Abstract
Low concentrations of the RNA polymerase inhibitor rifampin added to an exponentially growing culture of Bacillus subtilis led to an instant inhibition of growth. Survival experiments revealed that during the growth arrest the cells became tolerant to the antibiotic and the culture was able to resume growth some time after rifampin treatment. l-[35S]methionine pulse-labeled protein extracts were separated by two-dimensional polyacrylamide gel electrophoresis to investigate the change in the protein synthesis pattern in response to rifampin. The ςB-dependent general stress proteins were found to be induced after treatment with the antibiotic. Part of the oxidative stress signature was induced as indicated by the catalase KatA and MrgA. The target protein of rifampin, the β subunit (RpoB) of the DNA-dependent RNA polymerase, and the flagellin protein Hag belonging to the ςD regulon were also induced. The rifampin-triggered growth arrest was extended in a sigB mutant in comparison to the wild-type strain, and the higher the concentration, the more pronounced this effect was. Activity of the RsbP energy-signaling phosphatase in the ςB signal transduction network was also important for this protection against rifampin, but the RsbU environmental signaling phosphatase was not required. The sigB mutant strain was less capable of growing on rifampin-containing agar plates. When plated from a culture that had already reached stationary phase without previous exposure to the antibiotic during growth, the survival rate of the wild type exceeded that of the sigB mutant by a factor of 100. We conclude that the general stress response of B. subtilis is induced by rifampin depending on RsbP activity and that loss of SigB function causes increased sensitivity to the antibiotic.
The natural habitat of the gram-positive bacterium Bacillus subtilis is the soil—an environment in which stress and starvation are the rule. Not surprisingly B. subtilis has evolved a range of strategies to survive under these hostile conditions. The formation of stress-resistant spores, for example, guarantees long-term survival, while uptake of external DNA in the process of genetic competence may contribute to adaptation by recombination (12). Besides these time-consuming processes B. subtilis is able to quickly develop short-term protection of nongrowing cells against multiple stress conditions, including acidic, alkaline, osmotic, or oxidative stress, heat, or ethanol. A global regulator, the alternative sigma factor ςB, controls this general stress response. The ςB regulon contains about 150 genes, some of which have been shown to actively protect or repair macromolecules (see references 12 and 18 for reviews). Under laboratory conditions ςB is activated by two different groups of stimuli: physical stresses like heat, acid, or ethanol act via the RsbU-dependent pathway, while ςB activation after glucose or phosphate starvation is RsbP dependent (20, 21, 24).
An alternative sigma factor homologous to ςB of B. subtilis also exists in Mycobacterium tuberculosis and is named ςF (8). Small amounts of rifampin, a semisynthetic antibiotic of the rifamycin group (6), were shown to induce ςF of this pathogen (15). Rifampin blocks the β subunit of prokaryotic DNA-dependent RNA polymerase (5, 11) and by this mechanism inhibits RNA synthesis at an early stage of elongation.
Recently we started to investigate changes in the protein synthesis pattern of B. subtilis in response to various antibiotics. The obtained proteome signatures (19) might be used to predict the mode of action of new drugs. This study demonstrates that in B. subtilis the ςB-dependent general stress response is induced when cells are exposed to rifampin. Furthermore it is shown that the ςB response is involved in overcoming the rifampin-caused growth arrest.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used were B. subtilis wild-type 168 (trpC2) (1), sigB mutant ML6 (trpC2 sigB::ΔHindIII-EcoRV::cat) (13), B. subtilis wild-type BR16 (trpC2 lys) (10), rsbP mutant BCE17 (trpC2 lys yvfP::spec) (Specr) (10), and rsbU mutant BCE12 (trpC2 lys rsbU::aphA3) (Kmr) (10). B. subtilis strains were cultivated under vigorous agitation at 37°C in a synthetic medium described previously (2). For growth experiments with rifampin the antibiotic was added directly to the exponentially growing culture at an optical density at 500 nm (OD500) of 0.4 at a concentration of 0.03, 0.06, 0.12, or 0.24 μg/ml respectively (MIC = 0.06 μg/ml).
Survival experiments were carried out to investigate the acquisition of tolerance to rifampin. Dilutions of cultures were plated on rifampin-containing LB agar (Lennox L agar) plates (0.06 μg/ml). The number of surviving cells (CFU) was monitored along with the growth curve (OD500) after treatment with the antibiotic. A control culture was left untreated. The agar plates were incubated for 20 h at 37°C.
Preparation of the cytoplasmic l-[35S]methionine-labeled protein fraction.
Cells were labeled with l-[35S]methionine (10 μCi/ml) for 5 min at different time points after treatment with rifampin, as were untreated control cells at an OD500 of 0.4. l-[35S]methionine incorporation was stopped by the addition of chloramphenicol (1 mg/ml) and an excess of unlabeled l-methionine (10 mM), as well as by transferring the culture onto ice. Cells were disrupted by ultrasonic treatment, and the soluble cell fraction containing the soluble protein fraction was separated from insoluble cell remnants by centrifugation.
Analytical and preparative 2D PAGE.
Analytical two-dimensional polyacrylamide gel electrophoresis (2D PAGE) was performed using the immobilized pH gradient technique as described previously (4). Aliquots (50 μg) of crude protein extract were loaded on immobilized pH gradient strips (Amersham Pharmacia Biotech, Piscataway, N.J.) covering the pH range of 4 to 7. The gels were stained with silver nitrate using the protocol described by Bernhardt et al. (4). Afterwards gels were dried on filter paper. Exposure to Phosphor Screens (Molecular Dynamics) was followed by detection with the PhosphorImager SI (Molecular Dynamics). For identification of the proteins by mass spectrometry protein samples of 500 μg were separated by preparative 2D PAGE and the gels were stained with Sypro Ruby (Molecular Probes, Eugene, Oreg.).
Peptide mass fingerprinting.
In-gel digestion with trypsin (Promega, Madison, Wis.) was performed using a peptide collection device (16). Peptide solution (0.05 μl) was prepared with an equal volume of saturated α-cyano-4-hydroxy cinnamic acid solution in 50% acetonitrile-0.1% trifluoroacetic acid (vol/vol) and applied to a sample template of a matrix-assisted laser desorption ionization-time of flight mass spectrometer (Voyager DE-STR; PerSeptive Biosystems). Peptide mass fingerprints were analyzed using MS-Fit software (P. R. Baker and K. R. Clauser [http://prospector.ucsf.edu]).
Measuring incorporation of l-[35S]methionine into protein.
Aliquots of protein extracts were blotted on filter paper and precipitated with 10% trichloroacetic acid on ice. After washing twice with 5% trichloroacetic acid and once with 96% ethanol, the amount of incorporated l-[35S]methionine was measured in toluene scintillation cocktail in a Liquid Scintillation Counter Tri-Carb 1600TR (Packard).
Analysis of transcription.
The acidic phenol method (14) was used to isolate total RNA of B. subtilis 168. Samples were harvested 4 and 55 min after the addition of rifampin, while a control was taken at an OD500 of 0.4. Northern blotting was performed as described previously (23). For hybridization digoxigenin-labeled RNA probes specific for sigB (22), gspA (3), and tufA (C. Eymann, unpublished data) were synthesized in vitro with T7 RNA polymerase from T7 promoter-containing internal PCR products of the respective gene.
RESULTS
Induction of ςB-dependent general stress genes by rifampin.
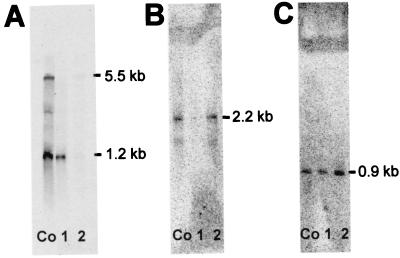
In M. tuberculosis the sigma factor ςF, which controls the general stress response, was shown to be induced in reaction to rifampin (15). The homologous alternative sigma factor in B. subtilis is ςB. To determine whether the ςB-dependent general stress response is induced on the transcriptional level in response to rifampin, Northern blot analyses for three genes were performed: the gene encoding the global regulator sigB itself, a solely ςB-dependent gene gspA, and, as a marker for exponential growth, the gene tufA, which encodes the elongation factor Tu (Fig. 1). Cells were harvested before as well as 4 and 55 min after addition of the antibiotic at a final concentration of half the MIC.
FIG. 1.
Northern blot analysis of B. subtilis 168 extracts of control cells (lanes Co) and cultures challenged with rifampin for 4 (lanes 1) and 55 (lanes 2) min were performed with the digoxigenin-labeled RNA probes tufA (1.2 kb, tufA monocistronic; 5.5 kb, ybxF-rpsL-rpsG-fusA-tufA transcript) (A), sigB (2.2 kb, rsbV-rsbW-sigB-rsbX transcript) (B), and gspA (0.9 kb, gspA monocistronic) (C).
While tufA mRNA was detectable in large amounts under control conditions, its level decreased rapidly after antibiotic treatment and was still low after 55 min, when the culture was still growth arrested.
In contrast, sigB and gspA mRNA levels were low in the control and that of sigB even lower 4 min after rifampin application, indicating mRNA degradation. But during the growth arrest at 55 min the sigB mRNA level returned to the level measured before rifampin application and that of gspA was even higher, indicating induction of this ςB-dependent gene.
ςB is involved in the resumption of growth.
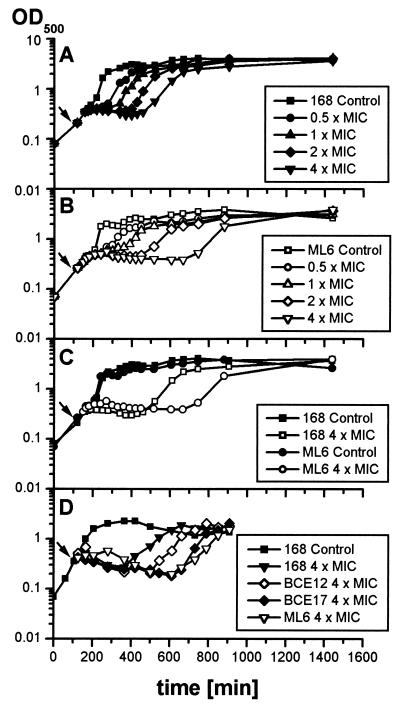
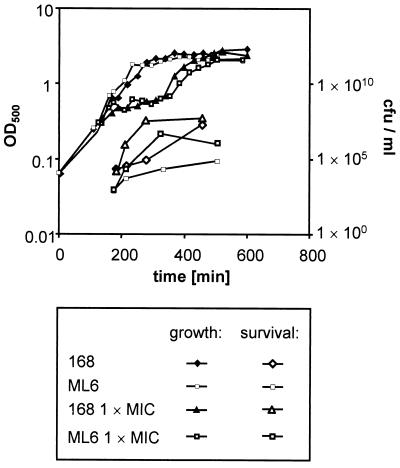
To investigate the role of the alternative sigma factor ςB in the tolerance of B. subtilis to rifampin, growth experiments with the wild type 168 (Fig. 2A) and the sigB mutant ML6 (Fig. 2B) were carried out. The application of rifampin at 0.03 to 0.24 μg/ml (0.5 to 4 times the MIC) to exponentially growing cultures instantly stopped cell growth of both strains. The cultures recovered from antibiotic treatment in a concentration-dependent manner and resumed growth. The higher the rifampin concentration applied was, the longer was the period of blocked growth (also described in reference 7). The sigB mutant culture was also able to overcome the growth arrest caused by the antibiotic, though the time from treatment with rifampin to the resumption of growth exceeded that of the wild type, and this difference became more accentuated at higher rifampin concentrations (Fig. 2C).
FIG. 2.
B. subtilis 168 (A), ML6 (sigB) (B), BCE12 (rsbU) (D), and BCE17 (rsbP) (D) were grown in synthetic medium to an OD500 of 0.4. The arrow indicates the time of addition of different concentrations of rifampin to the culture; the control was left untreated. (C) Comparison of the curves of B. subtilis 168 and ML6 treated with 0.24 μg of rifampin and untreated controls in one diagram.
An rsbP (BCE17) and an rsbU (BCE12) mutant strain were tested to identify the pathway by which ςB is activated under these conditions. While the rsbU mutant resumed growth in a time scale intermediate between the wild type and the sigB mutant, the rsbP mutant, like ML6, resumed growth with a distinct delay (Fig. 2D), indicating that the activation is RsbP dependent.
Pulse-labeling experiments along the growth curve were followed by 2D PAGE.
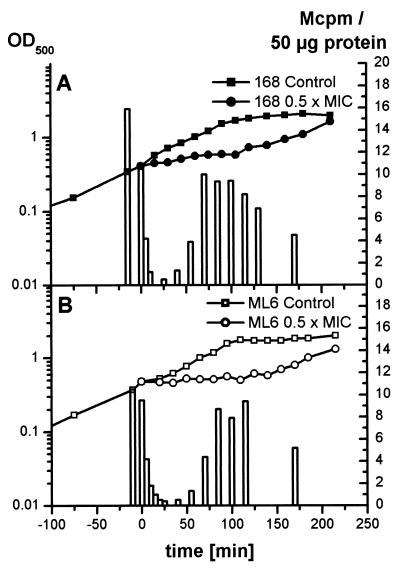
Changes in the protein synthesis rate were investigated by pulse-labeling experiments with l-[35S]methionine. At 10 min after the addition of rifampin at 0.03 μg/ml to an exponentially growing culture, the amount of l-[35S]methionine incorporation during 5 min of pulse-labeling was drastically reduced (Fig. 3A). Long before the resumption of growth, the incorporation of l-[35S]methionine began to increase.
FIG. 3.
The incorporation of l-[35S]methionine into 50 μg of B. subtilis 168 (A) and ML6 (sigB) (B) protein during 5 min of pulse-labeling was monitored parallel to the growth curve. The control sample was labeled immediately before rifampin application. Rifampin (0.03 μg/ml) was added at time zero.
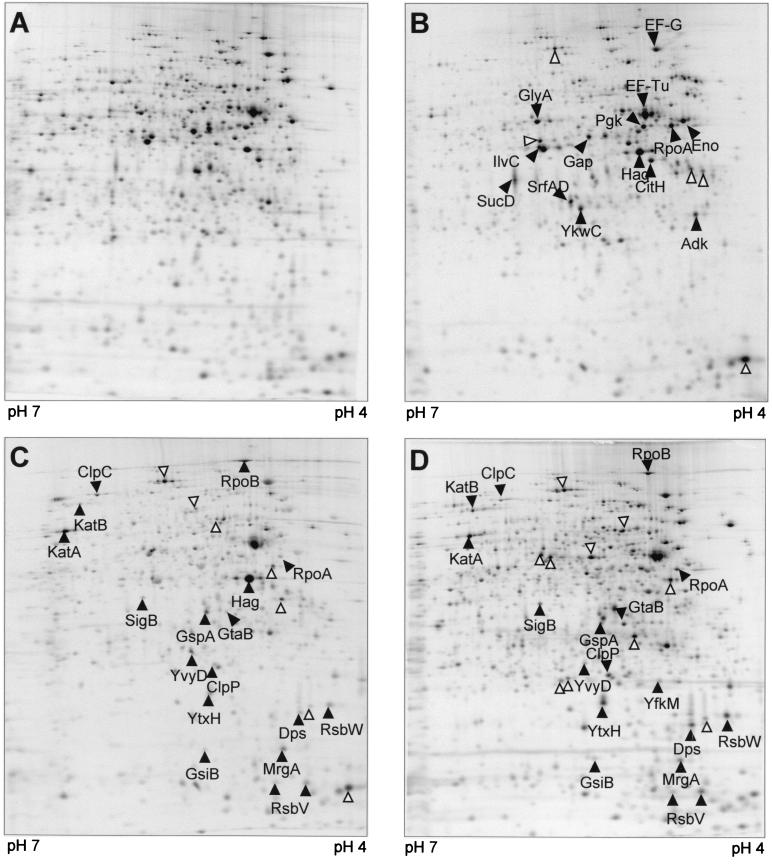
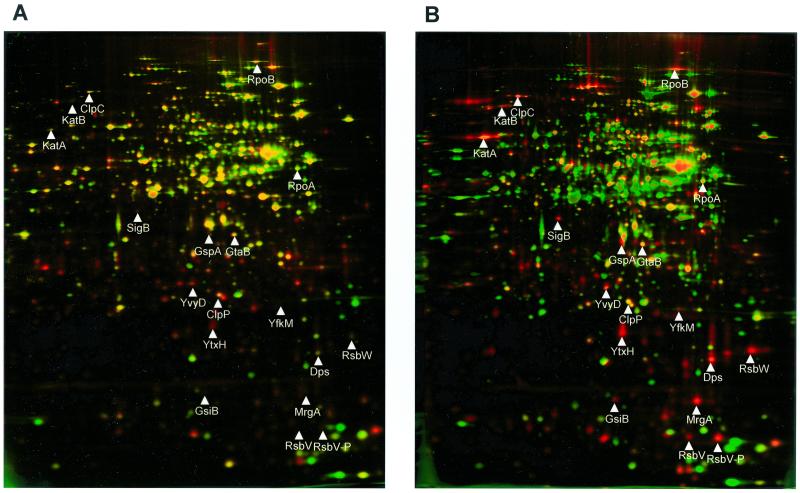
Crude protein extracts of cells pulse-labeled at different time points after antibiotic treatment were separated on 2D gels. The synthesis pattern of cytoplasmic proteins in the pI range of 4 to 7 was dramatically changed (Fig. 4). To identify newly synthesized or strongly induced proteins, dual-channel imaging (4) was used (Fig. 5 and 6). The autoradiographs (red) resemble protein synthesis during pulse-labeling, while silver nitrate-stained gels (green) represent total protein. Overlapping of both protein synthesis (red) and accumulated protein (green) results in yellow color, but newly induced proteins are colored red.
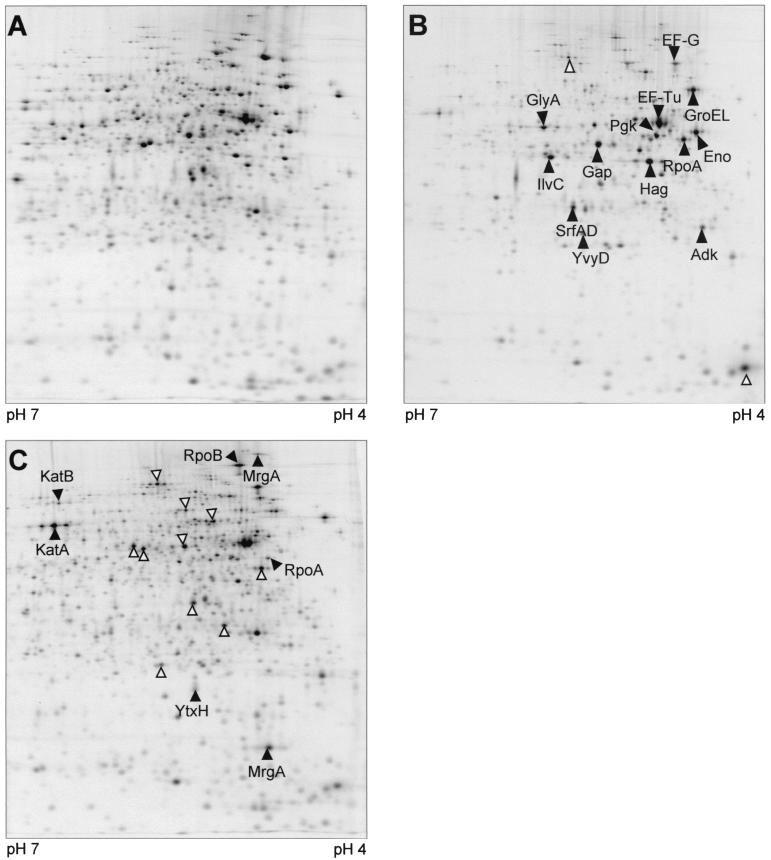
FIG. 4.
Autoradiographs of 2D gels, pH 4 to 7, of B. subtilis 168 l-[35S]methionine-labeled protein extracts. (A) Control; (B to D) 10 to 15 min, 40 to 45 min, and 70 to 75 min, respectively, after treatment with rifampin at 0.03 μg/ml. Arrowheads indicate proteins with increased relative synthesis rates; protein spots marked by empty arrows have not been identified.
FIG. 5.
Dual-channel images of 2D gels (4) produced with Delta2D Software (DECODON GmbH) illustrate the change in protein synthesis (red) and protein accumulation (green). B. subtilis 168 control extract (A); 55 min after rifampin application (B).
FIG. 6.
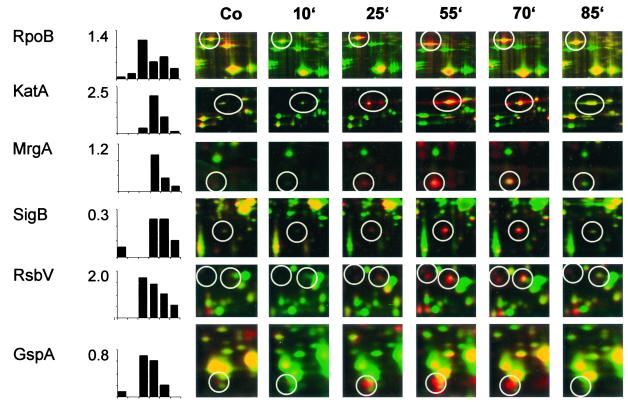
Details of dual-channel images of 2D gels are shown to illustrate the change in B. subtilis 168 protein synthesis (red) and protein accumulation (green) at various times (labels at top) after rifampin application. Percentages of total protein synthesis generated with the Delta2D Software (DECODON GmbH) are shown for proteins, including the target protein RpoB, proteins of the oxidative stress signature (KatA and the monomer of MrgA), and the ςB-dependent proteins SigB, RsbV, and GspA. Co, control.
Due to the great differences in l-[35S]methionine incorporation at the different time points we chose to apply equal amounts of total protein onto the gels. To obtain the best signal-to-background values, gels were exposed to photosensitive screens to almost full saturation of the major protein peaks. Quantitative data (Fig. 6) therefore resemble the percentage of total protein synthesis for the respective protein.
At 10 min after the addition of the antibiotic to the wild-type culture, the synthesis of most proteins decreased beyond the detection limit, which resulted in exposure times of several days (Fig. 4B). Only a few proteins were still being synthesized, though at significantly lower levels—among them RpoA, EF-G, EF-Tu, Hag, Gap, CitH, GlyA, SucD, Eno, Pgk, IlvC, Adk, SrfAD, and YkwC.
The DNA-dependent RNA polymerase β subunit RpoB and a subset of members of the oxidative stress response, namely KatA, KatB, and MrgA, were the first proteins to be induced 20 to 30 min after addition of the antibiotic. At 40 to 55 min after rifampin treatment the incorporation of l-[35S]methionine recovered, indicating increasing protein synthesis. At this time the ςB-dependent general stress response, including SigB itself, RsbW, RsbV, GspA, GtaB, YtxH, YvyD, Dps, ClpP, and ClpC (Fig. 4C), was induced. This is also shown by dual-channel imaging showing the newly induced proteins as red spots (Fig. 5).
Another protein, flagellin (Hag), was produced in comparably high amounts. It belongs to the ςD regulon, which mediates chemotaxis and motility.
In the autoradiograph of the wild-type cell extract labeled 70 to 75 min after rifampin treatment the oxidative stress and ςB response were still highly active (Fig. 4D).
In dual-channel images of rifampin-treated cells some of the newly induced ςB-dependent proteins (red color) and also KatA and MrgA turned yellow, indicating accumulation of these proteins (for examples, see Fig. 6).
Autoradiographs of cells labeled from min 100 to min 105 after rifampin application resembled that of control extracts, indicating that protein synthesis had recovered in preparation for the resumption of growth (data not shown).
In pulse-labeling experiments the sigB mutant showed the same pattern of methionine incorporation as the wild type (Fig. 3B). Protein synthesis reached a minimum shortly after challenge with the antibiotic, and a great number of proteins were no longer synthesized 10 min after rifampin treatment (Fig. 7). As in the wild type the relative synthesis of KatA, KatB, and MrgA, which all belong to the protein signature (9) obtained under oxidative stress conditions, increased in parallel during the growth arrest phase. The ςD-dependent Hag was one of the major protein spots 10 and 55 min after rifampin treatment. As expected the ςB-dependent general stress proteins were missing from the gels.
FIG. 7.
Autoradiographs of 2D gels, pH 4 to 7, of B. subtilis ML6 (sigB) l-[35S]methionine-labeled protein extracts. (A) control; (B and C) 10 to 15 min and 70 to 75 min, respectively, after application of rifampin (0.03 μg/ml). Solid arrowheads indicate proteins with increased relative synthesis rates; protein spots marked by empty arrows have not been identified.
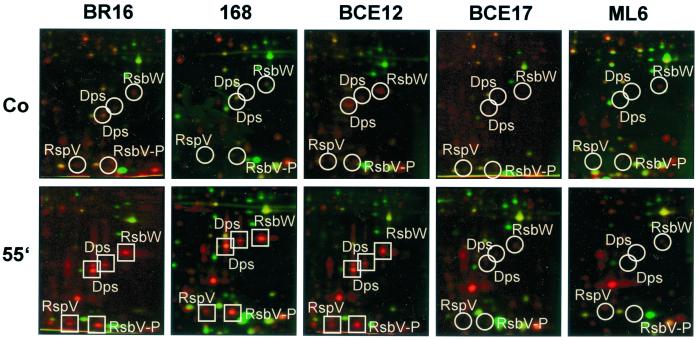
To investigate the role of RsbP and RsbU activity in activation of the general stress response in rifampin-treated B. subtilis cells, protein syntheses of the rsbU and rsbP mutants were compared with those of the corresponding wild types and the sigB mutant. The ςB-dependent proteins were synthesized 55 min after rifampin treatment in the wild type and the rsbU mutant but not in the sigB and rsbP mutants (Fig. 8).
FIG. 8.
Details of dual-channel images of 2D gels illustrate the difference in protein synthesis (red) and protein accumulation (green) after rifampin application among B. subtilis BR16, 168, BCE12 (rsbU), BCE17 (rsbP), and ML6 (sigB). The positions of induced proteins (squares) and the expected positions of proteins (circles) are indicated. Abbreviations: Co, control; 55′, 55 min.
The wild type survives better than the sigB mutant on rifampin-containing agar plates.
The growth experiments and 2D gels suggested that a phenotypic tolerance to rifampin is acquired by the cells in response to exposure to the antibiotic during exponential growth. The level of tolerance was measured by counting those CFU that were able to grow on rifampin-containing LB agar plates. At various time points after antibiotic treatment, aliquots of wild-type and sigB mutant cultures were plated on LB agar plates containing rifampin at the MIC. During exponential growth the survival of wild type cells was about 0.01% on rifampin agar compared to LB plates without antibiotic (not shown). The sigB mutant was 10 times more sensitive to plating on rifampin agar plates. In stationary phase 10% of the wild-type CFU were able to grow on rifampin-containing agar plates, but only 0.02% of the mutant cells could do so.
When the cells were treated with rifampin at 0.06 μg/ml during exponential growth, their survival on agar plates containing rifampin increased during growth arrest (Fig. 9). The sigB mutant, too, showed an adaptation to the antibiotic during the rifampin-triggered growth arrest. However, throughout the experiment the survival rate of the mutant stayed at 5 to 10% of that of the wild type.
FIG. 9.
Growth of B. subtilis 168 and ML6 (sigB) was monitored by measuring OD500. Both strains were challenged with rifampin (0.06 μg/ml) at an OD500 of 0.4. Aliquots of the cultures were plated on rifampin agar plates containing rifampin (0.06 μg/ml) to test the level of tolerance to the antibiotic.
During these experiments rifampin-resistant B. subtilis did not appear.
DISCUSSION
Rifampin, an antibiotic of the rifamycin group, induces ςF in M. tuberculosis. This alternate sigma factor is a homolog of ςB of B. subtilis, the stress sigma factor responsible for the general stress response (12, 18). In this study it is demonstrated that rifampin treatment also induces the ςB-dependent general stress response in B. subtilis.
Moderate concentrations of rifampin added to exponentially growing cells of B. subtilis stopped growth immediately (7). In parallel the incorporation of l-[35S]methionine into protein was drastically reduced because of the inhibition of early transcriptional elongation by rifampin. However, 40 to 50 min after the addition of rifampin l-[35S]methionine incorporation increased followed by the recovery of synthesis of vegetative proteins and finally by the resumption of growth. Clearly before this growth recovery, two main protein groups were induced which belong to the oxidative stress stimulon and to the general stress regulon. In addition to these main protein groups the direct target of rifampin, the β subunit of the DNA-dependent RNA polymerase (RpoB), was also synthesized early after rifampin treatment, as was flagellin (Hag). The induction of members of the oxidative stress stimulon such as KatA and MrgA just after rifampin treatment indicates that oxidative stress occurs in rifampin-treated cells. Hydrogen peroxide is known to be a strong inducer of this response, which includes proteins that are thought to protect cellular structures including proteins or DNA against oxidative damage. Only those proteins known to detoxify reactive oxygen species are induced after rifampin treatment. For strongly aerated cells continuous protein synthesis might be necessary to deal with oxygen radicals that may emerge as a side product of the reactions of the respiratory chain.
The induction of the ςB-dependent general stress response was delayed by 10 min with respect to the oxidative stress response but also preceded the resumption of growth by 100 min, suggesting a relationship between this induction and cell recovery from rifampin treatment. To address this problem, we analyzed the sigB mutant ML6, which was likewise able to overcome the growth arrest caused by rifampin, however, with a distinct delay. The differences in the duration of the growth recovery period between the wild type and the sigB mutant were even more pronounced at increasing rifampin concentrations. Survival experiments demonstrated that the mutant is more sensitive to the antibiotic after plating cells on rifampin-containing LB agar plates. The survival rate of the wild type exceeded that of the mutant by a factor of more than 10. The most dramatic effect, however, was obtained when stationary-phase cells without previous exposure to rifampin were plated. In this case the wild type showed a 500-times-higher survival rate than the sigB mutant. These findings suggest that the ςB-dependent general stress response known to be activated in the early stationary phase supports tolerance to rifampin. Wild-type cells treated with rifampin in the exponential growth phase showed 100-times-better survival. When the sigB mutant was similarly treated, the survival rate was also higher than in untreated cultures, though it did not reach the level of tolerance of the wild type. This result indicates a ςB-independent mechanism of developing tolerance to rifampin.
Our data strongly indicate that ςB-dependent general stress proteins are somehow involved in the growth recovery process of rifampin-treated cells. Amino acid starvation does not induce the ςB-dependent general stress response. After the addition of tryptophan to tryptophan-starved B. subtilis the resumption of growth is not accompanied by a parallel induction of ςB-dependent proteins (J. Bernhardt, personal communication), indicating that the induction of ςB is not a necessary prerequisite for the resumption of growth of stationary-phase cells. The question arises whether the higher level of tolerance to rifampin in the wild type compared with the sigB mutant is related to the protection of cellular structures in nongrowing cells through proteins belonging to the ςB regulon or whether some ςB-dependent proteins are able to inactivate rifampin. Two mechanisms of rifampin inactivation have so far been described for B. subtilis (7). The molecular basis for these mechanisms is not known. Whether either or both of these mechanisms are ςB dependent remains an open question. One possible candidate for a ςB-dependent mechanism is the putative multidrug resistance protein BmrU, previously shown to be under ςB control (17). This protein, however, has not yet been tested for export of rifampin.
ςB activity is regulated by two different pathways which both lead to activation of the anti-anti-sigma factor RsbV by dephosphorylation making use of two different phosphatases, RsbP and RsbU: a low cellular energy charge leads to ςB activation via the RsbP-dependent pathway, while physical stress such as heat shock or ethanol triggers the RsbU cascade (20, 21). In growth experiments the rsbU mutant recovered from rifampin treatment in a time scale intermediate between the wild type and sigB mutant but the rsbP mutant similar to the sigB mutant. 2D gels of the rsbP and rsbU mutants revealed that the ςB-dependent general stress response is induced only in rsbU mutants but not in rsbP mutants. This strongly suggests that ςB activation in response to rifampin depends on the energy-signaling RsbP phosphatase. It is unlikely that the energy charge decreases during the rifampin-caused growth arrest, and in silver-stained 2D gels only RsbV-P accumulates but not the dephosphorylated RsbV, indicating that RsbP phosphatase is not fully active. However, recently it was reported that RsbP is responsible for the basal activity of ςB during exponential growth (10), so we suggest that ςB is able to make better use of the rising transcription capacity than the housekeeping ςA when the effect of rifampin wears off. This is supported by the coincidence of induction of the ςB regulon and the resumption of l-[35S]methionine incorporation. For further discussion it would be helpful to study the in vitro affinity of both sigma factors to RNA polymerase core enzyme.
Taken together these results define a new phenotype of sigB. In addition to the multiple resistance against oxidative, salt, heat, ethanol, or alkaline stress (12, 18), the ςB-dependent stress response is also involved in acquiring tolerance to rifampin. We are currently investigating whether this effect is specific for rifampin or whether it also extends to other drugs.
Acknowledgments
We thank C. W. Price and U. Völker for generous gift of strains and R. Jack at the Institute for Medical Biochemistry of the University of Greifswald for carefully reading the manuscript. We especially thank H. Labischinski for helpful discussions.
This work was supported by a grant from the Fonds der chemischen Industrie to M. H. and the Bayer AG Wuppertal.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor ςB in Bacillus subtilis. J. Bacteriol. 179:7251–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., J. Bernhardt, R. Schmid, and M. Hecker. 1995. A gene at 333 degrees on the Bacillus subtilis chromosome encodes the newly identified SigB-dependent general stress protein GspA. J. Bacteriol. 177:3540–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhardt, J., K. Büttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225–2240. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. [DOI] [PubMed] [Google Scholar]
- 6.Cavalleri, B., M. Turconi, G. Tamborini, E. Occelli, R. Pallanza, R. Scotti, M. Berti, G. Romano, and F. Parenti. 1990. Synthesis and biological activity of some derivatives of rifamycin P. J. Med. Chem. 33:1470–1476. [DOI] [PubMed] [Google Scholar]
- 7.Dabbs, E. R., K. Yazawa, Y. Tanaka, Y. Mikami, M. Miyaji, S. J. Andersen, N. Morisaki, S. Iwasaki, O. Shida, H. Takagi, and K. Kadowaki. 1995. Rifampicin inactivation by Bacillus species. J. Antibiot. 48:815–819. [DOI] [PubMed] [Google Scholar]
- 8.DeMaio, J., Y. Zhang, C. Ko, and W. R. Bishai. 1997. The Mycobacterium tuberculosis sigF gene is part of a gene cluster organised like the Bacillus subtilis sigF and sigB operons. Tuber. Lung Dis. 78:3–12. [DOI] [PubMed] [Google Scholar]
- 9.Engelmann, S., and M. Hecker. 1996. Impaired oxidative stress resistance of Bacillus subtilis ςB mutants and the role of katA and katE. FEMS Microbiol. Lett. 145:63–69. [DOI] [PubMed] [Google Scholar]
- 10.Eymann, C., and M. Hecker. 2001. Induction of ςB-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis. FEMS Microbiol. Lett. 199:221–227. [DOI] [PubMed] [Google Scholar]
- 11.Halling, S. M., K. C. Burtis, and R. H. Doi. 1977. Reconstitution studies show that rifampicin resistance is determined by the largest polypeptide of Bacillus subtilis RNA polymerase. J. Biol. Chem. 252:9024–9031. [PubMed] [Google Scholar]
- 12.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35–91. [DOI] [PubMed] [Google Scholar]
- 13.Igo, M., M. Lampe, C. Ray, W. Schafer, C. P. Moran, Jr., and R. Losick. 1987. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J. Bacteriol. 169:3464–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA—a new approach for isolating DNA. BioTechniques 11:94–101. [PubMed] [Google Scholar]
- 15.Michele, T. M., C. Ko, and W. R. Bishai. 1999. Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob. Agents Chemother. 43:218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otto, A., B. Thiede, E. C. Müller, C. Scheler, B. Wittmann-Liebold, and P. Jungblut. 1996. Identification of human myocardial proteins separated by two-dimensional electrophoresis using an effective sample preparation for mass spectrometry. Electrophoresis 17:1643–1650. [DOI] [PubMed] [Google Scholar]
- 17.Petersohn, A., H. Antelmann, U. Gerth, and M. Hecker. 1999. Identification and transcriptional analysis of new members of the ςB regulon in Bacillus subtilis. Microbiology 145:869–880. [DOI] [PubMed] [Google Scholar]
- 18.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related Gram-positive bacteria, p.179–197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 19.VanBogelen, R. A., E. E. Schiller, J. D. Thomas, and F. C. Neidhardt. 1999. Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis 20:2149–2159. [DOI] [PubMed] [Google Scholar]
- 20.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the ςB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180–188. [DOI] [PubMed] [Google Scholar]
- 21.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Völker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Völker, R. Schmid, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741–752. [DOI] [PubMed] [Google Scholar]
- 23.Wetzstein, M., U. Völker, J. Dedio, S. Löbau, U. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 174:3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265–2275. [DOI] [PubMed] [Google Scholar]