of glucose-repressed genes (Carlson, 1999). ccSNF1 has the same role in C. carbonum as it does in yeast, that is, ccsnf1 mutants cannot express glucose-repressed genes even under derepressing conditions; as a result, ccsnf1 mutants grow poorly on alternative sugars or complex polysaccharides as sole carbon sources (Tonukari et al., 2000). To determine whether the phenotype of the hdc1 mutants was similar to that of the ccsnf1 mutants in this regard, independent hdc1 mutants were grown on various carbohydrates and polysaccharides. Growth of hdc1 mutants was the same as that of the wild type on glucose but was reduced slightly on sucrose and by 30 to 73% on the other carbohydrate sources tested (Table 1, Figure 10). The degree of growth reduction on the various substrates tested was similar for the ccsnf1 and the hdc1 mutants. For example, the growth of both mutants was least affected on glucose and was most severely affected on arabinose and maize cell walls (Table 1, Figure 10) (Tonukari et al., 2000).
Figure 10.
Growth Phenotype of hdc1 Mutants.
T702.1 through T702.5 are independent hdc1 knockout mutants (see Figure 2). Growth was measured on agar containing the indicated sole carbon source after 8 days for glucose and after 4 days for sucrose and maize cell walls.
The preferred growth temperature of C. carbonum is 25 to 27°C. At 30°C, the wild type and hdc1 mutant T702.1 grew less well to an equal extent (Table 1), and at 37°C, neither grew (data not shown). Therefore, mutation of HDC1 did not cause increased temperature sensitivity.
One possible explanation for the reduced growth of the hdc1 mutants on polysaccharides and not on glucose is that hdc1 is required for the expression of extracellular depolymerases, which C. carbonum makes in an abundant variety. To test this possibility, enzyme activities were measured in the culture filtrates of the mutant growing on appropriate inducing substrates. β-1,3-Glucanase, α-1,4-polygalacturonase, and β-1,4-xylanase activities in the culture medium of the hdc1 mutant T702.1 were reduced by ∼40, 40, and 80%, respectively, compared with those in the wild type. In the ccsnf1 mutants, reduced extracellular enzyme activity was attributable, at least in part, to decreased expression of the encoding genes (Tonukari et al., 2000). The steady state levels of mRNA of EXG1, encoding exo-β-1,3-glucanase (van Hoof et al., 1991), PGN1, encoding endopolygalacturonase (Scott-Craig et al., 1998), and XYL1 and XYL2, encoding endoxylanases (Apel-Birkhold and Walton, 1996), also were largely or entirely downregulated in the hdc1 mutants (Figure 11). Therefore, HDC1, like ccSNF1, is required for the expression of at least some glucose-repressed genes in C. carbonum.
Figure 11.
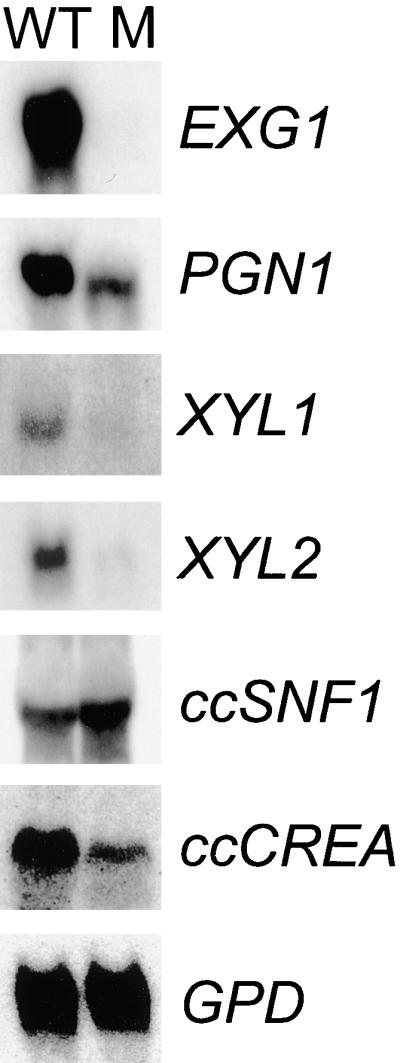
Gene Expression in an hdc1 Mutant.
Shown are blots of RNA extracted from the wild-type strain 367-2 (WT) and the hdc1 mutant T702 (M). The fungus was grown on maize cell walls as the sole carbon source for EXG1 expression, on pectin for PGN1, on xylan for XYL1 and XYL2, and on sucrose for ccSNF1, ccCREA, and GPD. EXG1 encodes exo-β-1,3-glucanase; PGN1 encodes endo-α-1,4-polygalacturonase; XYL1 and XYL2 encode endo-β-1,4-xylanases; ccSNF1 encodes a homolog of yeast SNF1; ccCREA encodes a homolog of CREA; and GPD encodes glyceraldehyde 3-phosphate dehydrogenase.
Mutation of HDC1 did not affect the expression of ccSNF1 or the housekeeping gene GPD (which encodes glyceraldehyde 3-phosphate dehydrogenase) (Figure 11). The repression of glucose-repressed genes in other filamentous fungi is mediated by CREA (which is equivalent to Mig1p in yeast) (Ebbole, 1998). One possible explanation for the requirement of HDC1 for the expression of extracellular depolymerase genes is that HDC1 is necessary for the repression of a repressor such as CREA. However, the mRNA abundance of the C. carbonum homolog of CREA (ccCREA) was reduced, not enhanced, in the hdc1 mutant (Figure 11).
DISCUSSION
In this report, we show that strains of the plant pathogenic fungus C. carbonum mutated in the HDC1 gene that encodes a putative HDAC are viable but have several significant phenotypes, the most significant of which is a strong reduction in virulence. The evidence suggests that the reduced virulence is attributable not to reduced conidial germination or appressorium formation in vitro or in vivo but to a decreased efficiency in penetration of the maize leaf epidermis. Because the lesions caused by the mutant have normal morphology, HDC1 does not appear to be important for ramification within the maize leaf.
C. carbonum probably breaches the maize epidermis by enzymatic action and not mechanical force (for discussion, see Horwitz et al., 1999; Tonukari et al., 2000), and, as shown in this study, HDC1 is required for the expression of at least some glucose-repressed extracellular enzymes that can depolymerize the plant cell wall. Thus, failure of the hdc1 mutants to penetrate leaves might be attributable to an inability to express appropriately some of these enzymes. In support of the idea that the critical factors controlled by HDC1 are extracellular wall depolymerases is the observation that the hdc1 mutant causes lesions only when many conidia are in close proximity on the leaf surface. However, this also would be consistent with HDC1 controlling some extracellular virulence factors other than depolymerases.
The HDC1 gene product is related to many characterized HDAC proteins and is most closely related to Hos2p of yeast. HOS2 itself has not been fully characterized as encoding a functional HDAC, although this conclusion is supported by several lines of genetic evidence (Edmondson et al., 1998; Watson et al., 2000; Wittschieben et al., 2000). The sequence similarity of Hdc1p and Hos2p and the reduction in total HDAC activity in hdc1 mutants further suggest that these genes encode HDACs, but biochemical confirmation is required.
The hdc1 mutant of C. carbonum was viable despite the fact that perturbation of a gene encoding a putative enzyme that modifies core histones might be expected to have large effects on global gene expression. However, in other systems, the perturbation of HDAC activity by inhibitors or by mutation often has been found to have subtle effects or phenotypes (Yoshida et al., 1990; Vidal and Gaber, 1991; Kijima et al., 1993; Rundlett et al., 1996; Edmondson et al., 1998; Kwon et al., 1998; Carmen et al., 1999; Watson et al., 2000; Wittschieben et al., 2000). HC-toxin, the HDAC inhibitor produced by C. carbonum, has been shown to have only subtle, nonlethal effects on protoplasts from hm1/hm1 maize leaves (Wolf and Earle, 1991). Filamentous fungi, including C. carbonum, A. nidulans, and N. crassa, have multiple predicted HDAC genes that sort into at least three classes that cluster with RPD3, HDA1, or HOS2 of yeast (Figure 1). In addition, N. crassa has a predicted homolog of yeast HOS3; therefore, A. nidulans and C. carbonum also might have a similar gene (Graessle et al., 2000; D. Baidyaroy and J.D. Walton, unpublished results; E.-M. Brandtner, S. Graessle, and G. Brosch, unpublished results; www.genome.wi.mit.edu/annotation/fungi/neurospora/). However, it cannot be assumed that the alignment clusters shown in Figure 1 actually reflect an underlying similarity in function, because, as shown here, at least HDC1 of C. carbonum and HOS2 of yeast have quite distinct biological roles despite the relatedness of their sequences (Watson et al., 2000).
In some cases, the mechanisms by which HDACs act as corepressors are understood in detail (Struhl, 1998; Bernstein et al., 2000; Watson et al., 2000; Wu et al., 2001). On the other hand, some studies suggest that HDACs also can be involved in gene activation. For example, mutation of RPD3 in yeast or Drosophila melanogaster causes enhanced silencing in heterochromatic regions such as subtelomeres, rDNA, and silent mating-type loci (De Rubertis et al., 1996; Kim et al., 1999; Sun and Hampsey, 1999). A recent microarray analysis found that an approximately equal number of yeast genes are upregulated as are downregulated in rpd3 mutants, and of the downregulated genes, ∼40% are near telomeres (Bernstein et al., 2000). In contrast to the situation with HDACs as corepressors, however, the mechanism by which HDACs upregulate gene expression is not understood for any specific case. HDC1 of C. carbonum apparently represents another example in which HDACs are required for gene expression, in this case of a specific subset of coregulated genes.
The phenotypes of the hdc1 and ccsnf1 mutants are similar in several regards, notably, a quantitative decrease in virulence, decreased growth on complex polysaccharides and sugars other than glucose and sucrose, and reduction in the expression of extracellular depolymerases (Tonukari et al., 2000). The decreased growth of both mutants on complex polysaccharides can be accounted for by decreased production of the polysaccharide depolymerases and/or enzymes needed for uptake or metabolism of alternative sugars, which in turn can be attributed to decreased expression of the encoding genes. As in the ccsnf1 mutant (Tonukari et al., 2000), there is not a strict correspondence between mRNA levels, enzyme activities, and growth on the corresponding substrates in the hdc1 mutants (Table 1, Figure 11). This can be explained by two phenomena. First, utilization of a complex polysaccharide such as xylan or maize cell walls requires, in addition to the extracellular depolymerases, enzymes for the uptake and metabolism of the released sugars. Thus, complete downregulation of any one critical enzyme such as a xylose uptake carrier would have a severe effect on the growth on xylan regardless of the expression levels of xylanase. Second, all known depolymerases are redundant in C. carbonum; therefore, xylanase genes such as XYL3 and XYL4 still might permit some growth on xylan despite the almost complete downregulation of XYL1 and XYL2 in the hdc1 mutants (Apel-Birkhold and Walton, 1996) (Figure 11, Table 1). The residual β-1,3-glucanase activity remaining in the hdc1 mutant also can be explained by redundancy, because EXG1 contributes only ∼55% of the total extracellular β-1,3-glucanase activity of C. carbonum (Schaeffer et al., 1994; see Tonukari et al., 2000, for further discussion of this point).
The overlap in the phenotypes of the hdc1 and ccsnf1 mutants suggests that the products of these two genes participate in the same or a related regulatory network in C. carbonum. In yeast, it has been shown that Tup1p, a repressor of glucose-repressed and other genes, recruits Hda1p to the promoters of TUP1-regulated genes (Wu et al., 2001). Tup1p or Tup1p-like proteins also have been shown to recruit other HDACs in yeast and HDACs in other systems (Chen et al., 1997; Edmondson et al., 1998; Guenther et al., 2000; Watson et al., 2000). Tup1p itself is recruited to promoters via Mig1p (Smith and Johnson, 2000; Wu et al., 2001). Filamentous fungi have homologs of TUP1 and MIG1 (in which it is known as CREA) (Ebbole, 1998). In yeast grown on glucose, therefore, genes required for growth on alternative sugar sources are repressed by a series of interacting proteins, Mig1p/Tup1p/Hda1p. Mig1p itself is regulated by Snf1p. In the absence of glucose, Snf1p phosphorylates Mig1p, thereby causing it to dissociate from the promoters of glucose-repressed genes (Carlson, 1999; Kuchin et al., 2000).
The logic of the yeast regulatory circuit, therefore, predicts that the mutation of HDAC genes that interact with Tup1p, such as HDA1, RPD3, and HOS2, should cause the derepression of glucose-repressed genes. This is observed in yeast, in which mutation of HOS2 (in an rpd3/hos1 background) causes derepression of SUC2, which encodes invertase. It is well established that SUC2 expression requires SNF1 (Carlson, 1999; Watson et al., 2000). In contrast, the logic of the yeast regulatory pathway is not consistent with our mutational analysis of HDC1. If HDC1 encodes a corepressing HDAC, its disruption would be predicted to result in the derepression of glucose-repressed genes, whereas the opposite was observed. This finding suggests a fundamentally different biological role for HDC1 compared with that of HOS2.
Previously, we and others hypothesized that the role of HC-toxin in allowing the establishment of a compatible interaction between maize and C. carbonum might be to act as a suppressor of induced plant defense responses through the inhibition of HDACs (Brosch et al., 1995; Ciuffetti et al., 1995). Since then, a large number of studies have shown that HDACs typically function as corepressors; therefore, maize defense or other genes that are controlled directly by HDAC activity would be expected to be overexpressed during infection, not repressed. In this report, we show that in at least one case, a putative HDAC gene is required for the induction of a set of strongly induced genes. In C. carbonum, these genes encode extracellular depolymerases, but perhaps a similar requirement for HDAC activity exists for the strongly induced defense genes of maize.
METHODS
Fungal Methods
Fungal strains, maintenance, transformation, DNA extraction, nucleic acid libraries, and nucleic acid blotting have been described previously (Apel-Birkhold and Walton, 1996; Pitkin et al., 1996; Tonukari et al., 2000). Cochliobolus carbonum strain 367-2 (Tox2+) was derived from SB111 (American Type Culture Collection [Rockville, MD] strain 90305). For enzyme activity assays, still cultures were grown for 7 days on xylan (for β-1,4-xylanase), oat bran cereal (for β-1,3-glucanase), or pectin (for α-1,4-polygalacturonase) as sole carbon sources. Activities in the culture filtrates against polysaccharide substrates were assayed independently four times with a reducing sugar assay and the appropriate substrates as described (Lever, 1972; van Hoof et al., 1991; Scott-Craig et al., 1998). Pathogenicity was assayed on 3-week old maize (Zea mays) plants of genotype hm1/hm1 grown in a greenhouse.
Isolation and Disruption of HDC1
Total C. carbonum RNA was used to prepare cDNA using Superscript reverse transcriptase (Life Technologies, Bethesda, MD) and a primer of oligo(dT17) according to the manufacturer's directions. The cDNA was used as a template for all polymerase chain reactions (PCR). For PCR amplification of a fragment of HDC1, two 64-fold degenerate 17mer primers were used. Forward primer OA was 5′-GGNCAYCCNATGAARCC-3′ based on the amino acid sequence GHPMKP, and reverse primer A1R was 5′-TCRTTNACRTARCARAA-3′ based on FCYVND (Graessle et al., 2000). PCR conditions were as follows: 95°C for 3 min, then 33 cycles of 95°C for 1 min, 53°C for 1 min, and 72°C for 50 sec, and finally 72°C for 5 min. A product of ∼380 bp was isolated and cloned into pGEM T-Easy (Promega).
A full-length genomic copy of HDC1 was obtained using cDNA PCR products to screen a λEMBL3 genomic library of C. carbonum DNA (Scott-Craig et al., 1992). A 6-kb SpeI fragment was subcloned into pBluescript KS+ (Stratagene), and 2 kb from the 5′ end, containing the complete HDC1 gene, was sequenced.
For amplification of the 3′ end of HDC1, the 3′ rapid amplification of cDNA ends (RACE) protocol of Frohman et al. (1988) was performed. Total C. carbonum RNA was used to prepare cDNA using reverse transcriptase and an oligo(dT17) adapter primer of sequence 5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′. The first PCR was performed with forward primer 5′-GACGCTCACCAAACAACTT-3′ and an adapter primer of sequence 5′-GACTCGAGTCGACATCGA-3′. The resulting ∼1500-bp product was cloned and sequenced. For nested PCR, primer 5′-GACTAGAGTACACGATGGA-3′ (forward) and dT4 adapter primer 5′-GACTCGAGTCGACATCGATTTT-3′ (reverse) were used. Amplified products were cloned into pGEM T-Easy.
To construct the HDC1 replacement vector pAJ63, pSP72 (Promega) was cut with SphI, blunted, and cut again with EcoRV to eliminate the multiple cloning sites between SphI and EcoRV. The resulting plasmid was cut with HindIII and ligated with a 1.3-kb HindIII fragment of HDC1. This plasmid was then cut with PstI and ClaI, and the deleted fragment was replaced with a PstI-ClaI fragment from pHYG4, which contains the hph gene for hygromycin resistance from pCB1004 (Carroll et al., 1994) subcloned into the SmaI site of pBluescript KS+.
HDAC Assay
HDAC activity was assayed using 3H-acetate–labeled chicken reticulocyte histones (Kölle et al., 1998; Lechner et al., 2000). Freeze-dried tissue (0.5 g) from mycelial mats grown in still culture for 7 days was ground in liquid nitrogen and resuspended by vortexing in 4.0 mL of extraction buffer (15 mM Tris-HCl, pH 7.3, 10 mM NaCl, 0.25 mM EDTA, 10% [v/v] glycerol, and 1 mM β-mercaptoethanol) containing one protease inhibitor tablet (Roche, Mannheim, Germany) per 30 mL of buffer. After centrifugation at 11,000g for 15 min, 3 mL of the supernatant was desalted by gel filtration (Econo-Pak 10 DG; Bio-Rad, Richmond, CA). Fifty microliters of protein extract and 5 μL of tritiated histones (40,000 dpm) were incubated for 2 hr at 23°C, 36 μL of 1 N HCl was added, and the released acetate was extracted twice with ethyl acetate, first with 0.8 mL (removing 0.6 mL) and then with 0.6 mL (removing 0.7 mL). The ethyl acetate fractions were combined and counted by scintillation spectroscopy.
Isolation of ccCREA
Degenerate PCR primers were synthesized based on conserved amino acid sequences of CREA homologs from Aspergillus niger, Neurospora crassa, Saccharomyces cerevisiae, Sclerotinia sclerotiorum, and Trichoderma reesei. The sequences of the primers were 5′-GARAARCCNCAYGCNTG-3′ (forward, based on the amino acid sequence EKPHAC) and 5′-GTRTGRTCNGGNGTNGG-3′ (reverse, based on PTPDHT), where N is any deoxynucleotide, R is A or G, and Y is C or T. The template was a C. carbonum cDNA library made in the yeast vector pMYR (Stratagene). Touchdown PCR (Don et al., 1991) conditions were as follows: initial denaturation at 94°C for 3 min, followed by 45 cycles of 1 min at 94°C, 1 min of annealing, and 2 min at 72°C. The annealing temperature ranged from 65 to 45°C with a decrease of 1°C every three cycles. This was followed by 10 cycles of 94°C for 1 min, 1 min at 45°C, and 2 min at 72°C. The 631-bp PCR product was cloned into pGEM T-Easy (Promega) and sequenced. From this sequence, a PCR primer of sequence 5′-CTCCTTCTCCAACTACTCTCCTG-3′ was synthesized and used with a reverse primer from the 3′-pMYR vector (5′-CGTGAATGTAAGCGTGACAT-3′). The 5′ end of ccCREA was then isolated (Frohman et al., 1988) using the 5′-RACE system, version 2.0 (Gibco BRL). First strand cDNA synthesis was primed using 5′-TCATGTGTGCAAGGTTAGATC-3′ and then was amplified by PCR using the nested primer 5′-GCTCCATGAGACCCGCCTGTA-3′. The resulting product (422 bp) was cloned and sequenced. The predicted amino acid sequence of ccCREA (GenBank accession number AF306571) was 40 to 46% identical to that of CREA genes from Aspergillus oryzae, Sclerotinia sclerotiorum, Gibberella fujikuroi, Neurospora crassa, and Trichoderma harzianum.
Microscopy
Plants were inoculated with 30 mL of a conidial suspension containing 105 conidia/mL in 0.1% Tween 20. For confocal microscopy, leaf samples were taken 48 hr after inoculation and fixed for 24 hr at 4°C in a solution consisting of 2.5% glutaraldehyde and 0.02 M sodium phosphate, pH 7.2, followed by vacuum infiltration for 10 min. The samples were then washed three times for 10 min each with 0.02 M sodium phosphate, pH 7.2, and dehydrated through an ethanol series of 25% ethanol for 30 min, 50% ethanol for 30 min, and 70% ethanol for 24 hr at room temperature. Afterward, samples were soaked in PBS (0.15 M NaCl and 0.01 M sodium phosphate, pH 7.2) for 2 min and then stained with 100 mg/mL fluorescein isothiocyanate (FITC)-labeled wheat germ agglutinin (Sigma) in PBS for 20 min at room temperature followed by 10 min of vacuum infiltration. Subsequently, the samples were washed with PBS for 10 min while shaking and then mounted in water on slides. The microscope was a Zeiss (Jena, Germany) 210 confocal laser scanning microscope using the 488-nm line of an argon ion laser and a 520- to 560-nm emission filter. The objective lenses were Plan Neofluar ×10 and ×40 with numerical apertures of 0.30 and 0.50, respectively.
For electron microscopy, samples were collected 24 hr after inoculation and fixed in 4% glutaraldehyde buffered with 0.1 M sodium phosphate, pH 7.4. The samples then were rinsed for 15 min in the buffer and dehydrated in a series of 25, 50, 75, and 100% ethanol, followed by three 15-min rinses in 100% ethanol. The dehydrated samples were dried using Blazer's critical point dryer with liquid carbon dioxide as the transitional fluid (five flushes with a 5-min diffusion time). After mounting on aluminum stubs, the samples were coated with gold (∼30 nm) in an Enscope (Los Gatos, CA) sputter coater and observed with a JEOL JSM-6400V scanning electron microscope.
Acknowledgments
We thank Ewa Danielewicz and Shirley Owens (Michigan State University Electron Optics Center) and Phillip Wharton (Michigan State University Department of Horticulture) for invaluable assistance with electron and confocal microscopy. This work was supported by the U.S. Department of Agriculture Competitive Grants Program and the U.S. Department of Energy Division of Energy Biosciences, and in part by grants from the Austrian Science Foundation (P13209) to G.B., and the Austrian National Bank (ÖNB-7415) to P.L., and an APART-fellowship from the Austrian Academy of Sciences to G.B.
References
- Apel-Birkhold, P.C., and Walton, J.D. (1996). Cloning, disruption, and expression of two endo-β1,4-xylanase genes, XYL2 and XYL3, from Cochliobolus carbonum. Appl. Environ. Microbiol. 62, 4129–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B.E., Tong, J.K., and Schreiber, S.L. (2000). Genome wide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97, 13708–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch, G., Ransom, R., Lechner, T., Walton, J.D., and Loidl, P. (1995). Inhibition of maize histone deacetylase by HC-toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7, 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch, G., Goralik-Schramel, M., and Loidl, P. (1996). Purification of histone deacetylase HD1-A of germinating maize embryos. FEBS Lett. 393, 287–296. [DOI] [PubMed] [Google Scholar]
- Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207. [DOI] [PubMed] [Google Scholar]
- Carmen, A.A., Rundlett, S.E., and Grunstein, M. (1996). HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271, 15837–15844. [DOI] [PubMed] [Google Scholar]
- Carmen, A.A., Griffin, P.R., Calaycay, J.R., Rundlett, S.E., Suka, Y., and Grunstein, M. (1999). Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA 96, 12356–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, A.M., Sweigard, J.A., and Valent, B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newslett. 41, 22. [Google Scholar]
- Chen, W.Y., Bailey, E.C., McCune, S.L., Dong, J.Y., and Townes, T.M. (1997). Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc. Natl. Acad. Sci. USA 94, 5798–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffetti, L.M., Kim, S.-D., Knoche, H.W., and Dunkle, L.D. (1995). Maize DNA alkylation and genotype-specific alterations in protein synthesis induced by the host-selective toxin produced by Cochliobolus carbonum. Physiol. Mol. Plant Pathol. 46, 61–70. [Google Scholar]
- De Rubertis, F., Kadosh, D., Henchoz, S., Pauli, D., Reuter, G., Struhl, K., and Spierer, P. (1996). The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384, 589–591. [DOI] [PubMed] [Google Scholar]
- Doetzlhofer, A., Rotheneder, H., Lagger, G., Koranda, M., Kurtev, V., Brosch, G., Wintersberger, E., and Seiser, C. (1999). Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19, 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K., and Mattick, J.S. (1991). ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole, D.J. (1998). Carbon catabolite repression of gene expression and conidiation in Neurospora crassa. Fungal Genet. Biol. 25, 15–21. [DOI] [PubMed] [Google Scholar]
- Edmondson, D.G., Zhang, W., Watson, A., Xu, W., Bone, J.R., Yu, Y., Stillman, D., and Roth, S.Y. (1998). In vivo functions of histone acetylation/deacetylation in Tup1p repression and Gcn5p activation. Cold Spring Harbor Symp. Quant. Biol. 63, 459–468. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessle, S., Dangl, M., Haas, H., Mair, K., Trojer, P., Brandtner, E.-M., Walton, J.D., Loidl, P., and Brosch, G. (2000). Characterization of two putative histone deacetylase genes from Aspergillus nidulans. Biochim. Biophys. Acta 1492, 120–126. [DOI] [PubMed] [Google Scholar]
- Grozinger, C.M., Hassig, C.A., and Schreiber, S.L. (1999). Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 96, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther, M.G., Lane, W.S., Fischle, W., Verdin, E., Lazar, M.A., and Schiekhattar, R. (2000). A core SMRT corepressor complex containing HDAC3 and TBL1, a WD-40 repeat protein linked to deafness. Genes Dev. 14, 1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Hassig, C.A., Tong, J.K., Fleischer, T.C., Owa, T., Grable P.G., Ayer, D.E., and Schreiber, S.L. (1998). A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. USA 95, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz, B.A., Sharon, A., Lu, S.-W., Ritter, V., Sandrock, T.M., Yoder, O.C., and Turgeon, B.G. (1999). A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 26, 19–32. [DOI] [PubMed] [Google Scholar]
- Imai, S., Armstrong, C.M., Kaeberlein, M., and Guarente, L. (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800. [DOI] [PubMed] [Google Scholar]
- Jennings, P.R., and Ullstrup, A.J. (1957). A histological study of three Helminthosporium leaf blights of corn. Phytopathology 47, 707–714. [Google Scholar]
- Kao, H.Y., Downes, M., Ordentlich, P., and Evans, R.M. (2000). Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- Kijima, M., Yoshida, M., Suita, K., Horinouchi, S., and Beppu, T. (1993). Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J. Biol. Chem. 268, 22429–22435. [PubMed] [Google Scholar]
- Kim, S., Benguria, A., Lai, C.Y., and Jazwinski, S.M. (1999). Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.B., Honda, A., Yoshida, M., and Horinouchi, S. (1998). phd1+, a histone deacetylase gene of Schizosaccharomyces pombe, is required for the meiotic cell cycle and resistance to trichostatin A. FEBS Lett. 436, 193–196. [DOI] [PubMed] [Google Scholar]
- Knoepfler, P.S., and Eisenman, R.N. (1999). Sin meets NuRD and other tails of repression. Cell 99, 447–450. [DOI] [PubMed] [Google Scholar]
- Kölle, D., Brosch, G., Lechner, T., Lusser, A., and Loidl, P. (1998). Biochemical methods for analysis of histone deacetylases. Methods 15, 323–331. [DOI] [PubMed] [Google Scholar]
- Kuchin, S., Treich, I., and Carlson, M. (2000). A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97, 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, H.J., Owa, T., Hassig, C.A., Shimada, J., and Schreiber, S.L. (1998). Depudecin induces morphological reversion of transformed fibroblasts via the inhibition of histone deacetylase. Proc. Natl. Acad. Sci. USA 95, 3356–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, J., Sutton, A., Tafrov, S.T., Heller, R.C., Stebbins, J., Pillus, L., and Sternglanz, R. (2000). The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 23, 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, T., Lusser, A., Pipal, A., Brosch, G., Loidl, A., Goralik-Schramel, M., Sendra, R., Wegener, S., Walton, J.D., and Loidl, P. (2000). RPD3-type histone deacetylases in maize embryos. Biochemistry 39, 1683–1692. [DOI] [PubMed] [Google Scholar]
- Lever, M. (1972). A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47, 273–279. [DOI] [PubMed] [Google Scholar]
- Luo, R.X., Postigo, A.A., and Dean, D.C. (1998). Rb interacts with histone deacetylase to repress transcription. Cell 92, 463–473. [DOI] [PubMed] [Google Scholar]
- Lusser, L., Brosch, G., Loidl, A., Hass, H., and Loidl, P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277, 88–91. [DOI] [PubMed] [Google Scholar]
- Miska, E.A., Karlsson, C., Langley, E., Nielsen, S.J., Pines, J., and Kouzarides, T. (1999). HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, G.M., and Maxwell, D.P. (1975). Penetration of Zea mays by Helminthosporium carbonum. Can. J. Bot. 53, 2872–2883. [Google Scholar]
- Ng, H.H., and Bird, A. (2000). Histone deacetylases: Silencers for hire. Trends Biochem. Sci. 25, 121–126. [DOI] [PubMed] [Google Scholar]
- Olsson, T.G.S., Ekwall, K., Allshire, R.C., Sunnerhagen, P., Partridge, J.F., and Richardson, W.A. (1998). Genetic characterisation of hda1+, a putative fission yeast histone deacetylase gene. Nucleic Acids Res. 26, 3247–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaccione, D.G., Scott-Craig, J.S., Pocard, J., and Walton, J.D. (1992). A cyclic peptide synthetase gene required for pathogenicity of the fungus Cochliobolus carbonum on maize. Proc. Natl. Acad. Sci. USA 89, 6590–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkin, J.W., Panaccione, D.G., and Walton, J.D. (1996). A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant pathogenic fungus Cochliobolus carbonum. Microbiology 142, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Rossi, V., Hartings, H., and Motto, M. (1998). Identification and characterisation of an RPD3 homologue from maize (Zea mays L.) that is able to complement an rpd3 null mutant of Saccharomyces cerevisiae. Mol. Gen. Genet. 258, 288–296. [DOI] [PubMed] [Google Scholar]
- Rundlett, S.E., Carmen, A.A., Kobayashi, R., Bavykin, S., Turner, B.M., and Grunstein, M. (1996). HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett, S.E., Carmen, A.A., Suka, N., Turner, B.M., and Grunstein, M. (1998). Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Schaeffer, H.J., Leykam, J., and Walton, J.D. (1994). Cloning and targeted gene disruption of EXG1, encoding exo-β-1,3-glucanase, in the phytopathogenic fungus Cochliobolus carbonum. Appl. Environ. Microbiol. 60, 594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake, T., Klehr-Wirth, D., Yoshida, M., Beppu, T., and Bode, J. (1994). Gene expression within a chromatin domain: The role of core histone hyperacetylation. Biochemistry 33, 4197–4206. [DOI] [PubMed] [Google Scholar]
- Scott-Craig, J.S., Panaccione, D.G., Pocard, J.A., and Walton, J.D. (1992). The cyclic peptide synthetase catalyzing HC-toxin production in the filamentous fungus Cochliobolus carbonum is encoded by a 15.7 kb open reading frame. J. Biol. Chem. 67, 26044–26049. [PubMed] [Google Scholar]
- Scott-Craig, J.S., Cheng, Y.Q., Cervone, F., De Lorenzo, G., Pitkin, J.W., and Walton, J.D. (1998). Targeted mutants of Cochliobolus carbonum lacking the two major extracellular polygalacturonases. Appl. Environ. Microbiol. 64, 1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker, E.U. (1998). Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 95, 9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R.L., and Johnson, A.D. (2000). Turning genes off by Ssn6-Tup1: A conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25, 325–330. [DOI] [PubMed] [Google Scholar]
- Struhl, K. (1998). Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Sun, Z.-W., and Hampsey, M. (1999). A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152, 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonukari, N.J., Scott-Craig, J.S., and Walton, J.D. (2000). The Cochliobolus carbonum SNF1 gene is required for cell wall– degrading enzyme expression and virulence in maize. Plant Cell 12, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Leykam, J., Schaefer, H.J., and Walton, J.D. (1991). A single β-1,3-glucanase secreted by the maize pathogen Cochliobolus carbonum acts by an exolytic mechanism. Physiol. Mol. Plant Pathol. 39, 259–267. [Google Scholar]
- Van Lint, C., Emiliani, S., and Verdin, E. (1996. a). The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expression 5, 245–253. [PMC free article] [PubMed] [Google Scholar]
- Van Lint, C., Emiliani, S., Ott, M., and Verdin, E. (1996. b). Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15, 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Vidal, M., and Gaber, R.F. (1991). RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, J.D. (1996). Host selective toxins: Agents of compatibility. Plant Cell 8, 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, A.D., Edmondson, D.G., Bone, J.R., Mukai, Y., Yu, Y., Du, W., Stillman, D.J., and Roth, S.Y. (2000). Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben, B.O., Fellows, J., Du, W., Stillman, D.J., and Svejstrup, J.Q. (2000). Overlapping roles for the histone acetyltransferase activities of SAGA and Elongator in vivo. EMBO J. 19, 3060–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, S.J., and Earle, E.D. (1991). Effects of Helminthosporium carbonum race 1 toxin on host and non-host cereal protoplasts. Plant Sci. 70, 127–137. [Google Scholar]
- Wu, J., Suka, N., Carlson, M., and Grunstein, M. (2001). TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7, 117–126. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., Kijima, M., Akita, M., and Beppu, T. (1990). Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179. [PubMed] [Google Scholar]