Abstract
Transcriptional analysis and disruption of five open reading frames (ORFs), ydiO, ydiP, ydiR, ydiS, and ydjA, in the prophage 3 region of the chromosome of Bacillus subtilis Marburg revealed that they are component genes of the intrinsic BsuM restriction and modification system of this organism. The classical mutant strain RM125, which lacks the restriction and modification system of B. subtilis Marburg, lacks the prophage 3 region carrying these five ORFs. These ORFs constitute two operons, the ydiO-ydiP operon and the ydiR-ydiS-ydjA operon, both of which are expressed during the logarithmic phase of growth. The predicted gene products YdiO and YdiP are the orthologues of cytosine DNA methyltransferases. The predicted YdiS product is an orthologue of restriction nucleases, while the predicted YdiR and YdjA products have no apparent paralogues and orthologues whose functions are known. Disruption of the ydiR-ydiS-ydjA operon resulted in enhanced transformation by plasmid DNA carrying multiple BsuM target sequences. Disruption of ydiO or ydiP function requires disruption of at least one of the following genes on the chromosome: ydiR, ydiS, and ydjA. The degrees of methylation of the BsuM target sequences on chromosomal DNAs were estimated indirectly by determining the susceptibility to digestion with XhoI (an isoschizomer of BsuM) of DNAs extracted from the disruptant strains. Six XhoI (BsuM) sites were examined. XhoI digested at the XhoI sites in the DNAs from disruptants with disruptions in both operons, while XhoI did not digest at the XhoI sites in the DNAs from the wild-type strain or from the disruptants with disruptions in the ydiR-ydiS-ydjA operon. Therefore, the ydiO-ydiP operon and the ydiR-ydiS-ydjA operon are considered operons that are responsible for BsuM modification and BsuM restriction, respectively.
The existence of an inherent BsuM restriction and modification system in Bacillus subtilis Marburg 168 was first suggested on the basis of the results of an experiment performed with phage φ105 and B. subtilis and Bacillus amyloliquefaciens host strains (28). The hsrM1 and nonB mutations (24, 36) which made host cells permissive to phage infection were isolated and mapped at around 50° on the B. subtilis chromosome (24), and it was thought that these mutations were mutations of the endonuclease gene of BsuM. On the other hand, strain RM125 (34) was constructed by transformation of wild-type B. subtilis Marburg 168 (YS11) with DNA from the related strain B. subtilis 202-5 (= IAM1169, a B. amyloliquefaciens strain), which lacked restriction activity against phage φ105. Strain RM125 constructed in this way turned out to be deficient in modification activity, as well as restriction activity. However, it was not known whether RM125 lost the same BsuM restriction activity as nonB and hsrM1 strains.
The target sequence of the BsuM restriction and modification system of B. subtilis was first predicted to be PyTCGAPu (7, 13), but in another study researchers determined that the BsuM target sequence was CTCGAG by performing a transformation analysis of plasmids carrying multiple PyTCGAPu target sequences (2). Therefore, XhoI is believed to be an isoschizomer of BsuM. Moreover, efficient restriction required the presence of multiple target sites for XhoI (BsuM) in plasmid DNA (2). On the other hand, cytosine DNA methyltransferase, which methylates a CTCGAG target, was purified from the B. subtilis Marburg strain (8), but its gene was not identified.
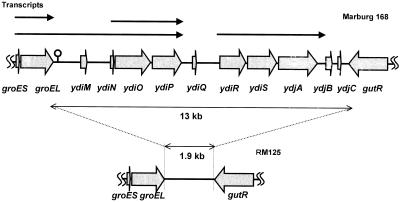
DNA sequencing of the B. subtilis chromosomal rrnE region revealed a 13-kb region with a lower G+C content and lower gene density between the groESL operon and the gut operon (14). This region was called prophage 3 (17) and contained five open reading frames (ORFs), ydiO, ydiP, ydiR, ydiS, and ydjA, as well as pseudogenes and five smaller ORFs, ydiM, ydiN, ydiQ, ydjB, and ydjC (14) (Fig 1). Although there are not any paralogue genes on the B. subtilis chromosome except mtbP of SPβ prophage, the predicted products of ydiO and ydiP exhibit high levels of similarity to DNA modification enzymes HgiDIIM from Herpetosiphon giganteus and MspI from members of the family Moraxellaceae (4, 18), respectively, and the predicted product of ydiS exhibits a high level of similarity to DNA restriction enzyme LlaI-2 of Lactococcus plasmid pTR2030 (20).
FIG. 1.
Transcriptional map of the prophage 3 region of B. subtilis Marburg 168 which is not present in the classical RM125 strain deficient in BsuM restriction and modification. The thin arrows indicate the transcripts identified by Northern analysis of this region. The thick arrows indicate the predicted ORFs. The transcription termination signal of the groESL operon is indicated by a lollipop.
In this study we tried to determine whether ORFs ydiO, ydiP, ydiR, ydiS, and ydjA are components of the intrinsic BsuM restriction-modification system of B. subtilis Marburg which are not present in the classical RM125 strain. Our results indicated that there are two operons, one consisting of ydiO and ydiP for DNA methylation and the other consisting of ydiR, ydiS, and ydjA for DNA restriction, in the prophage 3 region of the B. subtilis Marburg chromosome and that these two operons are not present in the classical RM125 strain. Furthermore, disruption of the rho-independent termination signal sequence of the groESL transcription region resulted in enhanced readthrough and enhanced transcription of the ydiO-ydiP operon.
MATERIALS AND METHODS
Bacterial strains and plasmids used.
The bacterial strains and plasmids used in this study are described in Table 1. B. subtilis Marburg 168 trpC2 (35) was used as a wild-type strain in all experiments.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype and/or relevant phenotypea | Reference and/or source |
|---|---|---|
| B. subtilis strains | ||
| 168 | trpC2 | 35 |
| RM125 | leuA8 argA15 | 34, M. Itaya |
| BSU1 | trpC2 ydiO::pET24b neo(Kmr) ydiS::pMUTIN2 (erm) | This study |
| BSU2 | trpC2 ydiP::pET24b neo(Kmr) ydiS::pMUTIN2 (erm) | This study |
| BSU3 | trpC2 ydiR::pMUTIN2 (erm) | This study |
| BSU4 | trpC2 ydiS::pMUTIN2 (erm) | This study |
| BSU5 | trpC2 ydjA::pMUTIN2 (erm) | This study |
| BSU6 | trpC2 ydiR::pMUTIN2 (Pspac-ydiR erm) | This study |
| BSU7 | trpC2 ydiO::pET24b neo(Kmr) ydiR::pMUTIN2 (Pspac-ydiR erm) | This study |
| BSU8 | trpC2 ydiP::pET24b neo(Kmr) ydiR::pMUTIN2 (Pspac-ydiR erm) | This study |
| BSU9 | trpC2 Δter(groESL) | This study |
| E. coli strains | ||
| C600 | thi-1 thr-1 leuB6 lacY1 tonA21 supE44 | 40, S. Yasuda |
| JM109 | recA1 hsdR17 Δ(lac-proAB) endA1 gyrA96 relA1 thi supE44 F′ [traD36 proAB+lacIqlacZΔM15] | 37, TaKaRa |
| Plasmids | ||
| pMUTIN2 | Emr (B. subtilis) Apr (E. coli) | 35 |
| pET24b neo | Kmr (B. subtilis) | 5 |
| pHV33 | Cmr (B. subtilis) Apr (E. coli), no XhoI site | 2, M. Itaya |
| pHV1401 | Cmr (B. subtilis) Apr (E. coli), three XhoI sites | 2, M. Itaya |
| pCP112 | Cmr (B. subtilis) Apr (E. coli) | 22 |
neo and erm, neomycin and erythromycin genes, respectively; Kmr, Apr, Emr, and Cmr, resistance to kanamycin, ampicillin, erythromycin, and chloramphenicol, respectively.
Media, reagents, and enzymes.
Cells of B. subtilis or Escherichia coli were grown in Luria-Bertani broth (25) or a complex sporulation medium (26). Reagents and enzymes were purchased from TaKaRa (Kyoto, Japan), Wako (Osaka, Japan), and Sigma (St. Louis, Mo.). AmpliTaq DNA polymerase (Applied Biosystems, Foster City, Calif.) and TaKaRa LA Taq DNA polymerase (TaKaRa) were used for PCR.
DNA manipulation.
Plasmid DNA purification, PCR amplification of DNA fragments, digestion of DNA with restriction enzyme, and DNA ligation were carried out as described elsewhere (25). Plasmid DNA was purified by polyethylene glycol precipitation of a sample of E. coli C600 cells lysed by alkali. The primers used for long accurate PCR amplification of the prophage 3 region were 5′-ATATGGGCGGCATGGGCGGTATGGGTGGAA (groELF30) and 5′-GCATAACAGCCGCTTTCATGTTTTGAGGTA (gutBR30).
For disruption and confirmation of the ORFs, a primer pair with HindIII or BamHI sites were used for PCR amplification of a portion of each ORF. The following primer pairs were used: for groES, 5′-GCCGAAGCTTCTAAAATTACATATTCA and 5′-CGCGGATCCGTATTTTGAGAAGATAA; for groEL, 5′-GCCGAAGCTTAAGAAATTAAGTTTAGT and 5′-CGCGGATCCGATTGTGATAACGCCGT; for ydiO, 5′-GCCGAAGCTTAACATAGAAAATTTTTA and 5′-CGCGGATCCCACCTAGTTTTTCAGCA; for ydiP, 5′-GCCGAAGCTTATCAGTCAGGACATACA and 5′-CGCGGATCCCCTTGTAGGCGCTCAGC; for ydiR, 5′-GCCGAAGCTTGCCGGAGTATCTAGTAC and 5′-CGCGGATCCATTTATGTTTTATCTTA; for ydiS, 5′-GCCGAAGCTTAGTATTCCTAGGAATAG and 5′-CGCGGATCCAAAGCGGTATCGAGCGG; for ydjA, 5′-GCCGAAGCTTAAAGGATTGTACGTGTA and 5′-CGCGGATCCTAATATCGGGTTCTAGT; for gutR, 5′-GCCGAAGCTTCCGTATCGCTTCCATAT and 5′-CGCGGATCCATGCTGGCACCTGGCGT; and for gutB, 5′-GCCGAAGCTTAAATTGCCGCTGTCGGA and 5′-CGCGGATCCTTTTCCGCCCCGGCGCA. These primers were custom made (Espec Oligo Service, Tsukuba, Japan) and were also used for PCR analysis of strain RM125.
Disruption of ORFs.
Disruption of ORFs with the integrative plasmid pMUTIN was carried out as described elsewhere (35, 39). The primer pairs used for PCR synthesis of ydiO, ydiP, ydiR, ydiS, and ydjA are described above.
Construction of IPTG-inducible ydiR-ydiS-ydjA operon on the chromosome.
An isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter was introduced into the promoter region of the ydiR-ydiS-ydjA operon by integration of plasmid pMUTIN carrying the N-terminal region of the ydiR gene with the Shine-Dalgarno sequence, which was synthesized by PCR performed with primers 5′-AAGAAGCTTGCTTGTGTGATTTTATGGGG and 5′-CGCGGATCCATTTATGTTTTATCTTA. The resulting strain carried Pspac-ydiR.
Transformation.
Competent B. subtilis cells were prepared in Spizizen’s minimal glucose medium, and transformation was carried out as described elsewhere (1, 15). Transformation of E. coli was performed as described elsewhere (6).
Construction of a deletion mutant with a mutation in the groESL transcription termination signal sequence.
A DNA segment lacking the transcription termination signal sequence was constructed by ligating two DNA fragments which were synthesized by PCR performed with primer pairs 5′-CTTCTGAATTCGACAGAG-5′-CGGTTAAAACATTGATGTATAAGGG and 5′-AAATCCCCAGTTGGGTTC-5′-ATCCATCAGGATTGATTCC, cloned into plasmid pCP112 (22), and then introduced into the chromosome of strain 168 by transformation. The inserted plasmid was cured from competent cells. The deletion, Δter(groESL), was confirmed by PCR.
Northern analysis.
RNA was extracted with glass beads from cells grown in sporulation medium, separated by electrophoresis, and detected with a digoxigenin (DIG)-labeled RNA probe as described elsewhere (39). A 20-μg RNA sample was used. The RNA probe was constructed with a PCR-synthesized DNA fragment containing the T7 promoter sequence (39). The primer pairs used each consisted of a forward primer and reverse primer T7rev (5′-TAATACGACTCACTATAGGGCGAAGTGTATCAACAAGCTGG). RNA probes for ydiO and ydiP were constructed with ydiO::pDX-CAT ydiS::pMUTIN2 and ydiP::pDX-CAT ydiS::pMUTIN2 double disruptants, in which the integrative plasmid pDX-CAT, provided by Y. Kasahara, carried the bgaB gene (41). A DIG-labeled RNA probe was synthesized with a DIG-UTP labeling kit, and DIG was detected with a DIG detection kit (Roche). The length of RNA detected was estimated by calibrating Northern signals with 16S rRNA, 23S rRNA, and RNA markers that were 9, 6, 5, 4, 3, 2.5, 2, 1.5, 1, and 0.5 kb long (Ambion, Inc., Austin, Tex.).
Southern analysis.
The degrees of cytosine methylation were estimated for six fragments containing only one XhoI site at nucleotide positions 106975 (9°, AGGCTCGAGTAT), 2633916 (54°, GCGCTCGAGACA), 015718 (172°, GCGCTCGAGCAG), 2800360 (239°, TCACTCGAGATT), 3105700 (265°, CTTCTCGAGGAT), and 3550939 (303°, TTTCTCGAGCTT) on the chromosome (17). Chromosomal DNA from each strain was double digested with XhoI and an appropriate restriction endonuclease, separated by agarose electrophoresis, and transferred to a membrane filter as described elsewhere (25). The DNAs were then hybridized to a DNA probe, which was synthesized by PCR and labeled in vitro with alkaline phosphatase (AlkPhos Direct; Amersham), and were detected with a CDP Star detection kit and Hyperfilm ECL (Amersham).
The following primer pairs were used for PCR: for xfrg9, 5′-CGGATCTACAAACGAAATGG and 5′-CCCTGTTATGGTCTATTCCC; for xfrg54, 5′-TATAACCACGGAAACTT and 5′-AAAATGTGTATTATGGT; for xfrg172, 5′-GCATCAATCAATCCTGCGAC and 5′-AGCTGTTTTTGGCACACGGC; for xfrg239, 5′-GTGATTAACTGCACTCAGGA and 5′-GGCTGTTCCTGCGACGGCTG; for xfrg265, 5′-ATGTTCTGGAATGAATTAAA and 5′-ATTCGTCAATGTGCAAAC; and for xfrg303, 5′-GCTGGATCATAGAAACCACC and 5′-GGGCGATTGAAAGTAACT.
RESULTS
Disruption of the ORFs located in the prophage 3 region.
Single-gene disruption was carried out with integrative plasmid pMUTIN2 carrying a portion of the target gene, as described elsewhere (35, 39). As the whole lacZ gene with the Shine-Dalgarno sequence is joined to the 5′ region of the disrupted gene, transcription of the disrupted gene is easily monitored by monitoring lacZ activity. As the inserted plasmid carries an IPTG-inducible Pspac promoter at the 3′ end of the plasmid, addition of IPTG induces transcription of the genes following the disrupted gene, thus avoiding a polar effect of any operon due to insertional disruption.
Single disruption of ydiR, ydiS, or ydjA was easily performed, while transformation of wild-type strain 168 to erythromycin resistance with pMUTIN2 carrying a portion of the ydiO or ydiP gene resulted in very tiny colonies on nutrient agar containing antibiotics. Disruption of the gene essential for growth by transformation with an integrative plasmid carrying a cloned target gene often resulted in very tiny transformants. Transformation of the strains in which ydiR, ydiS, or ydjA was already disrupted to kanamycin resistance with the pET24 neo plasmid carrying a portion of ydiO or ydiP yielded transformants (Table 2), indicating that disruption of ydiO and ydiP function was possible only when the ydiR, ydiS, or ydjA gene or a combination of these genes was already disrupted. Disruption of ydiO may have resulted in reduced expression of ydiP as pET24 neo does not have a Pspac promoter.
TABLE 2.
Disruption of ydiO or ydiPa
| Target gene | No. of kanamycin-resistant transformants (102)/μg of DNA with the following recipients:
|
|||
|---|---|---|---|---|
| 168 | BSU3 ydiR::pMUTIN2 | BSU4 ydiS::pMUTIN2 | BSU5 ydjA::pMUTIN2 | |
| ydiO | <3.0 | 580 | 570 | 670 |
| ydiP | <6.0 | 710 | 640 | 580 |
The pET24b neo plasmid carrying a portion of ydiO or ydiP was used to transform a disruptant strain to resistance to kanamycin (5 μg/ml) in order to disrupt ydiO or ydiP as described in Materials and Methods.
As the predicted products YdiO and YdiP are orthologues of DNA methylases and YdiS is an orthologue of DNA restriction enzymes, disruption of the ydiO or ydiP gene may result in defective methylation of the chromosome, leading to enhanced susceptibility to a restriction endonuclease which is predicted to be encoded by ydiS. Disruption of ydiS may result in a defect in the ability to digest the unmethylated chromosome of a ydiO- or ydiP-disrupted strain. The predicted products YdiR and YdjA have no orthologues; however, both of these proteins seem to be required for the function of YdiS, as disruption of either the ydiR gene or the ydjA gene was required for disruption of ydiO and ydiP function. YdiR, YdiS, and YdjA may constitute a restriction endonuclease protein complex that recognizes unmethylated BsuM sites.
Kanamycin-resistant transformation was caused by integration of plasmid pET24b neo carrying ydiO or ydiP at the ydiO or ydiP site but not at the pMUTIN plasmid site in the disruptant, as transformation resulted in few colonies when an unrelated ydhC disruptant with pMUTIN was used as the recipient (data not shown), and pET24b neo itself did not give transformants even in a ydiR-ydiS-ydjA disruptant (data not shown).
Restriction activity of disruptants with exogenous plasmid DNA.
As the restriction and modification system has been found to be a mechanism for defense against foreign phage DNA, disruption of the restriction and modification system also results in an increase in the ability to be transformed by exogenous plasmid DNA (29). As BsuM of B. subtilis has a weak effect on DNAs of phages such as φ105 (34), plasmids having multiple XhoI sites were used to characterize the BsuM system (2). The frequency of transformation with plasmid pHV1401, which had three XhoI sites, was much lower in the wild-type strain, while it was somewhat higher in mutant strains carrying disrupted ydiO, ydiP, ydiR, ydiS, or ydjA. The frequency of transformation with plasmid pHV33, which did not have a XhoI site, was comparable to or somewhat lower than the frequency of transformation with the wild-type recipient (Table 3). The ratio of the frequency of transformation with the plasmid without an XhoI site to the frequency of transformation with the plasmid with three XhoI sites was more than 10 times higher when the ydiR-ydiS-ydjA operon-disrupted strains were used. Plasmid DNAs were isolated from cells of E. coli C600 r+ m+, and they were susceptible to XhoI, indicating that the XhoI sites were not methylated. Disruption of only the ydiR-ydiS-ydjA operon was enough to result in defective digestion of plasmid DNA with three XhoI sites. The ydiO-ydiP function must be very weak, and it did not have a strong effect on incoming XhoI site methylation, as disruption of ydiO or ydiP had no effect on the transformation ratio. However, as described below, enhanced transcription of the ydiO-ydiP operon by increased readthrough from the groESL operon resulted in increased protection of pHV1401.
TABLE 3.
Transformation of the disruptant strains with plasmids carrying multiple XhoI sites
| Expt | Recipient straina | No. of Cmr transformants (102)/μg of DNA with:
|
No. of transformants with pHV1401/ no. of transformants with pHV33 | Ratio | |
|---|---|---|---|---|---|
| pHV33 | pHV1401 | ||||
| 1 | 168 | 226 | 24 | 0.11 | 1.0 |
| BSU1 (ydiO ydiS) | 75 | 324 | 4.32 | 40.7 | |
| BSU2 (ydiP ydiS) | 12 | 110 | 9.17 | 86.3 | |
| BSU3 (ydiR) | 38 | 128 | 3.37 | 31.7 | |
| BSU4 (ydiS) | 20 | 46 | 2.30 | 21.7 | |
| BSU5 (ydjA) | 13 | 54 | 4.17 | 39.1 | |
| 2 | 168 | 134 | 2 | 0.01 | 1.0 |
| BSU9 [Δter (groESL)] | 135 | 82 | 0.61 | 61.0 | |
| 3 | 168 | 1.00 | 0.063 | 0.063 | 1.0 |
| BSU1 (ydiO ydiS) | 4.75 | 4.93 | 1.04 | 16.5 | |
| BSU2 (ydiP ydiS) | 0.38 | 0.38 | 1.00 | 15.8 | |
| BSU3 (ydiR) | 3.50 | 3.87 | 1.11 | 17.6 | |
| BSU4 (ydiS) | 3.12 | 3.94 | 1.26 | 20.0 | |
| BSU5 (ydjA) | 8.75 | 9.69 | 1.11 | 17.6 | |
| BSU9 [Δter (groESL)] | 4.69 | 3.94 | 0.84 | 13.3 | |
Each strain was transformed to resistance to chloramphenicol (5 μg/ml) with plasmid pHV33 or pHV1401. The genotypes in parentheses or brackets indicate the disrupted genes.
Disruption of the smaller ORFs ydiM, ydiN, ydjB, and ydjC in the prophage 3 region did not affect restriction on plasmids (data not shown).
Chromosomal target of the restriction and modification system.
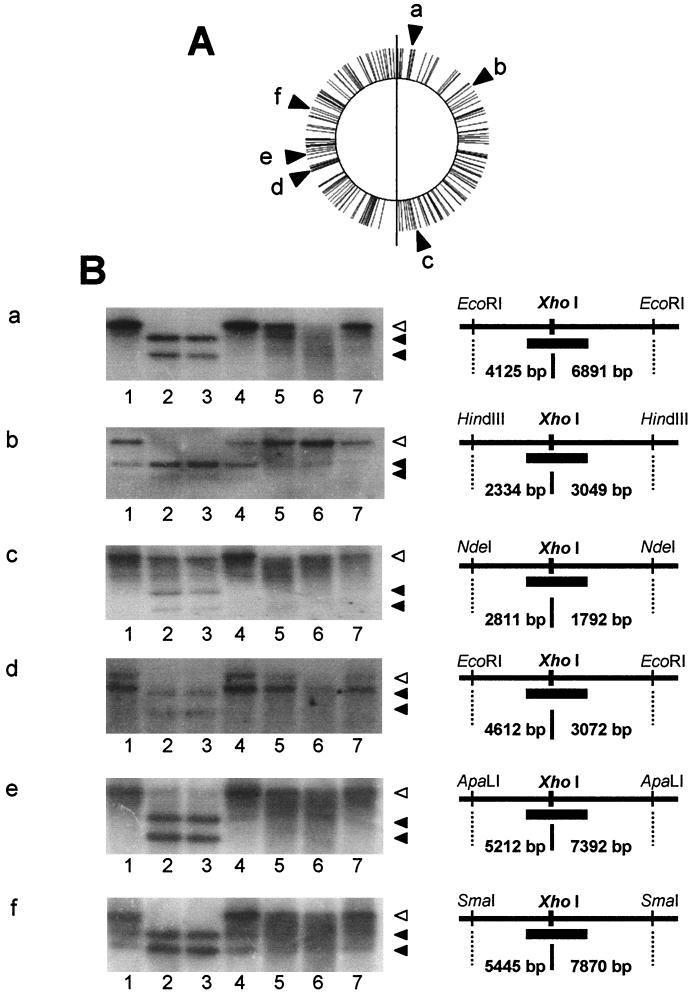
Restriction and modification activity against phage or plasmid DNA was weak (34), but a single disruption of ydiO and ydiP resulted in very tiny colonies on nutrient agar, which indicated that the restriction and modification system works well on the chromosome of B. subtilis. To determine whether this system works on the chromosome, methylation of six chromosomal restriction fragments carrying only one XhoI target sequence (17) was examined indirectly by digesting chromosomal DNAs with XhoI. Susceptibility to the XhoI enzyme was evaluated by Southern blot analysis.
As shown in Fig. 2B, gels a and e, at nucleotide positions 106975 (9°) and 3105700 (265°) DNA from the wild-type strain, the ydiP ydiS doubly disrupted strain, or the single disruptant with a disruption in the ydiR-ydiS-ydjA operon was not susceptible to XhoI digestion, while DNAs from RM125 and the ydiO ydiS doubly disrupted strain were susceptible. Thus, at least YdiO must be active in methylation of the XhoI target sequence, since the ydiO ydiS strain might have lost both YdiO and YdiP activities due to the probable polar effect of disruption by the pET24 neo plasmid without a Pspac promoter and the ydiP ydiS strain lost only YdiP activity. At nucleotide position 015718 (172°) (Fig. 2B, gel c), the results seem to be similar to those shown in Fig. 2B, gels a and e, although the XhoI site on this fragment was somewhat refractory to digestion.
FIG. 2.
Southern blot analysis of susceptibility to XhoI digestion of chromosomal DNAs extracted from disruptants. Chromosomal DNA was extracted from each disruptant, subjected to double digestion with XhoI and an appropriate restriction nuclease, separated by agarose electrophoresis, transferred to a membrane filter, and hybridized with alkaline phosphatase-labeled probe DNA (thick bars in the diagrams in the right portion of panel B). All XhoI sites in the chromosome of strain Marburg 168 are indicated by bars in panel A. The results of a Southern analysis for six XhoI sites on the chromosomes are shown in panel B. Lane 1, 168 DNA; lane 2, RM125 DNA; lane 3, BSU1 DNA; lane 4, BSU2 DNA; lane 5, BSU3 DNA; lane 6, BSU4 DNA; lane 7, BSU5 DNA. The positions of XhoI sites on the chromosome are as follows: gel a, nucleotide 106975 (9°, AGGCTCGAGTAT); gel b, nucleotide 2633916 (54°, GCGCTCGAGACA); gel c, nucleotide 015718 (172°, GCGCTCGAGCAG); gel d, nucleotide 2800360 (239°, TCACTCGAGATT); gel e, nucleotide 3105700 (265°, CTTCTCGAAGAT); and gel f, nucleotide 3550939 (303°, TTTCTCGAGCTT). The open arrowheads indicate the position of an undigested fragment. The solid arrowheads indicate the positions of digested fragments. The configuration of the restriction sites and the probe binding site for each blot is also shown in panel B.
At nucleotide position 2633916 (54°) (Fig. 2B, gel b), the XhoI enzyme did not digest chromosomal DNAs from the wild-type strain and strains with disruptions in ydiR, ydiS, or ydjA, while DNAs from classical restriction and modification system-deficient strain RM125 and ydiO ydiS doubly disrupted strains were digested. DNA from the ydiP ydiS doubly disrupted strain was 50% digested. These results suggest that YdiO and YdiP are both active methylases.
For nucleotide position 2800360 (239°) (Fig. 2B, gel d), the results were similar to those shown in Fig. 2B, gels a and e; however, there must be an unknown XhoI site very close to the XhoI site identified. The former site must be somehow refractory to methylation with active YdiO and YdiP and thus is susceptible to XhoI even when active YdiO and YdiP are present. Without YdiO and YdiP or in RM125, the XhoI site identified was more susceptible to digestion than the unknown site. At nucleotide position 3550939 (303°) (Fig. 2B, gel f), the XhoI site must be partially unmethylated, because DNAs from strains other than RM125 and ydiO ydiS strains were partially digested.
Transcription of the prophage 3 region.
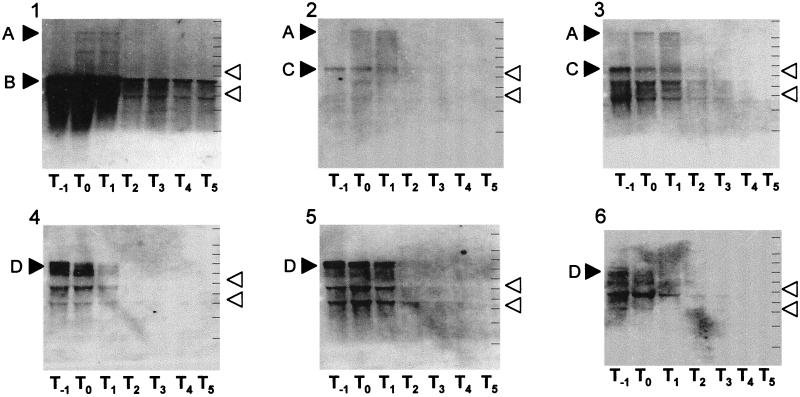
To observe transcription of the five ORFs, RNAs were extracted from cells grown at 37°C in sporulation medium. Northern analysis with five RNA probes for each of the five ORFs revealed two transcripts, which were 3 and 4 kb long (Fig. 3, bands C and D). The 3-kb transcript was detected by two probes for ydiO and ydiP, while the 4-kb transcript was detected by three probes for ydiR, ydiS, and ydjA. Both transcripts were detected in cells in the logarithmic phase of growth. They were not detected in cells collected during sporulation. Therefore, ydiO and ydiP constitute one operon, and ydiR, ydiS and ydjA constitute the other operon. We identified a very long 7.6-kb transcript (Fig. 3, band A) for the groESL operon that included the groESL and ydiO-ydiP operons, as well as a shorter 2.2-kb transcript (Fig. 3, band B).
FIG. 3.
Northern blot analysis of transcripts of the prophage 3 region. RNAs were prepared from cells grown in sporulation medium at 37°C, separated by electrophoresis, transferred to nitrocellulose membranes, and hybridized with DIG-labeled probe RNA. The RNA samples used were 20-μg samples. The open arrowheads indicate the positions of 16S rRNA (1.55 kb) and 23S rRNA (2.93 kb). The solid arrowheads indicate the positions of the transcripts detected, as follows: band A, 7.6 kb; band B, 2.2 kb; band C, 2.9 kb; and band D, 4.2 kb. Panel 1, groEL probe; panel 2, ydiO probe; panel 3, ydiP probe,; panel 4, ydiR probe; panel 5, ydiS probe; panel 6, ydjA probe. The lines on the right in each membrane indicate the positions of molecular weight markers (9, 6, 5, 4, 3, 2.5, 2, 1.5, 1, and 0.5 kb). The Tn values below each membrane indicate the numbers of hours (n) after the end of the logarithmic phase of growth.
Disruption of the terminal signal sequence for transcription of the groESL operon.
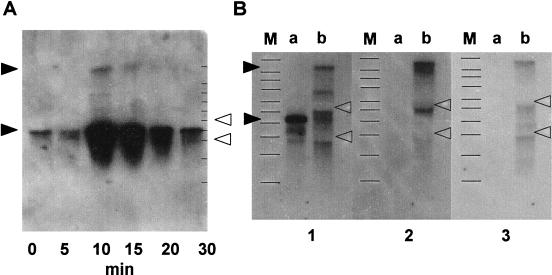
The long transcript must be a readthrough transcript of the groESL operon as heat treatment induced this transcript, as well as that of the groESL operon (27) (Fig. 4A). In order to determine the effect of readthrough from the groESL operon on ydiO-ydiP operon expression, we disrupted the terminal signal sequence for transcription of the groESL operon. The disrupted strain showed elevated readthrough from the groESL operon to the ydiO-ydiP operon (Fig. 4B). Enforced readthrough clearly revealed a 3.8-kb transcript, indicating that there is a transcription termination signal sequence between groESL and ydiO. The disrupted strain became permissive to plasmid transformation, carrying multiple XhoI sites that led to easy infection of unmethylated exogenous DNA (Table 2). This strain grew well in minimal glucose medium or sporulation medium and sporulated well (data not shown).
FIG. 4.
Heat induction and artificial derivation of readthrough transcript of groESL operon. (A) Wild-type cells grown at 37°C in Luria-Bertani broth to the early logarithmic phase were transferred to 48°C. At each of the times indicated, a portion of the culture was removed and RNA was prepared for Northern blot analysis. A DIG-labeled probe for groEL was used. Each lane contained 0.3 μg of RNA. The open arrowheads indicate the positions of 16S rRNA (1.55 kb) and 23S rRNA (2.93 kb). The lines on the right indicate the positions of the molecular weight markers described in the legend to Fig. 3. The solid arrowheads indicate the positions of 7.6- and 2.2-kb transcripts. (B) Cells of wild-type strain 168 or mutant strain BSU9, which has a deletion in the rho-independent transcription termination signal sequence of groESL, were grown at 37°C in sporulation medium to the end of logarithmic phase. RNA was prepared and subjected to Northern analysis. Each lane contained 5 μg of RNA. The open arrowheads indicate the positions of 16S rRNA (1.55 kb) and 23S rRNA (2.93 kb). The solid arrowheads indicate the positions of 7.6- and 2.2-kb transcripts. In panel 1, the DIG probe was groEL, lane a contained wild-type strain 168, and lane b contained a Δter(groESL) disruptant. In panel 2, the DIG probe was ydiO, lane a contained wild-type strain 168, and lane b contained a Δter(groESL) disruptant. In panel 3, the DIG probe was ydiP, lane a contained wild-type strain 168, and lane b contained a Δter(groESL) disruptant. Lanes M contained molecular weight markers.
Classical restriction and modification system-deficient mutant RM125 has an altered prophage 3 region.
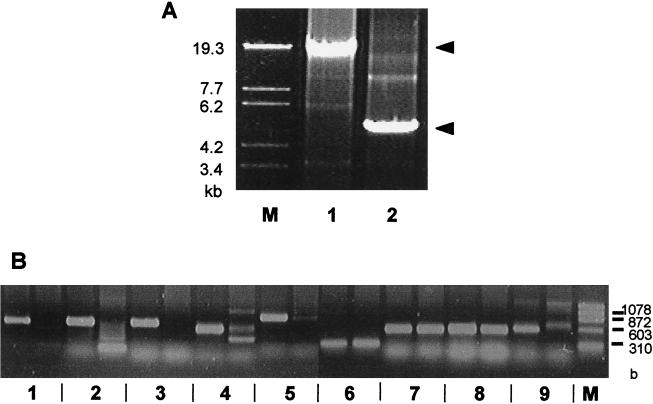
PCR DNA synthesis with the groEL and gutB primer pairs resulted in a fragment that was 15.7 kb long when wild-type chromosomal DNA was used as the template and in a fragment that was 5 kb long when the template DNA was from RM125 (34) (Fig. 5A). To determine whether the five ORFs of the prophage 3 region are in a 5-kb region in RM125, we performed PCR DNA synthesis with the primer pair for each ORF using chromosomal DNA from the wild-type strain or RM125. As shown in Fig. 5B, when the wild-type strain DNA was used, the PCR synthesized DNA of each ORF, while when RM125 DNA was used, no DNA was synthesized, although with both wild-type strain DNA and strain RM125 DNA PCR synthesized DNA of groES and groEL (27), as well as DNA of gutR and gutB (38). Therefore, the original B. subtilis-related strain from which the substituted chromosome was isolated, strain 202-5 (= IAM1169, a B. amyloliquefaciens strain), has no genes similar to those of BsuM found in the prophage 3 region of B. subtilis Marburg 168 and may have not been infected with the phage carrying BsuM system genes. In short, RM125 has a 1.9-kb insert instead of the 13-kb prophage 3 region. We did not analyze the substituted fragment further.
FIG. 5.
PCR analysis of prophage 3 region of the RM125 chromosome. (A) Long accurate PCR products obtained with groEL and gutB primers. Lane1, PCR with 168 DNA; lane 2, PCR with RM125 DNA. Lane M contained molecular weight markers. (B) PCR products obtained with primer pairs for each ORF in the prophage 3 region. For each pair of lanes the left lane contained a PCR mixture with strain 168 DNA, and the right lane contained a PCR mixture with strain RM125 DNA. Lanes 1, primer pair for ydiO; lanes 2, primer pair for ydiP; lanes 3, primer pair for ydiR; lanes 4, primer pair for ydiS; lanes 5, primer pair for ydjA; lanes 6, primer pair for groES; lanes 7, primer pair for groEL; lanes 8, primer pair for gutR; lanes 9, primer pair for gutB. Lane M contained molecular weight markers.
Effect of induction of the ydiR-ydiS-ydjA operon on cell growth.
To determine the balance between chromosome methylation and restriction, we placed the ydiR-ydiS-ydjA operon under the Pspac promoter and induced the operon. Addition of IPTG arrested the growth of the cells of only the ydiO or ydiP disruptant. The viability of the cells was lost rapidly (within 60 min). The concentration decreased from 732 × 106 to less than 1 × 106 CFU/ml for BSU7 and from 456 × 106 to 3 × 106 CFU/ml for BSU8. Even after addition of IPTG, growth was not arrested when the ydiO-ydiP operon was active (data not shown). The ydiO disruptant might have defective expression of both ydiO and ydiP, and the ydiP disruptant might have defective expression of only ydiP. This indicates that even when the ydiR-ydiS-ydjA operon is overexpressed, the presumed YdiS nuclease complex does not digest chromosomal DNA if there is expression of the ydiO or ydiP methylase gene.
DISCUSSION
As described above, the classical B. subtilis Marburg mutant strain RM125 (34) lacks the 13-kb prophage 3 region, which contains the genes of the intrinsic BsuM restriction-modification system of this organism. The classical hsrM1 and nonB (24, 36) mutations were not mapped in this study, but they may be mutations in the ydiR-ydiS-ydjA operon as there are no paralogue genes on the B. subtilis Marburg chromosome (17).
Instead, the RM125 strain carried a 1.9-kb chromosomal fragment derived from B. subtilis-related strain 202-5 (= IAM1169, a B. amyloliquefaciens strain), which did not have the restriction-modification system genes of B. subtilis Marburg BsuM. On the other hand, five restriction-modification systems have been found in B. subtilis Marburg-related bacteria (32). These systems have been introduced into the B. subtilis Marburg chromosome by transformation (10, 11, 12, 33). They are BsuB, BsuC, BsuE, BsuF, and BsuR, and BsuC, BsuF, and BsuR are allelic. BsuB, BsuF, and BsuR are known to have one restriction gene and one modification gene. These restriction-modification system genes must be introduced into each B. subtilis Marburg-related strain by horizontal transfer, such as E. coli P4 phage-mediated transfer of the EcoO109I restriction-modification system (16), or by transfer with plasmids such as Lactococcus lactis plasmid pTR2030 (20), which carries an LlaI restriction-modification system operon composed of llaIM, llaIC, llaI-1, llaI-2, llaI-3, and llaI-4. LlaIM is a DNA methyltransferase, and LlaI-2 is a homologue of the YdiS protein described above.
Chromosomal DNA from the ydiO-disrupted strain, which may have a defective ydiP gene due to a probable polar effect, was susceptible to XhoI digestion, suggesting that MtbP (17), a prophage SPβ-encoded modification methylase, recognizes a target sequence other than the XhoI site if the mtbP gene is active in the ydiO disruptant. Parental strain 168 harbors SPβ and the metbP gene.
YdiO and YdiP have many common conserved motif sequences (14, 21), suggesting that these genes had the same origin. However, YdiP has a higher level of similarity to MtbP than to YdiO, suggesting that YdiP may have originated from MtbP. Some B. subtilis phages carry a tandem pair of methyltransferase genes (reviewed in reference 32), and the L. lactis plasmid pTR2030 described above carries an operon consisting of a regulator gene, a methyltransferase gene, a restriction endonuclease gene, and other genes of unknown function that are necessary for endonuclease activity (20). It should be noted that YdiS and YdjA exhibit some homology with LlaI-2 and LlaI-3, respectively. Thus, it is not surprising that BsuM is composed of two operons, one operon consisting of duplicated methyltransferase genes and the other consisting of an endonuclease gene and its associated genes. Helicobacter pylori has multiple restriction-modification system genes (31); jhp0164 and jhp0165 are located in tandem and are homologous to ydiS and ydjA, respectively, and jhp0435, which is located far from jhp0164 and jhp0165, is homologous to ydiO. E. coli mcrB is homologous to ydiS and constitutes an operon with mcrC (3, 23); its product is presumed to be similar to YdjA.
Target sequence CTCGAG of BsuM is symmetrical. Predicted methyltransferases YdiO and YdiP seem to be active, and they are not identical. It is not known how a symmetrical target is recognized by two homologous but nonidentical methyltransferase genes. Determination of the site of methylation will require biochemical study with target DNA and purified YdiO and YdiP proteins, as well as with YdiR, YdiS, and YdjA proteins, which may form a restriction nuclease complex.
Transcriptional analysis of the restriction-modification system genes revealed that the ydiO-ydiP operon can be transcribed by readthrough from the groESL operon (27) upon heating. Heat treatment of B. subtilis Marburg conferred permissiveness to phage SP10 infection (9). This might be because of enhanced methyltransferase activity due to heat induction of the ydiO-ydiP operon. Disruption of a transcription termination signal sequence resulted in enhanced synthesis of long transcripts, which led to permissive infection of plasmid DNA with multiple XhoI sites. This clearly indicates that methylation of unmethylated incoming DNA is enhanced to protect it from attack by a restriction endonuclease.
The genes controlling the restriction-modification system genes, such as the C gene (30), are not found in the prophage 3 region, but there are some small genes (ydiM, ydiN, ydiQ, ydjB, and ydjC), and these small genes might be regulatory genes. However, they seem not to contribute to restriction-modification activity, as disruptants with disruptions in these genes showed wild-type transformation by plasmid DNAs.
Pseudogenes (14) are reminiscent of phage and exhibit homology to genes such as the integrase gene of bacteriophage T270 of Streptococcus pyogenes and the small subunit of the terminase gene of bacteriophage LL-H (19). There is another pseudogene of B. subtilis encoding the YhxB paralogue (17), a glycolysis phosphomannomutase, on the strand opposite ydiQ. B. subtilis Marburg harbors 10 prophages or regions reminiscent of phages. The prophage 3 region is the smallest, has the smallest number of ORFs, and has the lowest gene density. Therefore, the prophage 3 region may be the oldest region, where restriction-modification genes remain undestroyed. It may be reasonable to assume that the prophage 3 region, which has a lower G+C content, was integrated into the B. subtilis chromosome with a lot of active genes and lost most of them by mutational pressure. Only five ORFs coding for restriction and modification enzymes remain active. Inactivation of genes for restriction makes a cell permissive to invasion by foreign DNA, while inactivation of genes for modification leads to autodigestion of the chromosome by the remaining restriction activity. Thus, a set of restriction-modification system genes survived in the prophage 3 region of the B. subtilis chromosome.
Acknowledgments
We are grateful to R. H. Doi for critically reading the manuscript and to M. Itaya for providing bacterial strains and plasmids.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bron, S., L. Janniere, and S. D. Ehrlich. 1988. Restriction and modification in Bacillus subtilis Marburg 168: target sites and effects on plasmid transformation. Mol. Gen. Genet. 211:186–189. [DOI] [PubMed] [Google Scholar]
- 3.Dila, D., E. Sutherland, L. Moran, B. Slatko, and E. A. Raleigh. 1990. Genetic and sequence organization of the mcrBC locus of Escherichia coli K-12. J. Bacteriol. 172:4888–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duesterhoeft, A., and M. Kroeger. 1991. Cloning, sequence and characterization of m5C-methyltransferase-encoding gene, hgiDIIM (GTCGAC), from Herpetosiphon giganteus strain Hpa 2. Gene 106:87–92. [DOI] [PubMed] [Google Scholar]
- 5.Fujita, M., and Y. Sadaie. 1998. Rapid isolation of RNA polymerase from sporulating cells of Bacillus subtilis. Gene 221:185–190. [DOI] [PubMed] [Google Scholar]
- 6.Groth, D., R. Reszka, and J. A. Schenk. 1996. Polyethylene glycol-mediated transformation of Escherichia coli is increased by room temperature. Anal. Biochem. 240:302–304. [DOI] [PubMed] [Google Scholar]
- 7.Guha, S. 1985. Determination of DNA sequences containing methylcytosine in Bacillus subtilis Marburg. J. Bacteriol. 163:573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guha, S. 1988. DNA methyltransferase of Bacillus subtilis Marburg: purification properties and further evidence of specificity. Gene 74:77–81. [DOI] [PubMed] [Google Scholar]
- 9.Gwinn, D. D., and W. D. Lawton. 1968. Alteration of host specificity in Bacillus subtilis. Bacteriol. Rev. 32:297–301. [PMC free article] [PubMed] [Google Scholar]
- 10.Ikawa, S., T. Shibata, and T. Ando. 1979. Host-controlled modification and restriction in Bacillus subtilis. Bsu168-system and BsuR-system in B. subtilis 168. Mol. Gen. Genet. 170:123–127. [DOI] [PubMed] [Google Scholar]
- 11.Ikawa, S., T. Shibata, T. Ando, and H. Saito. 1980. Genetic studies on site-specific endodeoxyribonucleases in Bacillus subtilis: multiple modification and restriction systems in transformants of Bacillus subtilis 168. Mol. Gen. Genet. 177:359–368. [DOI] [PubMed] [Google Scholar]
- 12.Ikawa, S., T. Shibata, K. Matsumoto, T. Iijima, H. Saito, and T. Ando. 1981. Chromosomal loci of genes controlling site specific restriction endonucleases of Bacillus subtilis. Mol. Gen. Genet. 183:1–6. [DOI] [PubMed] [Google Scholar]
- 13.Jentsch, S. 1983. Restriction and modification in Bacillus subtilis: sequence specificities of restriction/modification systems BsuM, BsuE, and BsuF. J. Bacteriol. 156:800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasahara, Y., S. Nakai, N. Ogasawara, K. Yata, and Y. Sadaie. 1997. Sequence analysis of the groESL-cotA region of the Bacillus subtilis genome, containing the restriction/modification system genes. DNA Res. 4:335–339. [DOI] [PubMed] [Google Scholar]
- 15.Kawamura, F., H. Saito, and Y. Ikeda. 1980. Bacteriophage phi 1 as a gene-cloning vector in Bacillus subtilis. Mol. Gen. Genet. 180:259–266. [DOI] [PubMed] [Google Scholar]
- 16.Kita, K., J. Tsuda, T. Kato, K. Okamoto, H. Yanase, and M. Tanaka. 1999. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol. 181:6822–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst, F., N. Ogasawara, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. [DOI] [PubMed] [Google Scholar]
- 18.Lin, P. M., C. H. Lee, and R. J. Roberts. 1989. Cloning and characterization of the genes encoding the MspI restriction modification system. Nucleic Acids Res. 17:3001–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikkonen, M., and T. Alatossava. 1995. A group I intron in the terminase gene of Lactobacillus delbrueckii subsp. lactis phage LL-H. Microbiology 141:2183–2190. [DOI] [PubMed] [Google Scholar]
- 20.O’Sullivan, D. J., K. Zagula, and T. R. Klaenhammer. 1995. In vivo restriction by LlaI is encoded by three genes, arranged in an operon with llaIM, on the conjugative Lactococcus plasmid pTR2030. J. Bacteriol. 177:134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posfai, J., A. S. Bhagwat, G. Posfai, and R. J. Roberts. 1989. Predictive motif derived from cytosine methyltransferases. Nucleic Acids Res. 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price, C. W., M. A. Gitt, and R. H. Doi. 1983. Isolation and physical mapping of the gene encoding the major sigma factor of Bacillus subtilis RNA polymerase. Proc. Natl. Acad. Sci. USA 80:4047–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raleigh, E. A. 1992. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol. Microbiol. 6:1079–1086. [DOI] [PubMed] [Google Scholar]
- 24.Saito, H., T. Shibata, and T. Ando. 1977. Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol. Gen. Genet. 170:117–122. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schaeffer, P., J. Millet, and, J. P. Aubert. 1965. Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt, A., M. Schiesswohl, U. Volker, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, mapping, and transcriptional analysis of the groESL operon from Bacillus subtilis. J. Bacteriol. 174:3993–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata, T., and T. Ando. 1974. Host controlled modification and restriction in Bacillus subtilis. Mol. Gene. Genet. 131:275–280. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, T. 1979. Restriction of plasmid-mediated transformation in Bacillus subtilis 168. Mol. Gen. Genet. 175:235–237. [DOI] [PubMed] [Google Scholar]
- 30.Tao, T., J. C. Bourne, and R. M. Blumenthal. 1991. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 173:1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomb, J.-F., et al. 1979. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. [DOI] [PubMed] [Google Scholar]
- 32.Trautner, T. A., and M. Noyer-Weidner. 1993. Restriction/modification and methylation systems in Bacillus subtilis, related species, and their phages, p.539–552. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 33.Trautner, T. A., B. Pawick, S. Bron, and C. Anagnostopoulos. 1974. Restriction and modification in B. subtilis, biological aspects. Mol. Gen. Genet. 131:181–191. [DOI] [PubMed] [Google Scholar]
- 34.Uozumi, T., T. Hoshino, K. Miwa, S. Horinouchi, T. Beppu, and K. Arima. 1977. Restriction and modification in Bacillus species. Genetic transformation of bacteria with DNA from different species. Part I. Mol. Gen. Genet. 152:525–538. [DOI] [PubMed] [Google Scholar]
- 35.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097–3104. [DOI] [PubMed] [Google Scholar]
- 36.Yajima, Y., H. Saito, and Y. Ikeda. 1979. Mechanisms of nonpermissiveness in abortive infection of bacteriophage NR2 in Bacillus subtilis Marburg strain. J. Gen. Appl. Microbiol. (Tokyo) 26:291–298. [Google Scholar]
- 37.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the m13mp18 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]
- 38.Ye, R., S. N. Rehentulla, and S. L. Wong. 1994. Glucitol induction in Bacillus subtilis is mediated by a regulatory factor, GutR. J. Bacteriol. 176:3321–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida, K., I. Ishino, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573–579. [DOI] [PubMed] [Google Scholar]
- 40.Young, R. A., and R. W. Davis. 1983. Efficient isolation of genes by using antibody probes. Proc. Natl. Acad. Sci. USA 80:1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan, G., and S. L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the ςA-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]