Abstract
The Paracoccus denitrificans transcription factor FnrP has been characterized using artificial FNR-dependent promoter-lacZ fusion plasmids in Escherichia coli. FnrP can activate both class I and class II FNR-dependent promoters in response to anoxia but shows a marked preference for the class II promoter, where the FNR binding site is centered at −41.5 with respect to the transcription start site. FnrP was found to be inactive in an iscS mutant in vivo, demonstrating a requirement for cysteine desulfurase activity to assemble an iron-sulfur cluster in FnrP. Accordingly, an iron-sulfur cluster could be reconstituted into the purified protein in vitro using cysteine desulfurase, ferrous ions, and cysteine. Thus, FnrP is a true orthologue of FNR from E. coli and switches on target genes in response to anoxia. Inactivation of FnrP by oxygen very likely involves the oxidative disassembly of an iron-sulfur cluster. Possible ligands for the iron-sulfur cluster were identified by substituting each of the seven cysteine residues with serine and characterizing the altered proteins in vivo. Four substituted proteins showed activities less than 5% of the wild type, and so identify the four cysteines (Cys-14, Cys-17, Cys-25, and Cys-113) that are most likely to be involved in cluster ligation. The effects of N-oxides, NO-releasing compounds and a nitrosating agent on FNR and FnrP activity were investigated in vivo using the reporter system. Both proteins are very sensitive to the inclusion of sodium nitroprusside (a source of NO+) in defined growth media but are only moderately sensitive to those sources of NO that were tested.
The Escherichia coli FNR protein is a transcription factor that is active only under anaerobic conditions and regulates the expression of genes involved in anaerobic respiration and carbon metabolism (24). The active form of FNR is a homodimer that contains one [4Fe-4S]2+ cluster per monomer. Exposure of the protein to oxygen in vitro causes its inactivation, transformation of the cluster to the [2Fe-2S]2+ form, and monomerization of the protein (11, 12). FNR purified from aerobic cultures is monomeric and does not contain an iron-sulfur cluster. A cluster can be reconstituted into this form of the protein in vitro using cysteine desulfurase, the product of the nifS gene of Azotobacter vinelandii, and Fe2+ ions (7). Recently, it has been shown that assembly of the Fe-S cluster into FNR in vivo requires the cysteine desulfurase encoded by the E. coli iscS gene (22). The Fe-S cluster is believed to be ligated by four cysteine residues in each subunit, three in an N-terminal cluster (Cys-20, Cys-23, and Cys-29) and a fourth central residue, Cys-122 (6). There is an additional cysteine residue close to the N terminus of FNR that is dispensable for activity (6). FNR belongs to a large family of transcriptional regulators, the members of which are related in primary structure but have different signal recognition mechanisms (24). The closest relatives to FNR are characterized by the presence of N-terminal cysteine clusters and are therefore frequently assumed to be structurally and functionally analogous to FNR itself. Examples are the CydR protein of A. vinelandii and HlyX from Actinobacillus pleuropneumoniae, which have N-terminal cysteine clusters very similar to that of FNR itself and are capable of assembling [Fe-S] centers in vitro (5, 30). In both cases, the spacing of cysteine residues in the N-terminal cluster is the same as in FNR itself. The CydR protein is reported to be 10-fold more sensitive to oxygen than FNR, which is consistent with the role of CydR in the physiology of A. vinelandii (30). The structural basis for variations in oxygen sensitivity among FNR-like proteins is not known. There is a group of FNR-like proteins from members of the α-proteobacteria (such as Paracoccus denitrificans, Rhodobacter sphaeroides, and Bradyrhizobium japonicum) which have N-terminal cysteine clusters and the conserved central cysteine, but with a different spacing between the cysteines in the N-terminal region (Fig. 1). While there is good reason to believe that all of these proteins are functionally analogous to FNR in that they are activated by anoxia or low-oxygen tension, none has been proven to contain an Fe-S cluster. Furthermore, it would be of some interest to determine whether the altered spacing in the cysteine cluster of these proteins results in changes in the biochemical properties and physiological roles of the proteins. As a first step in addressing these questions, the FnrP protein of P. denitrificans has been characterized.
FIG. 1.
Alignment of the N-terminal regulatory domains of FNR from E. coli (23), HlyX from A. pleuropneumoniae (5), CydR from A. vinelandii (30), FnrL from R. sphaeroides (31), FixK from B. japonicum (1), and FnrP from P. denitrificans (25). The alignment is not intended to be exhaustive but rather to show examples of proteins from the gamma and alpha subdivisions of the proteobacteria for which some biological information is available. Cysteine residues of FNR known to be essential for activity are indicated (+), as are the cysteine residues of FnrP shown in this study to be essential (*). Residues of FNR that can be substituted with effects on oxygen sensitivity and that are discussed in the text are indicated (§).
During anaerobic growth, P. denitrificans cells respire using nitrogen oxides and oxyanions as terminal electron acceptors, reducing nitrate to dinitrogen in the pathway known as denitrification (33). At least two transcription factors regulate the expression of the denitrification genes in P. denitrificans. The nar (nitrate reductase) promoter is activated by FnrP, most likely in response to anoxia (25), while the nir (nitrite reductase) and nor (nitric oxide reductase) promoters are coordinately regulated by NNR (9, 21). Both transcription factors belong to the FNR superfamily (24, 25), although NNR is activated by nitric oxide (10, 26) and does not have the conserved cysteines that ligate an Fe-S cluster in FNR. FnrP is not only involved in denitrification, since it also regulates expression of oxidases and cytochrome c peroxidase (19, 25). FnrP and NNR recognize the same DNA sequence, yet activate their regulons independently (25). It will be necessary to develop in vitro systems for the study of FnrP- and NNR-mediated activation in order to address the question of how specificity is built in to the two regulatory circuits. FnrP shows 21% identity to the E. coli FNR protein and contains seven cysteine residues compared to five in E. coli FNR (25). In sequence alignments, it is not obvious which of the cysteine residues of FnrP have equivalent roles to those in FNR, partly because there are two Cys-X-X-Cys motifs in FnrP. The alignment shown in Fig. 1 is based upon the data reported in this paper. In this study, a system for measuring FnrP activity in E. coli is reported, which demonstrates that FnrP is an oxygen sensor and a true orthologue of FNR. The reporter system is used to characterize FnrP proteins in which the cysteine residues have been substituted with serine. It is demonstrated that an Fe-S cluster can be reconstituted into purified FnrP and that activity of FnrP in vivo requires the product of the iscS gene.
MATERIALS AND METHODS
Bacterial strains and plasmids and DNA manipulations.
E. coli strain DH5α [φ80dlacZΔM15 recA endA gyrA thi hsdR(rK− mK+) supE relA deoR Δ(lacZYA-argF)U169] was used for all routine DNA manipulations, JRG1728 [ΔlacX74 galU galK rpsL Δ(ara-leu) Δ(tyrR-fnr-rac-trg)] was used as the host for the reporter system (10), and BL21(λDE3) [ompT hsdSB(rB− mB−) dcm gal (λDE3)] was used for overexpression of a glutathione S-transferase (GST)-FnrP fusion protein. The iscS gene was disrupted by the replacement of codons 2 through 394 with a kanamycin resistance gene. The mutation was constructed in strain DH10B [mcrA Δ(mrr hsdRMS mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA endA araD Δ(ara leu) galU galK λ rpsL nupG] transformed with pKD46 (which expresses the Red recombinase) by using the method described by Datsenko and Wanner (3). It was found that microgram quantities of the PCR product were required to successfully transform the host strain by electroporation. Once constructed in strain DH10B, the mutation was subsequently transferred to JRG1728 by P1 transduction (10). E. coli strains were grown in L broth (tryptone, 10 g liter−1; yeast extract, 5 g liter−1; NaCl, 5 g liter−1) for routine manipulations. For some β-galactosidase assays, M9 minimal medium (18) was used, supplemented as appropriate to satisfy growth requirements of strains, and with glucose (0.5% [wt/vol]) as the carbon source and nitrate (50 mM), nitrite (2 mM), sodium nitroprusside (100 μM), S-nitroso-N-acetylpencillamine (100 μM), and S-nitrosoglutathione (50 μM), as indicated. The plasmids used were pGEX-KG (8), FF/pRW2A (15), and pGS24 (23). The fnrP gene was amplified twice from P. denitrificans genomic DNA by using two different 5′ primers, FnrP1 (5′-CCATGGCACATGACGCCGCCCTCTG-3′) and FnrP2 (5′-GAATTCATGCCACATGACGCCGCCCT-3′), and a reverse primer, FnrPREV (5′-CCTAGCTGAGCGGCCCGCCGTCCG-3′). The PCR products were cloned into pUC18 and sequenced on both strands (by MWG-Biotech). The first PCR product was subcloned into pGEX-KG, using an engineered 5′ NcoI site, to make plasmid pSAD105 for overexpression of a GST-FnrP fusion protein. The second product was cloned in pUC18 to generate pFnrP, in which fnrP is expressed from the lac promoter. This plasmid was used for the in vivo studies of FnrP activity and as the template for mutagenesis of the fnrP gene. The techniques used for PCR mutagenesis have been described previously (10). Assay of β-galactosidase was according to Miller (18).
Purification of FnrP.
Cultures (100 ml) of BL21 (pSAD105) were grown by shaking at 30°C to an optical density at 600 nm of approximately 0.5, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and grown for a further 2 h at 30°C. Cells were harvested, washed twice in purification buffer (25 mM Tris, pH 8.0, 2.5 mM CaCl2, 100 mM NaCl, 100 mM NaNO3), and then sonicated on ice in 10 ml of purification buffer containing 23 μg of phenylmethylsulfonyl fluoride/ml. Cell lysates were clarified by low-speed centrifugation at 5,000 rpm for 10 min. Lysates were applied immediately to a 2 ml of glutathione-Sepharose Fast Flow column (Amersham Pharmacia Biotech) preequilibrated with 50 volumes of purification buffer. The column was washed with 100 volumes of purification buffer and then incubated in an anaerobic glovebox with 10 U of human plasma thrombin (Sigma-Aldrich) in 2 ml of purification buffer (mixed to a slurry with the Sepharose). After 2 h at 30°C, the 2 ml of buffer (containing most of the purified FnrP) was collected and the column was further eluted with 10 ml of purification buffer. The purified FnrP was then used immediately in reconstitution reactions.
Reconstitution of iron-sulfur cluster into FnrP.
The reconstitution was carried out essentially as described by Green et al. (7) except that 5 mol of (NH4)2Fe(SO4)2 was used per mol of protein, and the reaction mixture was incubated at 37°C. The reaction was performed under strictly anaerobic conditions with cysteine as a source of sulfur, dithiothreitol (DTT), (NH4)2Fe(SO4)2, and the NifS protein of A. vinelandii, which was purified from E. coli cells transformed with pDB551 according to Zheng et al. (32). FnrP samples were then transferred from the anaerobic cabinet in a sealed cuvette, and spectra were recorded every 5 min in a Hewlett Packard 8453 spectrophotometer at 37°C.
RESULTS
Correction of FnrP sequence.
The sequence of the two independent fnrP clones made in this study differed from the published sequence of the fnrP gene (25). Deletion of nucleotide C at coordinate 188 and an insertion of T after nucleotide 203 in the published sequence resulted in a change in the primary sequence from ARWSPALA to ASVVAGMA at positions 62 to 69 in the FnrP sequence. This revision strengthens the similarity of FnrP to its related proteins, such as FixK and FnrL (Fig. 1).
Activation of FNR-dependent promoters in E. coli.
In previous work, an E. coli reporter system was used to study the activation of the P. denitrificans NNR protein by N-oxides in vivo and also to characterize a number of altered NNR proteins (10). In this work, the same reporter system was utilized to study the activity of FnrP. FnrP activated the artificial FF-melR class II promoter (with an FNR-binding site centered at −41.5) in response to anoxia, as is the case for E. coli FNR (Table 1). When cultures were grown anaerobically, the promoter was activated 46-fold in L broth and 18-fold in minimal medium by FnrP, compared to aerobically grown cultures. Thus, FnrP behaves qualitatively in a similar fashion to the E. coli FNR protein. It is not clear why higher levels of FnrP-mediated gene expression were observed with minimal medium than with rich medium. An artificial class I FNR-dependent promoter with a binding site centered at −61.5, was activated only twofold by FnrP (results not shown), suggesting that, like E. coli FNR (28) and P. denitrificans NNR (10), FnrP has a strong preference for class II binding sites. Interestingly, an artificial FNR-repressible promoter in which the FNR binding site is centered at −30.5 was not repressed by FnrP (results not shown). This promoter was designed to demonstrate that FNR can mediate simple repression by binding to DNA and blocking RNA polymerase binding (27). One possible explanation for the failure of FnrP to repress this promoter may be that the level of expression of FnrP, or its affinity for the FF site, may not be high enough to mediate repression. This may also explain why FnrP activates the FF-melR promoter to a lesser extent than FNR itself (Table 1). Nevertheless, these results clearly demonstrate that FnrP can function in E. coli as an oxygen-sensitive activator of transcription. Since NNR can also activate the FF-melR promoter (10), these results provide further evidence that FnrP and NNR have the same, or very similar, DNA-binding specificities.
TABLE 1.
β-Galactosidase activities directed by the FF-melR promoter in the presence of plasmids expressing FnrP (pFnrP) and FNR (pGS24)
| Strain | β-Galactosidase activitya
|
||||
|---|---|---|---|---|---|
| Plasmid | Rich medium
|
Minimal medium
|
|||
| Aerobic | Anaerobic | Aerobic | Anaerobic | ||
| JRG1728 | pFnrP | 14 ± 2 | 647 ± 13 | 95 ± 7 | 1,705 ± 119 |
| JRG1728 iscS::kan | pFnrP | 4 ± 1 | 24 ± 2 | 6 ± 1 | 323 ± 55 |
| JRG1728 | pGS24 | 507 ± 39 | 3,211 ± 260 | 440 ± 19 | 3,085 ± 75 |
| JRG1728 iscS::kan | pGS24 | 77 ± 10 | 1,813 ± 68 | 77 ± 10 | 1,641 ± 258 |
Activities were measured in duplicate on at least three independently grown cultures; means ± standard errors are shown. Growth of cultures was aerobic or anaerobic, in either L broth, or M9 minimal medium, as indicated. E. coli JRG1728(pRW2A/FF) and JRG1728 iscS::kan (pRW2A/FF) were transformed with plasmids expressing FnrP (pFnrP) and FNR (pGS24). Units of β-galactosidase are as defined by Miller (18).
Activity of FnrP in an iscS mutant.
Recently, it was shown that the cysteine desulfurase encoded by the E. coli iscS gene is required for efficient in vivo Fe-S cluster formation in a variety of proteins (22). It was shown that the FNR protein has a greatly reduced activity in an iscS mutant background, demonstrating that cysteine desulfurase is required for de novo synthesis of the Fe-S cluster in FNR (22). To provide further evidence that FnrP senses oxygen via an iron-sulfur cluster, an iscS::kan derivative of JRG1728 was constructed and the activity of the FF-melR promoter under the control of either FnrP or FNR was measured (Table 1). In both rich and minimal medium (supplemented with thiamine and nicotinic acid to satisfy the auxotrophies of the iscS strain), the activity of FNR was about half of that seen in the parent strain under anaerobic growth conditions. The effect of the iscS mutation was smaller than that reported previously (22), which may reflect the fact that FNR was expressed from a multicopy plasmid in these experiments. FNR had a relatively high activity in aerobic cultures under these conditions (Table 1), as has been seen previously (e.g., reference 10), perhaps because of the fnr copy number and the fact that a strong consensus FNR binding site was used. Interestingly, this aerobic activity of FNR was substantially reduced in the iscS background (Table 1), indicating that cysteine desulfurase can insert a cluster into FNR in an aerobic cell, as was previously observed in experiments with a variant of FNR in which the cluster is less sensitive to oxygen (22). In the case of FnrP, anaerobic activity in the iscS mutant was almost completely abolished in rich medium (4% of the parent strain) and greatly reduced in minimal medium (19%). The greater effect of the iscS mutation on FnrP than on FNR may reflect differences in the levels of expression of the two proteins. The greatly reduced activity of FnrP in the absence of IscS strongly supports the hypothesis that active FnrP contains an iron-sulfur cluster, and the pattern of FnrP-mediated activation suggests that the cluster serves as a sensor for oxygen.
Anaerobic reconstitution of iron-sulfur cluster into purified FnrP.
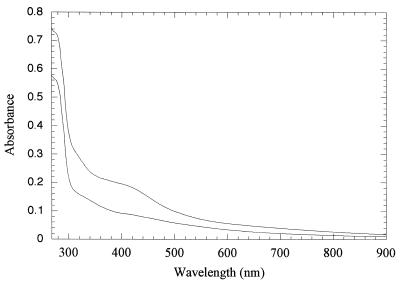
The fnrP gene was cloned into the vector pGEX-KG (8) and a GST-FnrP fusion protein was overexpressed in strain BL21(λDE3). The fusion protein was purified on a glutathione-Sepharose column, and purified FnrP was isolated by cleaving the fusion protein with thrombin while it remained bound to the column. The purified FnrP protein proved to be very prone to aggregation and could not be concentrated above 0.3 mg ml−1. Inclusion of 0.1 M sodium nitrate in purification buffers (29) was found to stabilize the protein enough to attempt anaerobic reconstitution of an iron-sulfur cluster, without causing too much interference with the spectral properties of the protein. FnrP was reconstituted in the absence of oxygen in a reaction mixture containing DTT, the cysteine desulfurase NifS from A. vinelandii, and cysteine as a source of sulfur plus ferrous ammonium sulfate (7). Reconstituted protein showed a broad absorption band at 420 nm (Fig. 2), which increased in intensity during the course of the reaction. Appearance of this band is consistent with the presence of iron-sulfur clusters in the protein (7, 11). These results demonstrate that FnrP can be reconstituted with an iron-sulfur cluster, which provides a likely mechanism for redox sensing in vivo and is consistent with the finding that FnrP activity is lost in the iscS strain (Table 1).
FIG. 2.
Reconstitution of an iron-sulfur cluster into purified FnrP. FnrP was purified from a GST fusion and was treated with cysteine desulfurase (NifS, from A. vinelandii) under anaerobic conditions in the presence of cysteine, DTT, and ferrous ammonium sulfate. FnrP concentration was approximately 14 μM. The lower spectrum was recorded immediately after the reaction was set up, and the upper spectrum was recorded after a reaction time of 95 min.
Effects of nitric oxide on FNR and FnrP activity.
Recently, it has been reported that the CydR (FNR) protein of A. vinelandii is as sensitive to NO in vitro as it is to oxygen (30). The possible interaction of NO with FNR-like proteins may be of particular significance for organisms, such as P. denitrificans that make appreciable amounts of NO endogenously as an intermediate of denitrification (33). Furthermore, there is evidence that E. coli can make NO endogenously during nitrate respiration (10), so the interaction of NO with FNR itself may have physiological significance. Indeed, FNR activity is consistently significantly lower in cultures grown anaerobically in media amended with nitrate than in the absence of nitrate (10; Table 2). Since little is known about the in vivo effects of NO and other reactive N species on FNR-like proteins, the activities of FNR and FnrP were assayed in cultures of the E. coli reporter strain grown in media containing different N-oxyanions, NO-generating agents and the nitrosating agent sodium nitroprusside (SNP). The effects of the NO-generating compounds on FnrP and FNR were examined in a defined medium, since thiol groups and other organic species in rich media may act as a sink for reactive N species. FnrP activity was reduced to various degrees by the inclusion of nitrate, nitrite, SNP, or the S-nitrosothiols S-nitroso-N-acetylpencillamine (SNAP) and S-nitrosoglutathione (GSNO) in growth media (Table 2). Since E. coli may generate some NO during nitrate respiration (10), at least some of the inhibition by nitrate may be due to NO production. However, the negligible effect of nitrite is not consistent with this idea, unless it is only the reduction of nitrate to nitrite that is accompanied by the formation of NO or the lower concentrations of nitrite used in growth media prevent accumulation of NO to inhibitory levels. The most marked effect was observed when the NO+-generating agent SNP was added to growth media. SNP at concentrations low enough to have no effect on growth reduced FnrP activity to 6% of that seen in unamended medium and FNR activity to 15%. In contrast, SNP had little effect in rich media (data not shown), perhaps because SNP reacts nonspecifically with components of rich media reducing its effective toxicity. The NO generators SNAP and GSNO reduced activation by 50 and 10%, respectively. The relatively small effect of GSNO may be due to its rapid degradation by the E. coli GSNO reductase (14). These results should be interpreted with caution, because of the possible secondary effects of adding different sources of NO and reactive N species to growth media.
TABLE 2.
Effect of N-oxides and oxyanions on β-galactosidase activities directed by the FF-melR promoter in the presence of plasmids expressing FnrP (pFnrP) and FNR (pGS24)
| Addition | FNR
|
FnrP
|
||
|---|---|---|---|---|
| β-Galactosidasea | % Activation | β-Galactosidase | % Activation | |
| None | 3,085 ± 75 | 100 | 1,705 ± 119 | 100 |
| Nitrite | 3,451 ± 50 | 112 | 1,571 ± 14 | 92 |
| Nitrate | 2,367 ± 138 | 77 | 964 ± 82 | 57 |
| SNP | 464 ± 45 | 15 | 101 ± 26 | 6 |
| SNAP | 2,318 ± 39 | 75 | 887 ± 166 | 52 |
| GSNO | 3,239 ± 152 | 105 | 1,555 ± 218 | 91 |
Activities were measured in duplicate on at least three independently grown cultures; means ± standard errors are shown. Growth of cultures was anaerobic, in M9 minimal medium, supplemented as indicated. E. coli JRG1728(pRW2A/FF) was transformed with plasmids expressing FnrP and FNR. Units of β-galactosidase are as defined by Miller (18).
Substitution of cysteine residues of FnrP.
In an attempt to identify the essential cysteine residues of FnrP, each was substituted with serine and the ability of altered proteins to activate transcription was assayed in vivo in E. coli cells. Substitutions of Cys-14, Cys-17, Cys-25, and Cys-113 led to the most severe defects in activity (Table 3), implicating these residues as analogues of the four essential cysteines of FNR. Like FNR, FnrP has two essential cysteines in a Cys-X-X-Cys motif, but the spacing of the third essential residue from this motif differs in FnrP (Fig. 1). Of the other three altered proteins, C8S had 8%, C28S had 77%, and C114S had 27% of wild-type activity (Table 3). The retention of significant activities in the C28S and C114S proteins argues against the residues having an essential role in FnrP activity. The low activity of the C8S protein is difficult to explain, and the possibility that this residue has some role in protein stability or as an Fe-S cluster ligand cannot be ruled out at this stage.
TABLE 3.
β-Galactosidase activities directed by the FF-melR promoter in the presence of plasmids expressing wild-type and altered FnrP
| Protein | β-Galactosidasea | % Activity |
|---|---|---|
| Wild type | 1,705 ± 119 | 100 |
| C8S | 144 ± 6 | 8 |
| C14S | 47 ± 2 | 3.1 |
| C17S | 52 ± 1 | 2.8 |
| C25S | 59 ± 11 | 3.4 |
| C28S | 1,316 ± 303 | 77 |
| C113S | 31 ± 6 | 1.8 |
| C114S | 462 ± 25 | 27 |
Activities were measured in duplicate on at least three independently grown cultures; means ± standard errors are shown. Growth of cultures was anaerobic, in M9 minimal medium, as indicated. E. coli JRG1728(pRW2A/FF) was transformed with plasmids expressing FnrP and its altered derivatives. Units of β-galactosidase are as defined by Miller (18).
DISCUSSION
This study has shown that FnrP from P. denitrificans can activate an artificial E. coli FNR-dependent promoter in response to anoxia. The evidence shows that FnrP contains an oxygen-labile iron-sulfur cluster. The ligands for the 4Fe-4S cluster are most likely provided by Cys-14, Cys-17, Cys25, and Cys-113. Thus, it can be concluded that FnrP is a true orthologue of FNR, which senses and responds to anoxia in a mechanistically similar fashion. It will be interesting to determine whether the different primary structure of FnrP (particularly in the N-terminal region) is reflected in different biochemical properties of the protein, for example, its sensitivity to oxygen. The extreme oxygen sensitivity of CydR (30) compared to that of FNR indicates that changes in the sequences of FNR-type proteins can have profound effects on their responses to oxygen. Both CydR and FNR repress expression of the high-affinity cytochrome bd oxidase (4, 30), whereas FnrP activates the expression of a high-affinity oxidase (19). In the long term, it will be of interest to determine the ranges of oxygen tension over which these regulatory proteins are activated, both in vivo and in vitro. Some preliminary insights into the relationship between primary structure and oxygen sensitivity have come from the isolation of mutant FNR proteins that have altered sensitivities to oxygen, and in the context of the present work, it is those substituted in the region of the N-terminal cysteine cluster that are of greatest interest. The L28H mutant of FNR incorporates an oxygen-insensitive Fe-S cluster, perhaps because of the influence of the histidine residue on the protein environment of the cluster (2). Other mutations that decrease the oxygen sensitivity of the protein in vivo have not been characterized biochemically but include D22G, D22S, and Q27R (13, 16, 17). Interestingly, all these mutations that decrease the oxygen sensitivity of FNR affect residues that are neither conserved nor conservatively substituted in FnrP (Fig. 1). Nevertheless, it seems clear that the sequence of FNR-like proteins in the vicinity of the N-terminal cysteine cluster has a role to play in determining oxygen sensitivity, and in this context, the different spacing of cysteine residues in FnrP may also be important. Recently, it has been shown that some FNR proteins in which cysteine residues have been substituted (specifically, C20S, C23G, and C29G) retain the ability to incorporate an iron-sulfur cluster in vitro and to bind to DNA, suggesting that cluster acquisition alone is not sufficient to promote transcription activation (20). Clearly, much is still to be learned about the role of the Fe-S cluster and its cysteine ligands in this group of proteins.
Previous work demonstrated that NNR could also activate the class II FNR-dependent promoter in E. coli (10), providing further evidence that FnrP and NNR recognize identical binding sites at their target promoters. The mechanistic basis for the lack of cross talk between the two regulons in P. denitrificans remains unresolved, but it is unlikely to be at the level of DNA binding. The possibility that NNR and FnrP have a differential preference for class I and class II promoters is perhaps unlikely, since both show good activity at class II promoters, at least in E. coli. This preference is consistent with what is known about the architecture of P. denitrificans promoters. The NNR-activated nor promoter is class II, since the NNR binding site is centered 43.5 bp upstream of the transcription start site (9). Likewise, the NNR-activated nirIX promoters are also class II, since the single NNR binding site in the intergenic region is centered at −41.5 with respect to both transcription start sites (21). The start site of a P. denitrificans promoter activated by FnrP has yet to be mapped. The ability of FNR to activate class II promoters involves amino acids in activating region 1 (AR1) and AR3, which make contacts with the alpha subunit of RNA polymerase. Two of the key residues in AR1 are Thr-118 and Ser-187 (29), which are not conserved in FnrP. Thus, the ability of FnrP to activate E. coli RNA polymerase may involve contacts that are different from those made by FNR.
Acknowledgments
We are grateful to Steve Busby, Barry Wanner, Dennis Dean, and Jeff Green for supplying strains and plasmids and to Rob van Spanning for helpful discussions.
This work was supported by the Biotechnology and Biological Sciences Research Council, through research grants to S.S. and A.J.T. and support of the Centre for Metalloprotein Spectroscopy and Biology at UEA. We are grateful to the Society for General Microbiology for funding a Vacation Studentship to B.J.T.
REFERENCES
- 1.Anthamatten, D., B. Scherb, and H. Hennecke. 1992. Characterization of a fixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J. Bacteriol. 174:2111–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, D., C. V. Popescu, N. Khoroshilova, K. Vogt, H. Beinert, E. Münck, and P. J. Kiley. 2000. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 275:6234–6240. [DOI] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govantes, F., J. A. Albrecht, and R. P. Gunsalus. 2000. Oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon: roles of multiple promoters and the Fnr-1 and Fnr-2 binding sites. Mol. Microbiol. 37:1456–1469. [DOI] [PubMed] [Google Scholar]
- 5.Green, J., and M. L. Baldwin. 1997. HlyX, the FNR homologue of Actinobacillus pleuropneumoniae, is a [4Fe-4S]-containing oxygen-responsive transcription regulator that anaerobically activates FNR-dependent class I promoters via an enhanced AR1 contact. Mol. Microbiol. 24:593–605. [DOI] [PubMed] [Google Scholar]
- 6.Green, J., A. D. Sharrocks, B. Green, M. Geisow, and J. R. Guest. 1993. Properties of FNR proteins substituted at each of the 5 cysteine residues. Mol. Microbiol. 8:61–68. [DOI] [PubMed] [Google Scholar]
- 7.Green, J., B. Bennett, P. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic anaerobic switch in vitro. Biochem. J. 316:887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262–267. [DOI] [PubMed] [Google Scholar]
- 9.Hutchings, M. I., and S. Spiro. 2000. The nitric oxide-regulated nor promoter of Paracoccus denitrificans. Microbiology 146:2635–2641. [DOI] [PubMed] [Google Scholar]
- 10.Hutchings, M. I., N. Shearer, S. Wastell, R. J. M. Van Spanning, and S. Spiro. 2000. Heterologous NNR-mediated nitric oxide signalling in Escherichia coli. J. Bacteriol. 182:6434–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoroshilova, N., C. Popescu, E. Munck, H. Beinert, and P. J. Kiley. 1997. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. USA 94:6087–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiley, P. J. and H. Beinert. 1999. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341–352. [DOI] [PubMed] [Google Scholar]
- 13.Kiley, P. J., and W. S. Reznikoff. 1991. Fnr mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 173:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, L., A. Hausladen, M. Zeng, L. Que, J. Heltman, and J. S. Stamler. 2001. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410:490–494. [DOI] [PubMed] [Google Scholar]
- 15.Lodge, J., R. Williams, A. Bell, B. Chan, and S. Busby. 1990. Comparison of promoter activities in Escherichia coli and Pseudomonas aeruginosa: use of a new broad-host-range promoter-probe plasmid. FEMS Microbiol. Lett. 67:221–226. [DOI] [PubMed] [Google Scholar]
- 16.Melville, S. B., and R. P. Gunsalus. 1990. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J. Biol. Chem. 265:18733–18736. [PubMed] [Google Scholar]
- 17.Melville, S. B., and R. P. Gunsalus. 1996. Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc. Natl. Acad. Sci. USA 93:1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Otten, M. F., D. M. Stork, W. N. Reijnders, H. V. Westerhoff, and R. J. van Spanning. 2001. Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur. J. Biochem. 268:2486–2497. [DOI] [PubMed] [Google Scholar]
- 20.Ralph, E. T., C. Scott, P. A. Jordan, A. J. Thomson, J. R. Guest, and J. Green. 2001. Anaerobic acquisition of [4Fe-4S] clusters by the inactive FNR(C20S) variant and restoration of activity by second-site amino acid substitutions. Mol. Microbiol. 39:1199–1211. [DOI] [PubMed] [Google Scholar]
- 21.Saunders, N. F. W., E. N. G. Houben, S. Koefed, S. de Weert, W. N. M. Reijnders, H. V. Westerhoff, A. P. N. De Boer, and R. J. M. Van Spanning. 1999. Transcription regulation of the nir gene cluster encoding nitrite reductase of Paracoccus denitrificans involves NNR and NirI, a novel type of membrane protein. Mol. Microbiol. 34:24–36. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz, C. L., O. Djaman, J. A. Imlay, and P. J. Kiley. 2000. The cysteine desulfarase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:9009–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw, D. J., and J. R. Guest. 1982. Nucleotide sequence of the fnr gene and primary structure of the Fnr protein of Escherichia coli. Nucleic Acids Res. 10:6119–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiro, S. 1995. The FNR family of transcriptional activators. Antonie Leeuwenhoek 66:23–36. [DOI] [PubMed] [Google Scholar]
- 25.van Spanning, R. J. M., A. P. N. De Boer, W. N. M. Reijnders, H. J. Westerhoff, A. H. Stouthamer, and J. Van der Oost. 1997. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcription activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol. Microbiol. 23:893–907. [DOI] [PubMed] [Google Scholar]
- 26.van Spanning, R. J. M., E. Houben, W. N. M. Reijnders, S. Spiro, H. V. Westerhoff, and N. Saunders. 1999. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J. Bacteriol. 181:4129–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams, S. M., H. J. Wing, and S. J. W. Busby. 1998. Repression of transcription initiation by Escherichia coli FNR protein: repression by FNR can be simple. FEMS Microbiol. Lett. 163:203–208. [DOI] [PubMed] [Google Scholar]
- 28.Wing, H. J., S. M. Williams, and S. J. W. Busby. 1995. Spacing requirements for transcription activation by Escherichia coli FNR protein. J. Bacteriol. 177:6704–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wing, H. J., J. Green, J. R. Guest, and S. J. Busby. 2000. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J. Biol. Chem. 275:29061–29065. [DOI] [PubMed] [Google Scholar]
- 30.Wu, G. H., H. Cruz-Ramos, S. Hill, J. Green, G. Sawers, and R. K. Poole. 2000. Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr)-sensitivity to oxygen, reactive oxygen species, and nitric oxide. J. Biol. Chem. 275:4679–4686. [DOI] [PubMed] [Google Scholar]
- 31.Zeilstra-Ryalls, J. H., and S. Kaplan. 1995. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J. Bacteriol. 177:6422–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]