Abstract
Swarming motility, a flagellum-dependent behavior that allows bacteria to move over solid surfaces, has been implicated in biofilm formation and bacterial virulence. In this study, light and electron microscopic analyses and genetic and functional investigations have shown that at least 50% of Aeromonas isolates from the species most commonly associated with diarrheal illness produce lateral flagella which mediate swarming motility. Aeromonas lateral flagella were optimally produced when bacteria were grown on solid medium for ≈8 h. Transmission and thin-section electron microscopy confirmed that these flagella do not possess a sheath structure. Southern analysis of Aeromonas reference strains and strains of mesophilic species (n = 84, varied sources and geographic regions) with a probe designed to detect lateral flagellin genes (lafA1 and lafA2) showed there was no marked species association of laf distribution. Approximately 50% of these strains hybridized strongly with the probe, in good agreement with the expression studies. We established a reproducible swarming assay (0.5% Eiken agar in Difco broth, 30°C) for Aeromonas spp. The laf-positive strains exhibited vigorous swarming motility, whereas laf-negative strains grew but showed no movement from the inoculation site. Light and scanning electron microscopic investigations revealed that lateral flagella formed bacterium-bacterium linkages on the agar surface. Strains of an Aeromonas caviae isolate in which lateral flagellum expression was abrogated by specific mutations in flagellar genes did not swarm, proving conclusively that lateral flagella are required for the surface movement. Whether lateral flagella and swarming motility contribute to Aeromonas intestinal colonization and virulence remains to be determined.
Aeromonas bacteria (aeromonads) are ubiquitous aquatic organisms that are also found in many foods. At least 14 species are now recognized, and there is substantial evidence that some strains (particularly of the species Aeromonas hydrophila HG1, A. caviae HG4, and A. veronii biovar sobria HG8/10) are primary gastrointestinal pathogens (18). However, it is still not possible to identify these virulent strains definitively, as Aeromonas pathogenic mechanisms are not well understood.
Efficient colonization of the human intestine is likely to be critical for virulence. Type IV pilus adhesins which mediate enterocyte adhesion have been described (19), and lipopolysaccharide and carbohydrate-reactive outer membrane proteins have also been implicated as Aeromonas intestinal colonization factors (25, 32). Colonization of mucosal surfaces is a complex process, however. For most microbial infections it is now thought to involve biofilm formation. A biofilm is an accumulation of organisms embedded in a polysaccharide matrix of their own making and adherent to a surface. Bacteria in biofilms are more resistant to host defenses and antimicrobial agents than they are as free-floating planktonic cells. They may also express more virulent phenotypes as a result of gene activation through bacterial communication (“quorum sensing”) or gene transfer (8, 12, 42).
An important component in biofilm formation is the ability to move over and colonize surfaces after the initial attachment. Such movement may be mediated in a variety of ways, including twitching motility (mediated by type IV pili) and swarming motility (mediated by flagella) (8, 13, 31, 37). Swarming has been studied extensively in Proteus mirabilis, in which it is characterized by differentiation of short, motile, vegetative cells at the colony margin into elongated, hyperflagellated swarm cells that assemble into multicellular rafts and migrate away from the colony (3, 9). Other members of the family Enterobacteriaceae have also been shown to exhibit the swarming phenomenon (1, 14, 47). For these organisms, hyperflagellation is due to increased expression of their peritrichous flagella, although the number of flagella per unit area of the cell surface is smaller than the number observed on P. mirabilis (twice as many flagella compared to 10 to 50 times more flagella, respectively). Members of the family Vibrionaceae (e.g., Vibrio alginolyticus and V. parahaemolyticus) produce numerous, unsheathed, lateral flagella for swarming (24). These flagella are separate and distinct from the sheathed, polar flagellum used for swimming.
Mesophilic Aeromonas strains usually show polar, monotrichous flagellation, but it is known that they can also express multiple lateral flagella. Shimada and colleagues (1985), using Leifson’s staining method and light microscopy, described multiple flagella on ≈18% of 186 mesophilic Aeromonas strains grown on solid medium for 18 h at 22°C (39). Another report described different patterns of flagellation on three A. hydrophila fish isolates (29). More recently, lateral flagella were purified from a diarrhea-associated isolate of A. caviae (strain Sch3) (40). The molecular weight and N-terminal amino acid sequence of the structural protein differed from those of the structural protein of the polar flagellum but were homologous to LafA of V. parahaemolyticus. Moreover, the predicted amino acid sequence from the A. caviae lateral flagellin genes had 63% identity and 75% similarity to the V. parahaemolyticus LafA protein (10, 40). Virtually nothing has been published regarding the function(s) of Aeromonas lateral flagella. Swarming motility has been reported in Aeromonas spp., but this has not been well characterized (13, 27).
To shed more light on their role in intestinal colonization, lateral flagella and swarming motility in Aeromonas spp. were investigated in this study. Light and electron microscopic analyses were used to examine expression and structure of lateral flagella. The distribution of laf genes among Aeromonas species from different sources and geographic regions was determined using a probe designed to detect the structural flagellin subunit genes (lafA1 and lafA2) of A. caviae strain Sch3 (Fig. 1). An Aeromonas swarming assay was established, and selected isolates as well as polar and lateral flagellar mutant strains were investigated for their swarming abilities. This report presents convincing evidence that Aeromonas lateral flagella are required for swarming motility and could serve as important colonization factors for those strains able to express them.
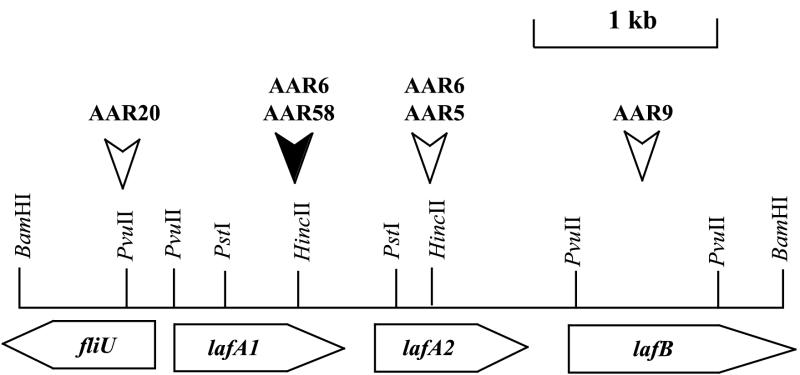
FIG. 1.
Schematic representation of the genetic organization of the A. caviae Sch3 lateral flagellin locus. Flagellar genes and open reading frames (ORFs) are indicated by shaded horizontal arrows and indicate the direction of transcription. ORFs are named after their homologues in other bacterial species. The open vertical arrows indicate the site of insertion of the kanamycin resistance cassette in the single mutants. The solid vertical arrow shows the site of insertion of the kanamycin cassette in the single mutant AAR58 and the site of insertion of the chloramphenicol resistance cassette in the tandem mutant AAR6.
MATERIALS AND METHODS
Bacterial strains.
Aeromonas hybridization group (HG) reference strains examined for laf genes were obtained from M. Altwegg, University of Zurich, Zurich, Switzerland. They were A. hydrophila ATCC 7966 (HG1), A. hydrophila CDC 9533-76 (HG2), A. hydrophila CDC 0434-84 (HG3), A. caviae ATCC 15468 (HG4), A. media CDC 0862-83 (HG5a), A. media ATCC 23212 (HG5b), A. eucrenophila ATCC 23309 (HG6), A. sobria CIP 7433 (HG7), A. veronii biovar sobria CDC 0437-80 (HG8), A. jandaei CDC 0787-80 (HG9), A. veronii biovar veronii ATCC 35624 (HG10), Aeromonas sp. strain CDC 1306-83 (HG11), A. schubertii ATCC 43700 (HG12), and A. popoffii LMG 17541 (HG17). (A. trota ATCC 49657 [HG14] was not obtained.)
A. caviae strain Sch3 was isolated in 1991 at the Sheffield Children’s Hospital, Sheffield, United Kingdom, from the diarrheal feces of a child (44). It was the strain from which lateral flagella were purified (40), and the lateral flagellin gene locus was analyzed (Fig. 1) (10). This strain was included as a positive control in all of the laf gene hybridization experiments. A spontaneous nalidixic acid-resistant mutant of this strain (A. caviae Sch3N) was used to construct insertion mutations in polar and lateral flagellar genes, as described elsewhere (10, 33). In brief, a kanamycin resistance cassette with an outward-reading promoter that ensures the transcription of downstream genes was inserted into each gene.
Single mutants were constructed for each of the genes of the polar flagellin locus; the flagellins (AAR269, flaA::Kmr, and AAR27, flaB::Kmr); flaG (AAR150, flaG::Kmr), which encodes a protein of unknown function; and flaH (AAR59, flaH::Kmr) and flaJ (AAR8, flaJ::Kmr), which encode a putative HAP2 capping protein and a flagellin chaperone protein, respectively. For the lateral flagellin locus, single mutations were constructed in the flagellins (AAR58, lafA1::Kmr, and AAR5, lafA2::Kmr) and in lafB (AAR9, lafB::Kmr) and fliU (AAR20, fliU::Kmr), which encode a flagellar capping protein and a flagellar protein of unknown function, respectively (Fig. 1). To generate the tandem polar flagellin knockout mutant AAR31 (flaA::Cmr flaB::Kmr) and the tandem lateral flagellin knockout mutant AAR6 (lafA1::Cmr lafA2::Kmr), two insertions were required and a second antibiotic resistance cassette which gave resistance to chloramphenicol was used.
Other mesophilic Aeromonas strains used in the study (n = 84) were from varied geographic regions (principally Australia, Bangladesh, United Kingdom, Spain, and Poland). They comprised 55 clinical isolates, predominantly (98%) from diarrheal feces, 11 isolates from food and water, 10 from fish or amphibians, and 8 of unknown source. Species were determined by conventional phenotypic (7, 20, 30, 45) and/or genetic methods (6, 17, 21). Four strains of the psychrophilic fish pathogen Aeormonas salmonicida were also examined. A P. mirabilis clinical isolate and two V. parahaemolyticus strains (strains ACM 2776 and NCTC 10884) were used as controls.
Short-term storage of isolates was in minimal maintenance medium at room temperature (20). Isolates were stored long-term in glycerol-peptone (1:4, glycerol–1% [wt/vol] bacteriological peptone [Oxoid, Basingstoke, United Kingdom]) at −70°C.
Growth conditions.
Aeromonas strains were routinely grown on tryptone soy agar (1.5%, wt/vol) supplemented with 0.6% (wt/vol) yeast extract (Oxoid) (TSAY). For propagation and isolation of the chromosomal DNA, Aeromonas strains were grown in tryptone soy broth with yeast extract (TSBY) at 37°C for 16 to 18 h. Luria-Bertani broth (LB; Oxoid) and brain heart infusion broth (BHIB; Oxoid) were also used to propagate (37°C, 16 to 18 h) Aeromonas strains and V. parahaemolyticus for the swarming assay and to prepare swarm plates by the addition of bacteriological agar (Oxoid) or Difco (Difco Laboratories, Detroit, Mich.) or Eiken (Eiken Chemical Co., Ltd., Tokyo, Japan) to a final concentration of 0.5 to 0.7% (wt/vol).
For swim plates (prepared with TSBY or LB), the final agar concentration was 0.3% (Oxoid). Swim plates were inoculated by stabbing into the center of the agar with growth from LB plates and incubated at 30°C for 16 to 18 h. Growth from the edge of the swimming zone within the agar of these plates provided the inoculum for the swarming assay. Swarm plates were inoculated on the surface of the agar in the center. When glucose (D-form; BDH, Victoria, Australia) was added to swarm plates, it was at concentrations of 0.5 and 1.5% (wt/vol). The iron chelator deferoxamine mesylate (Sigma, St. Louis, Mo.) was used at a final concentration of 200 μM.
Light microscopic detection of flagella.
Flagella were detected using Leifson’s staining method (26, 39) or flagella stain droppers (Difco, Becton Dickinson Microbiology Systems). Slides used for flagella staining were thoroughly cleaned in chromic acid (several hours to days), followed by washing 5 to 10 times in Milli-Q water. Bacteria cultured on agar slant or plate cultures were overlaid with 1 to 2 ml of phosphate-buffered saline (PBS). After ≈15 min the PBS was carefully removed, and a few drops were applied to a number of cleaned slides. These were tilted to allow the drops to run down the length of the slide. Slides were air dried (not heat fixed). Multiple slides were prepared for each organism, as background staining and reproducibility were limitations of the method. They were stained for ≈12 min (Leifson’s stain) or ≈8 min (Difco stain), then gently rinsed in running water and allowed to air dry.
A modification of the above procedure, used with the Difco stain, resulted in better preservation of lateral flagella on Aeromonas species. Glutaraldehyde (≈3 ml, 1% [wt/vol] in distilled water) was placed directly on the bacterial growth on the agar surface. The excess liquid was removed, and the plate was left at room temperature for 5 min. Bacteria from the culture were then carefully immersed in a drop of water on the cleaned slide (≈2 min) before being processed for staining. At least 100 cells on each stained slide were examined under oil immersion (1,000×).
TEM.
For examination of polar flagella by transmission electron microscopy (TEM), bacteria from mid-log-phase broth cultures (TSBY, 37°C, 6 h) were adsorbed onto Formvar-coated copper grids (10 min) and fixed on a drop of 0.5% glutaraldehyde in 0.1 M PIPES buffer (piperazine-N,N′-bis[2-ethanesulfonic acid]), pH 7.3, for 2 min. The grids were then washed three times in PIPES buffer for 10 s each wash and negatively stained with either 1% (wt/vol) uranyl acetate, 2% (wt/vol) ammonium molybdate, or 0.5 or 2% (wt/vol) phosphotungstic acid for time intervals varying between 10 and 30 s. Staining with phosphotungstic acid (0.5% for 10 s) gave the most consistent results for visualization of flagellum structure. For examination of lateral flagella, the grids were wetted on drops of PIPES buffer and then applied directly to bacterial growth on the agar surface (TSAY, 37°C, 8 h) before fixing and staining as above.
FESEM.
Field emission scanning electron microscopy (FESEM) was performed at the Centre for Electron Microscopy and Microstructure Analysis, University of Adelaide, Adelaide, South Australia, Australia. Bacteria on agar surfaces were recovered and fixed as for TEM above. Samples were dehydrated through a graded acetone series and dried to critical point. Specimens were mounted on stubs, coated with gold, and examined with a Philips XL30 FESEM microscope at 15 kV.
Preparation of bacterial DNA.
Chromosomal DNA for Southern hybridization experiments was prepared from the Aeromonas strains using proteinase K digestion and cetyltrimethylammonium bromide extraction as described by Ausubel et al. (4).
Preparation of a laf gene probe.
In A. caviae Sch3, the two lateral flagellin genes, lafA1 and lafA2, are found on a ≈4.0-kb BamHI fragment (Fig. 1). The two genes are very similar, encoding proteins that have 87% amino acid identity. The nucleotide sequences have been submitted to GenBank under accession number AF348135. A 1.0-kb PstI fragment which contained the last three-quarters of the lafA1 gene (C-terminal sequence) and the first quarter of the lafA2 gene (N-terminal sequence) was ligated into the cloning vector pUC19 to give pARLA20. This fragment was successfully used as a probe in hybridization experiments to detect laf structural genes.
The laf probe was prepared by PCR from pARLA20 using M13 forward and reverse universal primers. An initial denaturation step (96°C for 10 min) was followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 55°C for 40 s, and extension at 72°C for 1 min on a PTC100 thermal cycler with a hot bonnet (MJ Research, Boston, Mass.). Digoxigenin-labeled dUTP (Roche Diagnostics Australia, Pty. Ltd., New South Wales, Australia) was incorporated into the probe during this amplification procedure. The probe would have contained 40 bp of pUC19 at the forward primer end and 25 bp at the reverse primer end. (DNA of pUC19 does not cross-react with Aeromonas DNA [J. G. Shaw, unpublished observation].) Prior to hybridization, the laf probe was incubated at 100°C for 5 min. After addition to the hybridization solution, it was incubated at 65°C for 20 min.
Southern analysis of laf gene distribution in Aeromonas spp.
Chromosomal DNA from each strain tested was digested with the restriction endonuclease PstI (Promega Corporation, Madison, Wis.), and the DNA fragments were electrophoresed in a 0.8% agarose gel. The fragments were transferred to a nylon membrane (Magnacharge MSI membrane; Geneworks, Adelaide, Australia) using the standard Southern protocol (36). Membranes were rinsed for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and baked at 120°C for 30 min. They were then incubated in prehybridization solution (1% blocking reagent [Roche], 5× SSC, 0.1% [wt/vol] sodium lauroyl sarcosine, 0.02% sodium dodecyl sulfate [SDS], and 50% formamide) at 42°C for 2 h.
Hybridization of the labeled probe (≈25 ng) to the membranes was carried out at 42°C overnight. Following hybridization, the membranes were washed twice in 2× SSC–0.1% SDS for 5 min at room temperature and twice in 0.5× SSC–0.1% SDS for 15 min at 65°C. The bound probe was visualized using a chemiluminescent detection kit (Roche) according to the manufacturer’s directions. After the final step, excess liquid was removed and the membranes were incubated in plastic bags (37°C, 20 min) before being exposed to X-ray film (Cronex medical X-ray film; Sterling Diagnostic Imaging Inc., Newark, N.J.) for 30 min to 24 h.
RESULTS
Light microscopic detection of lateral flagella.
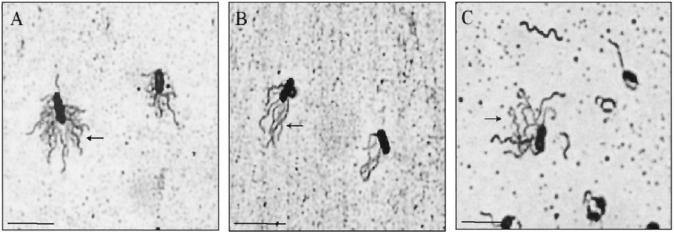
The Leifson staining method was established using control strains P. mirabilis and V. parahaemolyticus (Fig. 2A and 2B). A. caviae strain Sch3 was shown to express lateral flagella on >60% of the bacterial population when grown on solid medium (TSAY) at 37°C for 6 h (Fig. 2C). Growth conditions favoring expression of lateral flagella were investigated using this strain. Bacteria were grown in liquid medium (TSBY) and on solid medium (TSAY) at 22 and 37°C and stained for flagella at 6, 8, 12, and 24 h. No lateral flagella were seen in cultures grown in liquid medium. More than 50% of the bacterial cells grown on solid medium for 6 to 8 h at 37°C expressed lateral flagella. After this time, expression of lateral flagella declined dramatically, and by 24 h they were rarely seen. At lower temperatures, lateral flagella were also seen optimally at 6 to 8 h and continued to be expressed on >20% of the population at 24 h.
FIG. 2.
Light microscopic detection of flagella. Bacteria were grown on TSAY at 37°C for 6 h and stained with Leifson’s stain. (A) P. mirabilis, showing peritrichous flagella (arrow). (B) V. parahaemolyticus strain ACM 2776, showing lateral flagella (arrow). (C) A. caviae strain Sch3, showing lateral flagella (arrow). Bar, 5 μm.
Culture in iron-deficient media (TSAY and TSBY containing the iron chelator deferoxamine mesylate at 200 μM) did not affect the expression of lateral flagella. Investigations with a second clinical strain (A. veronii biovar sobria strain CA25, a Tasmanian diarrheal fecal isolate) confirmed that lateral flagella were not expressed in liquid medium, but were expressed when the bacteria were grown on solid medium for <8 h. Seven of 12 (58%) additional strains of Aeromonas spp. (3 each of A. hydrophila and A. caviae, and 6 of A. veronii biovar sobria, predominantly [10 of 12] clinical isolates) expressed lateral flagella under these conditions.
Electron microscopic examination of Aeromonas flagella.
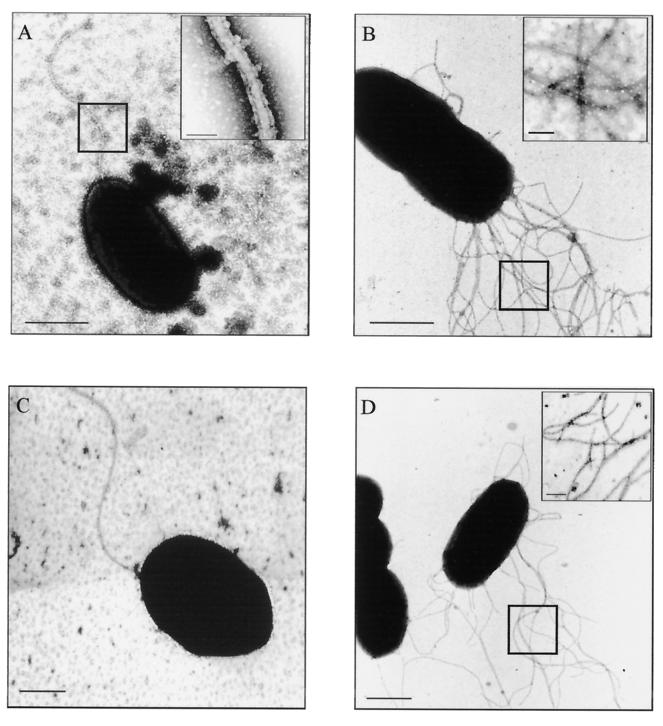
Electron microscopic staining methods were optimized to detect the polar flagellar sheath of V. parahaemolyticus. The sheath was demonstrated by an increase in the density of the stain, and the width of the flagellum (25 to 30 nm) (Fig. 3A) and its double membrane structure could be seen at high magnifications (inset, Fig. 3A). By contrast, the lateral flagellum of V. parahaemolyticus lacked this membrane structure and was ≈15 nm wide (Fig. 3B). A. caviae strain Sch3 and 10 other Aeromonas strains (A. hydrophila [n = 1], A. caviae [n = 2], and A. veronii biovar sobria [n = 7]) were similarly examined after growth on TSAY at 37°C for 6 h. Seven of these strains had been shown by Southern hybridization to possess laf genes (see below); the remaining three strains were laf negative. The laf-negative strains had only a single polar flagellum, while multiple flagella were seen on A. caviae strain Sch3 and all seven laf-positive strains. No Aeromonas flagella possessed a sheath structure; all flagella were ≈15 nm in width (Fig. 3C and 3D). Thin-section electron microscopic comparisons (ruthenium red and tannic acid staining) also confirmed these findings (images not shown).
FIG. 3.
Transmission electron microscopy of flagella (negative staining; 0.5% phosphotungstic acid, 10 s). Bars, 1 μm. (A) Sheathed polar flagellum of V. parahaemolyticus. The inset is a high-power magnification (boxed section) showing the double membrane structure of the flagellar sheath. Inset bar, 50 nm. (B) Numerous unsheathed lateral flagella of V. parahaemolyticus strain NCTC 10884. Inset bar, 100 nm. (C) Unsheathed polar flagellum of A. veronii biovar sobria strain BC88. (D) Numerous unsheathed lateral flagella of A. veronii biovar sobria strain CA25. Inset bar, 100 nm.
Distribution of laf genes in Aeromonas spp.
Aeromonas reference strains (HG1 to HG12 and HG17), 84 mesophilic Aeromonas strains, and four A. salmonicida strains were examined for the presence of laf structural genes by Southern hybridization analysis with the laf gene probe.
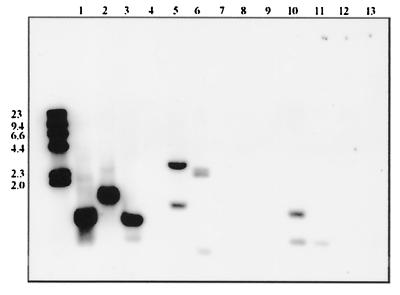
Only five of the reference strains, A. caviae HG4, A. media, HG5a, A. eucrenophila HG6, A. jandaei HG9, and A. schubertii HG12, hybridized with the probe (tested on three separate occasions). For the mesophilic strains, 58% showed positive hybridization. There was no marked species association in laf gene distribution (Table 1, Fig. 4). The majority of laf-positive strains (≈90%) reacted strongly with the probe, while ≈10% showed a weak reaction after longer exposure of the membranes. Most negative strains were retested twice with the same result. Forty-nine percent (27 of 55) of the clinical isolates were laf positive. Seventy percent (7 of 10) of the mesophilic strains isolated from fish and amphibians hybridized with the probe, as did all of four A. salmonicida isolates tested.
TABLE 1.
Hybridization of laf gene probe with mesophilic Aeromonas strains from varied sources and geographic regions
| Species | No. of strains tested | No. positivea | % Positive |
|---|---|---|---|
| A. hydrophila (HG1, HG2, and HG3) | 27 | 18 | 67 |
| A. caviae (HG4) | 16 | 7 | 44 |
| A. veronii biovar sobria (HG8/10) | 41 | 24 | 59 |
Includes all strains that reacted with the laf probe. Approximately 90% of strains were strongly reactive.
FIG. 4.
Southern blot (2-h exposure) of PstI-digested chromosomal DNA of Aeromonas isolates probed with the 1.0-kb PstI digoxigenin-labeled fragment of the lateral flagella genes lafA1 and lafA2. Lanes 1 to 13 are A. hydrophila strain DJ188, A. veronii biovar sobria FA132, A. caviae strain Sch3, and A. veronii biovar sobria strains BC88, CA17, CA110, BC96, CA112, CA113, EA49, EA57, EA42, and EA60, respectively. Isolates were from diarrheal feces except for strain FA132, which was isolated from a chicken carcass rinse, and strains EA49, EA57, EA42, and EA60, which were isolated from water samples. Positions of molecular size standards (λ HindIII digest) are indicated on the left (in kilodaltons).
Aeromonas spp. exhibit swarming motility.
Swarming is an organized surface motility (0.5 to 2% agar) that is dependent on extensive flagellation and cell-cell contact. Swarmer cells move in a group parallel to their long axis and maintain close contact with other cells (13, 15, 34). This movement is distinct from swimming, which is movement through water-filled channels within the agar (0.2 to 0.4% agar). An assay was established in which Aeromonas spp. consistently showed surface movement on swarm agar plates. P. mirabilis and V. parahaemolyticus swarm assays (TSAY 0.6% Oxoid agar and BHIB, and 1.5% NaCl–1.5% Oxoid agar, respectively) were used throughout as positive controls.
The source of the agar and its concentration greatly affected Aeromonas swarming. Eiken agar consistently supported a strong swarming response. No swarming occurred on Difco Bacto-agar, whereas swarming was occasionally seen with Oxoid agar. The optimal agar concentration for Aeromonas swarming was 0.5% (wt/vol). At higher agar concentrations, the proportion of strains exhibiting motility decreased, and the zone of swarming decreased in those strains that showed movement. Difco nutrient broth was better than LB broth in supporting motility on swarm plates. Manipulation of swarm plate composition (for example, by addition of glucose [0.5 and 1.5%] or iron depletion [200 μM deferoxamine mesylate]) did not result in an improved swarm response on swarm plates prepared with TSBY and Oxoid and Difco agar.
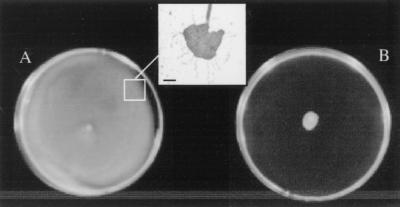
The method of inoculum propagation, the surface moisture of the swarm agar, and the temperature of incubation of inoculated swarm plates also influenced swarming motility. For optimal swarming, freshly poured and dried swarm agar plates (Eiken agar, 0.5% in Difco broth) were inoculated from swim agar plates and incubated at 30°C. Under these conditions, zones of surface translocation of ≥1.5 cm from the inoculation site were seen within 2 h, and bacteria completely covered the plate (zones of 5 to 8 cm) after overnight (16 to 18 h) incubation (Fig. 5A). Strains which did not swarm grew at the site of the initial inoculum but showed no movement (Fig. 5B).
FIG. 5.
Aeromonas swarming on Eiken agar. (A) Strong swarming response. (B) No swarming, but growth at the inoculum site. The inset (boxed section) shows a raft of bacteria surrounded by flagella taken from the swarm edge (glutaraldehyde-fixed sample, Difco flagella stain). Bar, ≈1 μm.
Cells from the periphery of spreading zones formed a pattern of whirls and bands and exhibited vigorous movement (phase-contrast microscopy) comparable to the micromorphological pattern and movement seen in P. mirabilis swarming (15, 46). The presence of “slime” preceding the colony spread was also noted. Flagella could be seen forming links between bacterial cells or surrounding rafts of cells (accumulations of bacteria aligned along their long axes and hyperflagellated) at the edge of the spreading zone (inset, Fig. 5A). More than 50% of bacteria near the periphery expressed multiple flagella (light and electron microscopic analyses). Elongation of bacteria from this area was also observed, but the extent and proportion of cells exhibiting it varied between strains, and it was not as marked as the elongation seen in the P. mirabilis swarm control.
FESEM analysis of bacteria on agar.
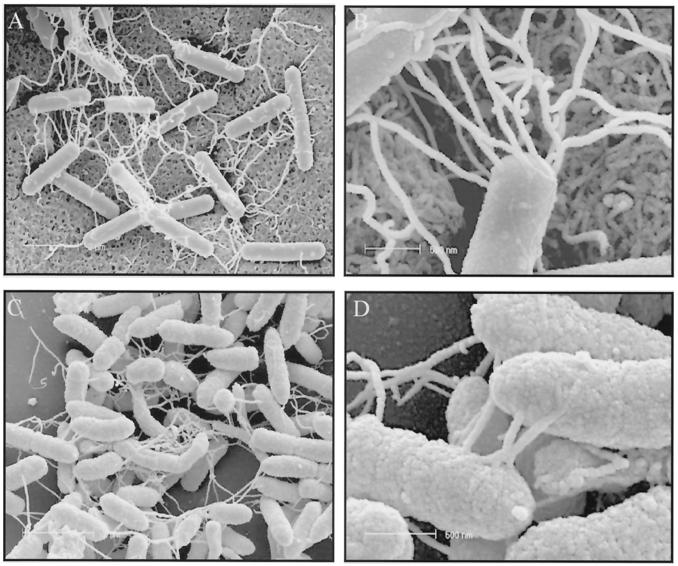
FESEM studies provided further evidence that lateral flagella form linkages between bacteria. When agar cores of V. parahaemolyticus cultures (TSAY, 37°C, 6 h) were examined by FESEM, numerous filamentous structures (lateral flagella from light and electron microscopic studies above) were seen linking bacteria on the agar surface (Fig. 6A and 6B). Similar filamentous structures were seen linking A. caviae strain Sch3 grown on agar as for V. parahaemolyticus (Fig. 6C and 6D) and when adherent to Henle intestinal 407 cells (not shown).
FIG. 6.
FESEM. (A and B) V. parahaemolyticus (strain ACM 2776) on TSAY (37°C, 6 h), showing intertwining networks of lateral flagella linking bacteria on the agar surface. (C and D) A. caviae strain Sch3 on agar as above, showing similar filamentous structures linking bacteria. Bars: (A and C) 2 μm; (B and D) 500 nm.
Correlation of presence of laf genes with swarming motility.
Thirteen strains (mixed species selection) which reacted strongly with the laf probe, eight strains that did not react with it all, and three strains which reacted weakly were examined in the swarm assay. Of the 13 strongly laf-positive strains, all showed vigorous surface movement with swarming zones after overnight incubation, as in Fig. 5A. The laf-negative and weakly positive strains showed no surface movement at all under the same conditions, only growth at the inoculation site, as in Fig. 5B.
Mutations in flagellar genes that affect lateral flagellum expression abolish swarming motility.
Definitive proof that lateral flagella are required for swarming motility was obtained when a selection of polar and lateral flagellar mutant strains of A. caviae strain Sch3 were examined. The preparation and characterization of the mutant strains are described in detail elsewhere (10, 33). Expression of flagella on these strains was determined by TEM and Western blot analysis (summarized in Table 2). For the polar flagellar mutant strains, mutation in flaH (AAR59), flaJ (AAR8), or both flagellin genes, flaAB (AAR31), resulted in the loss of both the polar and lateral flagella. All the lateral flagellar mutant strains expressed the polar flagellum, but only mutants with single mutations in lafA1 (AAR58), lafA2 (AAR5), and fliU (AAR20) had lateral flagella (10).
TABLE 2.
Effect of mutation of polar and lateral flagellar genes of A. caviae Sch3N on lateral flagellum expression and swarming motilitya
| Strain | Mutation | Polar flagellab | Lateral flagellab | Swimming phenotypec | Swarming phenotyped |
|---|---|---|---|---|---|
| Wild type | |||||
| A. caviae Sch3 | + | + | ++ | ++ | |
| Polar flagellar mutants | |||||
| AAR269 | flaA::Kmr | + | + | + | + |
| AAR27 | flaB::Kmr | + | + | ++ | + |
| AAR31 | flaA::CmrflaB::Kmr | − | − | − | − |
| AAR150 | flaG::Kmr | + | + | ++ | ++ |
| AAR59 | flaH::Kmr | − | − | − | − |
| AAR8 | flaJ::Kmr | − | − | − | − |
| Lateral flagellar mutants | |||||
| AAR58 | lafA1::Kmr | + | + | + | + |
| AAR5 | lafA2::Kmr | + | + | + | + |
| AAR9 | lafB::Kmr | + | − | + | − |
| AAR20 | fliU::Kmr | + | + | + | + |
| AAR6 | lafA1::CmrlafA2::Kmr | + | − | + | − |
Strains were tested in duplicate in at least two separate experiments, and the wild-type strain, A. caviae Sch3, was included in all experiments.
Flagellum expression was determined by TEM and Western blotting (10).
Swimming was determined as migration from the source of inoculation in LB broth containing 0.3% agar (Oxoid) after 16 to 24 h at 30°C. Symbols: ++, spreading zone ≥7 cm from central point of inoculation; +, spreading zone ≥2 cm but <7 cm from the inoculation point; −, growth at the inoculum site but no movement.
Swarming was determined as surface migration on Eiken agar swarm plates (0.5% agar in Difco nutrient broth) after 16 to 24 h at 30°C. Symbols: ++, surface motility zone ≥8 cm; +, surface motility zone ≥3 cm but <8 cm; or −, growth at the inoculum site but no surface movement.
Mutant strains were compared with the wild-type strain for their swimming and swarming phenotypes in this study (Table 2). Swimming and swarming motility zones showed strong correlation with the expression of polar and lateral flagella, respectively. All strains unable to express lateral flagella showed no movement on Eiken agar swarm plates, whereas this surface motility was seen in all mutant strains which expressed lateral flagella (Table 2).
DISCUSSION
This study has characterized the expression and distribution of lateral flagella in Aeromonas species. We found that only 50 to 60% of mesophilic aeromonads from the species most commonly associated with diarrhea are able to express lateral flagella when exposed to a solid surface. This restricted distribution of lateral flagella in Aeromonas spp. was supported by the close agreement between the light and electron microscopic expression studies, the laf gene Southern analysis, and swarming motility phenotypes. The validity of the Southern analysis was further shown by the fact that the laf gene probe reacted strongly with strains of all of the above three species, as well as with reference strains of A. media, A. eucrenophila, A. jandaei, and A. schubertii, despite being designed from the laf structural gene sequence of A. caviae strain Sch3. Strains that did not hybridize with the probe were negative on repeat testing. The small proportion (<10%) of strains shown to react weakly with the probe on longer exposure included some strains with probable, nonspecific, false-positive reactions, as shown from the expression and functional studies. As flagellar sequences are similar, these cross-reactions were possibly due to cross-reaction of the probe with polar flagellar gene sequences or with the vestigial remains of redundant laf genes. Hence, the overall percentage of strains that possessed laf genes was closer to 50 than 60%. Electron microscopic analysis confirmed that Aeromonas lateral flagella are unsheathed.
This study also documents the conditions influencing Aeromonas swarming motility and the role of lateral flagella in this behavior. Eiken agar was required to demonstrate consistent swarming motility by Aeromonas spp. Swarming by Escherichia coli and Salmonella enterica serovar Typhimurium has also been reported to be critically dependent on the source of the agar (Eiken or Difco) (14). Analytical data on the physical and chemical characteristics of these agars, however, showed no significant differences (14). It seems likely that some parameter that affects the surface, such as superior wettability, is responsible for the permissive quality of Eiken agar (41). Our microscopic analyses established that the motility seen on this agar was swarming and not just a result of the surfactant properties of this particular agar.
Only Aeromonas strains showing strong positive hybridization with the laf probe exhibited swarming motility. The laf-negative and weakly positive strains expressed only polar flagella and showed no surface movement. Moreover, strains of A. caviae Sch3 in which lateral flagellum expression was abrogated by mutation of polar or lateral flagellar genes which resulted in the loss of surface motility. These results show conclusively that lateral flagella are required for Aeromonas swarming motility.
It is of interest that in A. caviae, not only the flaH and flaJ polar flagellar mutants but also the tandem flaAB polar flagellin mutant were unable to produce both polar and lateral flagella. This suggests that the polar flagellar system controls the synthesis of the Aeromonas lateral flagella (10). Hence, regulation of the lateral flagella in Aeromonas spp. differs from that in V. parahaemolyticus, in which the two flagellum systems are distinct and genetic interference with polar function triggers constitutive expression of lateral flagella (23).
The role of lateral flagella in pathogenesis has not yet been established for any organism. Swarmer cell differentiation is an adaptation that is thought to facilitate colonization of inanimate or host cell surfaces by allowing movement on surfaces and through highly viscous environments (22). The bacterium-bacterium linkages on the agar surface were similar to linkages observed for the lateral flagella of swarming V. parahaemolyticus (5). Preliminary studies with A. caviae strain Sch3 suggested that such linkages were also present on bacteria adherent to Henle intestinal cell surfaces. These observations suggest that lateral flagella could have roles in addition to swarming motility which contribute to intestinal colonization.
At first the limited distribution of lateral flagella among clinical isolates of Aeromonas spp. appears to argue against their having a significant role in Aeromonas colonization and virulence. However, a spectrum of disease presentations are associated with Aeromonas gastrointestinal infection, ranging from watery diarrhea accompanied by a mild fever (approximately three-quarters of reported cases) to dysentery-like cases with blood and mucus present in the stools (approximately one-quarter of cases). In more than one-third of patients, diarrhea may last for over 2 weeks (11, 16). Cases longer than 17 months in duration have been documented (35). It seems likely that different combinations of virulence determinants may lead to these different clinical manifestations, analogous to the various pathotypes described for E. coli (18, 28).
Biofilms are a known to be a particular feature of persistent infections, and swarming motility has been linked to the formation of biofilms in a number of bacterial infections (8, 47). Lateral flagella facilitate biofilm formation by Aeromonas spp. on borosilicate glass at 37°C (unpublished observations) and in microtiter plates (10). Swarming motility has also been linked to invasive ability, for example, in P. mirabilis, which causes serious kidney infections that can involve invasion of host urothelial cells (2). Invasion by Aeromonas spp. may account for the dysenteric presentation (43). Lateral flagella potentiate the adherence of A. caviae strain Sch3 to HEp-2 cells (10), and this strain can invade HEp-2 and Caco-2 cells (38). Hence, it is possible that Aeromonas strains that are able to produce lateral flagella may be more frequently associated with persistent or dysenteric infection. Further studies are required to evaluate the role(s) and significance of lateral flagella in Aeromonas-host interactions.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia (981530), University of Tasmania ARC Small Grant, the Clifford Craig Foundation, and the Royal Hobart Hospital Research Foundation.
We thank Kathleen Shaw and Hooshang Lahooti for their contributions to the light microscopic studies and Southern analysis, respectively, and Rasika Harshey, University of Texas at Austin, Austin, Tex., for helpful discussion regarding swarming in gram-negative bacteria.
REFERENCES
- 1.Alberti, L., and R. M. Harshey. 1990. Differentiation of Serratia marcescens 274 into swimmer and swarmer cells. J. Bacteriol. 172:4322–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, C., N. Coleman, P. L. Jones, and C. Hughes. 1992. The ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect. Immun. 60:4740–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, C., and C. Hughes. 1991. Bacterial swarming: an example of prokaryotic differentiation and multicellular behaviour. Sci. Progress Edinburgh 75:403–422. [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, and R. E. Kingston. 1994. Current protocols in molecular biology, 2nd ed. John Wiley and Sons Inc., New York, N.Y.
- 5.Belas, M. R., and R. R. Colwell. 1982. Scanning electron microscope observation of the swarming phenomenon of Vibrio parahaemolyticus. J. Bacteriol. 150:956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrell, N., S. G. Acinas, M-J. Figueras, and A. J. Martinez-Murcia. 1997. Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S RNA genes. J. Clin. Microbiol. 35:1671–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnahan, A. M., S. Behram, and S. W. Joseph. 1991. Aerokey II: a flexible key for identifying clinical Aeromonas species. J. Clin. Microbiol. 29:2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opinion Microbiol. 2:630–635. [DOI] [PubMed] [Google Scholar]
- 10.Gavín, R., A. A. Rabaan, S. Merino, J. M. Tomas, I. Gryllos, and J. G. Shaw. 2001. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol., in press. [DOI] [PubMed]
- 11.Gracey, M., V. Burke, and J. Robinson. 1982. Aeromonas-associated gastroenteritis. Lancet ii:1304–1306. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg, E. P. 1999. Quorum sensing in gram-negative bacteria. ASM News 63:371–377. [Google Scholar]
- 13.Harshey, R. M. 1994. Bees aren’t the only ones: swarming in Gram-negative bacteria. Mol. Microbiol. 13:389–394. [DOI] [PubMed] [Google Scholar]
- 14.Harshey, R. M., and T. Matsuyama. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. USA 91:8631–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrichsen, J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations and unanswered questions. Clin. Infect. Dis. 27:332–344. [DOI] [PubMed] [Google Scholar]
- 17.Kaznowski, A. 1997. Numerical taxonomy and DNA-DNA hybridizations of Aeromonas strains isolated from human diarrhoeal stool, fish and environment. Syst. Appl. Microbiol. 20:458–467. [Google Scholar]
- 18.Kirov, S. M. 1997. Aeromonas and Plesiomonas, p.265–287. In M. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. American Society for Microbiology, Washington, D.C.
- 19.Kirov, S. M., L. A. O’Donovan, and K. Sanderson. 1999. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect. Immun. 67:5447–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirov, S. M., B. Rees, R. C. Wellock, J. M. Goldsmid, and A. D. van Galen. 1986. Virulence characteristics of Aeromonas spp. in relation to source and biotype. J. Clin. Microbiol. 24:827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinetti Lucchini, G., and M. Altwegg. 1992. rRNA gene restriction patterns as taxonomic tools for the genus Aeromonas. Int. J. Syst. Bacteriol. 42:384–389. [DOI] [PubMed] [Google Scholar]
- 22.McCarter, L. 1999. The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1:51–57. [PubMed] [Google Scholar]
- 23.McCarter, L., M. Hillman, and, M. Silverman. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351. [DOI] [PubMed] [Google Scholar]
- 24.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057–1062. [DOI] [PubMed] [Google Scholar]
- 25.Merino, S., X. Rubires, A. Anguillar, J. F. Guillot, and J. M. Tomás. 1996. The role of O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Aeromonas hydrophila serogroup O:34. Microb. Pathog. 20:325–333. [DOI] [PubMed] [Google Scholar]
- 26.Meynell, G. G., and E. Meynell. 1970. p.167–168 In Theory and practice of experimental bacteriology, 2nd ed. Cambridge University Press, London, United Kingdom.
- 27.Müller, H. E., and W. Lenz. 1975. Swarming phenomenon of an Aeromonas species. Zentbl. Bakteriol. Hyg. I Abt. Orig. A 231:451–465. (In German.) [PubMed] [Google Scholar]
- 28.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nzeako, B. C. 1991. Variation in Aeromonas hydrophila surface structures. Bull. Eur. Assoc. Fish Pathol. 1:176–179. [Google Scholar]
- 30.Oakey, H. J., J. T. Ellis, and L. F. Gibson. 1996. A biochemical protocol for the differentiation of current genomospecies of Aeromonas. Zentbl. Bakteriol. 284:32–46. [DOI] [PubMed] [Google Scholar]
- 31.O’Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for the Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304. [DOI] [PubMed] [Google Scholar]
- 32.Quinn, D. M., H. M. Atkinson, A. H. Bretag, M. Tester, T. J. Trust, C. Y. F. Wong, and R. F. L. P. Flower. 1994. Carbohydrate-reactive, pore-forming outer membrane proteins of Aeromonas hydrophila. Infect. Immun. 62:4054–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabaan, A. A., I. A. Gryllos, J. M. Tomás, and J. G. Shaw. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect. Immun. 69:4257–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rautelin, H., M. L. Hänninen, A. Sivonen, U. Turunen, and V. Valtonen. 1995. Chronic diarrhea due to a single strain of Aeromonas caviae. Eur. J. Clin. Microbiol. Infect. Dis. 14:51–53. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Semmler, A. B. T., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863–2873. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, J. G., J. P. Thornley, I. Palmer, and I. Geary. 1995. Invasion of tissue culture cells by Aeromonas caviae. Med. Microbiol. Lett. 4:324–331. [Google Scholar]
- 39.Shimada, T., R. Sakazaki, and K. Suzuki. 1985. Peritrichous flagella in mesophilic strains of Aeromonas. Jpn. J. Med. Sci. Biol. 38:141–145. [DOI] [PubMed] [Google Scholar]
- 40.Thornley, J. P., J. G. Shaw, I. A. Gryllos, and A. Eley. 1997. Virulence properties of clinically significant Aeromonas species: evidence for pathogenicity. Rev. Med. Microbiol. 8:61–72. [Google Scholar]
- 41.Toguchi, A., M. Siano, M. Burkhart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling in Pseudomonas aeruginosa. Emerg. Infect. Dis. 4:551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson, I. M., J. O. Robinson, V. Burke, and M. Gracey. 1985. Invasiveness of Aeromonas spp. in relation to biotype, virulence factors and clinical features. J. Clin. Microbiol. 22:48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilcox, M. H., Cook, A. M., Eley, A., and R. C. Spencer. 1992. Aeromonas spp. as a potential cause of diarrhoea in children. J. Clin. Pathol. 45:959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilcox, M. H., A. M. Cook, K. J. Thickett, A. Eley, and R. C. Spencer. 1992. Phenotypic methods for speciating clinical Aeromonas isolates. J. Clin. Pathol. 45:1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, F. D. 1978. Nature of the swarming phenomenon in Proteus. Annu. Rev. Microbiol. 32:101–122. [DOI] [PubMed] [Google Scholar]
- 47.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]