Abstract
Recently, we have shown the first evidence for allelic exchange in Leptospira spp. By using the same methodology, the cloned recA of Leptospira biflexa was interrupted by a kanamycin resistance cassette, and the mutated allele was then introduced into the L. biflexa chromosome by homologous recombination. The recA double-crossover mutant showed poor growth in liquid media and was considerably more sensitive to DNA-damaging agents such as mitomycin C and UV light than the wild-type strain. The efficiency of plating of the recA mutant was about 10% of that of the parent strain. In addition, microscopic observation of the L. biflexa recA mutant showed cells that are more elongated than those of the wild-type strain. Fluorescent microscopy of stained cells of the L. biflexa wild-type strain revealed that chromosomal DNA is distributed throughout most of the length of the cell. In contrast, the recA mutant showed aberrant nucleoid morphologies, i.e., DNA is condensed at the midcell. Our data indicate that L. biflexa RecA plays a major role in ensuring cell viability via mechanisms such as DNA repair and, indirectly, active chromosome partitioning.
The RecA protein is thought to be ubiquitous in bacteria and possesses a highly conserved primary amino acid sequence. The bacterial RecA protein plays a central role in homologous recombination and in induction of the error-prone DNA repair mechanism. In Escherichia coli, RecA catalyzes strand exchanges between homologous DNA molecules via RecA-single-stranded DNA (ssDNA) complexes. RecA also plays a key role in signal transduction following DNA damage. RecA binds ssDNA generated by DNA damage. The activated RecA complex then induces the SOS repair functions (17).
Studies of recA mutants suggest that RecA may promote and participate to many other functions. For instance, RecA is required for plasmid maintenance in Streptococcus pneumoniae (15), adherence and colonization of Vibrio cholerae (13), DNA segregation in Bacillus subtilis (21), and phenotypic switching in Pseudomonas tolaasii (22). The complex and pleiotropic effects of RecA suggest that it has several biochemical activities and can be involved in different stress pathways.
No recA mutant of spirochetes has been reported. 16S rRNA analysis indicates that the phylum of spirochetes has a deep branching lineage (25) and that they differ considerably from well-studied gram-negative and gram-positive bacteria. Stamm et al. (23) have shown that both recombination functions and SOS response were restored in an E. coli recA mutant by the Leptospira biflexa recA gene. No other study concerning DNA repair and recombination mechanisms has been undertaken in any member of the order Spirochaetales. The sequences of the genomes of the spirochetes Treponema pallidum and Borrelia burgdorferi showed a minimal set of genes for DNA repair and recombination mechanisms in comparison with other bacterial genomes (4).
Recently, we have shown the feasability of performing allelic replacement in Leptospira spp. by homologous recombination. Allelic exchange was only obtained when a suicide vector containing the L. biflexa flaB gene disrupted by a kanamycin resistance marker was UV irradiated or alkali denatured prior to electroporation (18). This study suggested that introduction of altered DNA into L. biflexa may recruit the recombination and/or DNA repair machinery for initiating homologous recombination. Since no data for spirochetes are available on these mechanisms, we decided to create an L. biflexa recA mutant by allelic exchange. The mutant was assessed for RecA functions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L.biflexa serovar patoc strain Patoc 1 (National Reference Center, Institut Pasteur, Paris, France) was grown at 30°C in EMJH (6, 10) liquid medium or on EMJH 1% agar plates. E. coli XL10-Gold (Stratagene) was grown at 37°C on solid or liquid Luria-Bertani (LB) medium. When necessary, kanamycin and catalase (bovine liver catalase; Sigma) were used at 50 μg/ml and 150 U/ml, respectively.
DNA amplification.
Amplification was achieved using one cycle of denaturation (94°C, 5 min), followed by 35 cycles of amplification consisting of denaturation (94°C, 30 s), annealing (55°C, 30 s), and primer extension (72°C, 1 min), and a final cycle of extension of 10 min at 72°C.
Construction of an L. biflexa recA mutant.
The 1,313-bp fragment of the L. biflexa recA gene was amplified by PCR with primers RCA (5′-GTT CAT CCT TGA GAG AGT CG-3′) and RCB (5′-AAC CCC GAA CCG GTC AAT GG-3′). After purification on a minicolumn (Qiaquick gel extraction kit; Qiagen), the recA fragment was blunt ended and cloned into PvuII-cut pUC19; the resulting vector was called pRca, and its insert was sequenced. The flanking DNA of the L. biflexa recA was deduced by ligation-mediated PCR (LM-PCR) (19) using EcoRI adaptors as described previously (18) and primers RAD (5′-GAC TAG GTA GGG GAC TTG GC-3′) and RAC (5′-GAC CGA AGC TGA TGG AGA CA-3′), directed upstream and downstream from recA, respectively.
The Enterococcus faecalis kanamycin resistance cassette was amplified from plasmid pAT21 (24) with primers Kan3 (5′-ATC GGC TCC GTC GAT ACT AT-3′) and Kan5 (5′-GTA GTC TCA TAC CTG TCA ACG CCT ACA T-3′). The resulting 1,062-bp amplified product and the ClaI-cut pRca (ClaI cuts once in recA of pRca) were blunt ended and ligated. The extent of homologous L. biflexa DNA present on either side of the Kmr marker of pRca was 0.55 and 0.75 kb. After deletion of the ampicillin resistance cassette of pUC19 by ScaI and AatII digestion, blunt-ending, and religation, the resulting suicide vector pKRc was subjected to 10 mJ of UV irradiation cm−2 as described previously (18) and used to deliver the inactivated recA in L. biflexa.
L. biflexa cells were prepared for electroporation as previously described (20). Approximately 10 μg of plasmid DNA and 100 μl of competent cells were used for electroporation. Primers RAA (5′-TTC CAC AGG CTC TTT GGA-3′) and RAB (5′-GAG TCG ATG ACA ATG AGG TC-3′) were used for the detection of double-crossover mutants.
Southern blot analysis.
Genomic DNA was extracted, digested with EcoRI, subjected to electrophoresis overnight in a 1% agarose gel, and transferred onto nylon membranes as previously described (18). The recA gene was amplified with primers RAA and RAB, radiolabeled with [α-33P]dATP using a commercial kit (Megaprime, Amersham). Membranes were hybridized overnight at 65°C in Rapid hybridization buffer (Amersham) and then washed as previously described (18).
Sensitivity to UV and mitomycin C.
Susceptibility to mitomycin C and UV irradiation was investigated with exponential-phase cultures of L. biflexa. For mitomycin C sensitivity, diluted cultures were incubated in EMJH liquid medium supplemented with various concentrations of mitomycin C (Sigma) at 30°C for 24 h in a dark room, and samples were plated on EMJH agar plates. For UV irradiation, cells were spread at appropriate dilutions on EMJH agar plates and irradiated under UV light (254 nm, 400 μW/cm2) for various time periods. Experiments were carried out under minimal light, and plates were covered with foil to prevent photoreactivation. UV and mitomycin C sensitivity was evaluated by colony counting. Untreated cells served as a control.
Microscopy.
Cells were analyzed by dark-field microscopy, fluorescent microscopy, and electron microscopy. For fluorescent microscopy, cells were harvested from culture media by centrifugation at 2,000 × g for 4 min, resuspended in 3% paraformaldehyde for 15 min, and washed once in phosphate-buffered saline (PBS). The cell suspension was then spread on slides and air-dried at room temperature. Approximately 10 μl of 4′,6-diamidino-2-phenylindole (DAPI; Sigma) at a final concentration of 1 μg/ml was added to the cells, cover slips were positioned, and slides were viewed by fluorescent microscopy. For electron microscopy, cells were washed once in PBS, resuspended in 10 mM Tris-HCl (pH 8), and then placed on a glow-discharged carbon-coated grid for 1 min and negatively stained with 2% uranyl acetate or 1% phosphotungstate.
Nucleotide sequence accession number.
The GenBank accession number for the L. biflexa recA nucleotide and amino acid sequences is AF410431.
RESULTS
Construction of an L. biflexa recA mutant.
The recA gene from the saprophytic species L. biflexa was isolated previously by complementation of an E. coli recA mutant (23). It has to be noted that the strain used in our study, Patoc 1, is different from the strain studied by Stamm et al. (23). Indeed, a recent study has demonstrated that the strain maintained in the United States (studied by Stamm et al.) belongs to the saprophytic Leptospira meyeri species, and not to L. biflexa (2). However, we were able to amplify the recA locus from our L. biflexa strain by using primers designed from the recA sequence of L. meyeri.
The sequence determined was not identical to that reported by Stamm et al. (23); the L. biflexa recA and the transcribed open reading frame (ORF) exhibit 84 and 97% identity with the nucleotide and amino acid sequences of L. meyeri, respectively. The ORF of L. biflexa recA encodes a protein of 387 amino acids, which exhibits 72 and 71% identity with RecA proteins of Leptospira interrogans and the proteobacterium Aeromonas salmonicida, respectively. L. biflexa RecA shows 58% identity with RecA from E. coli. The 2-kb nucleotide sequence of the L. biflexa recA locus does not contain known genes other than recA, and no ORF overlaps the recA coding sequence.
The recA coding region was interrupted at its unique ClaI site by insertion of a kanamycin resistance marker gene, resulting in plasmid pKRc. The insertion site of the Kmr marker corresponds to the monomer-monomer (M-M) 2 region of E. coli RecA. On the basis of the E. coli RecA three-dimensional polymeric structure, seven segments, M-M1 to M-M7, were assigned. Residues found in M-M surfaces are conserved across all RecA sequences, and these interfaces are involved in RecA monomer polymerization (11).
A gene replacement protocol (18) in which plasmid pKRc was treated by UV irradiation prior electroporation was used to interrupt the chromosomal recA in L. biflexa. Electroporation of L. biflexa with UV-treated plasmid pKRc resulted in four Km-resistant colonies. The number of transformants was considerable lower than observed for the previously inactivated genes flaB and metY of L. biflexa (18). Screening of transformants was performed by PCR amplification with primers flanking the ClaI site used for insertion of the Kmr cassette (data not shown) and Southern blot analysis. Hybridization with an L. biflexa recA probe resulted in a single band of approximately 2 kb for the parental recA+ strain (Fig. 1, lane 1). In transformants, the increase in size of the hybridizing fragment by 1.1 and 4.3 kb is due to the insertion of the Kmr cassette (double-crossover event) and to plasmid pKRc (single-crossover event), respectively, into the recA+ locus (Fig. 1). Southern blot analysis confirmed the results obtained by PCR. The recA disruption was stable in the absence of selection.
FIG. 1.
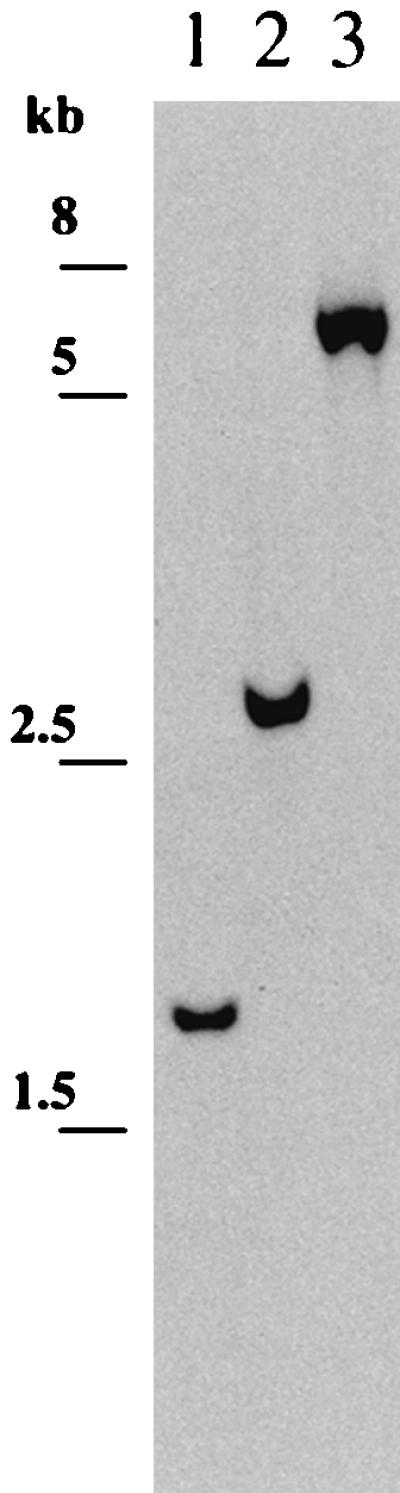
Southern blot analysis of representative clones resulting from homologous recombination. Lane 1, L. biflexa wild-type strain; lane 2, Kmr colony resulting from a double crossing-over with the recA locus; lane 3, Kmr colony resulting from a single crossing-over with the recA locus. Genomic DNA was digested with EcoRI and hybridized with a recA probe. Sizes of the fragments of the molecular mass marker are indicated on the left.
Increased sensitivity to DNA-damaging agents of the L.biflexa recA mutant.
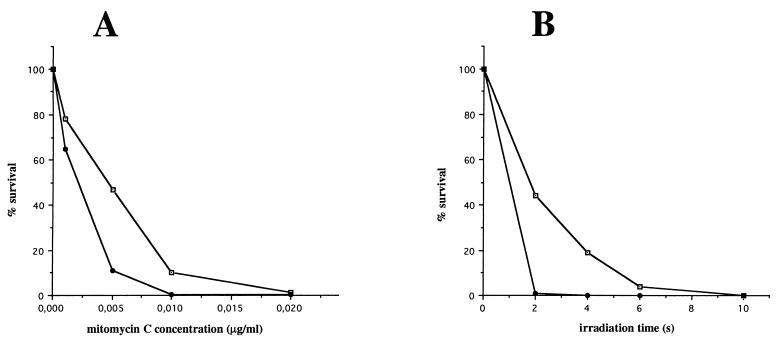
The sensitivity of the wild-type strain, the recA single-crossover mutant (recA+/recA::Km), and the recA double-crossover mutant (recA::Km) to DNA-damaging agents was evaluated by using mitomycin C and UV light. Both treatments had a significantly greater effect on mortality of the recA double-crossover mutant than on the wild-type strain (Fig. 2) and the recA single-crossover mutant (data not shown). For instance, the survival rate of the recA mutant decreased dramatically at low UV irradiation dosage: less than 1% mutant survivors were detected after 2 s of irradiation, compared to about 50% survival for the parental strain (Fig. 2) and the recA single-crossover mutant (data not shown). At 5 ng of mitomycin C per ml, the recA mutant survival rate was 11%, while the survival rate of the wild-type strain was 47%. For each strain and in all conditions, similar results were obtained in three independent experiments.
FIG. 2.
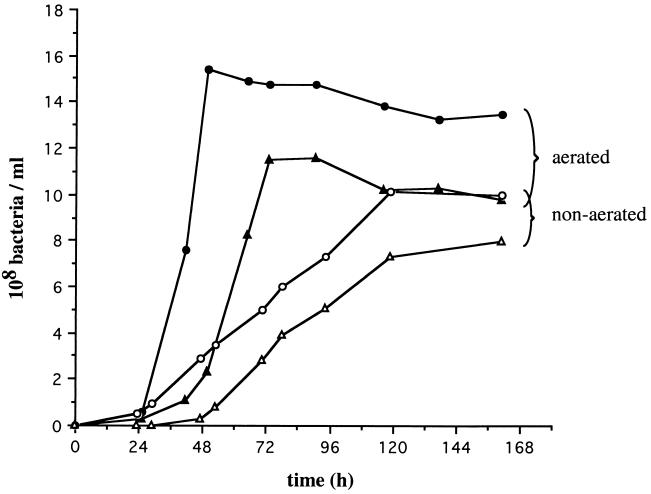
Growth of aerated and nonaerated cultures of L. biflexa. Parental L. biflexa (solid circles, aerated cultures; open circles, nonaerated cultures) and the recA mutant (solid triangles, aerated cultures; open triangles, nonaerated cultures). Growth was determined by measuring the OD at 420 nm.
Low growth of L. biflexa recA mutant in liquid and solid media.
Cell counts of the wild-type strain and the recA double-crossover mutant (or recA mutant) were compared during growth without and with vigorous aeration (Fig. 3). In nonaerated cultures, the recA mutant exhibited a lag period longer than that of the wild-type strain. Although growth of recA mutant cells was similar to that of the wild-type strain at the exponential phase (doubling time for both strains was 30 to 34 h), they did not reach the plateau of the wild-type strain (Fig. 3). In aerated cultures, a 24-h delay was observed before entry of the recA mutant cells into exponential phase by comparison with the wild-type strain. The doubling time was approximately 9 h for both the wild-type strain and recA mutant (Fig. 3).
FIG. 3.
Survival after mitomycin C treatment (A) and UV irradiation (B) of the parental L. biflexa (open squares) and the recA mutant (solid circles). The number of viable cells was determined by plating dilutions of treated and untreated cells.
Aeration does not have a negative effect on exponential growth of the recA mutant. For the wild-type strain and the recA mutant, a plateau was achieved after 2 and 3 days, respectively. Again, recA mutant cells were unable to reach the maximum value of the wild-type strain. Survival of the recA mutant was not significantly affected in stationary-phase culture. The recA single-crossover mutant behaved essentially like the wild-type strain (data not shown).
When cells were grown to exponential phase and then plated, the recA mutant presented a plating defect compared to the wild-type strain and the recA single-crossover mutant. The plating efficiency of an exponentially growing recA mutant was about 10% (8 to 12% in four independent experiments) of that seen in the parental strain and in the recA single-crossover mutant. The term plating efficiency is used here to mean the ratio of CFU to optical density (OD) at 420 nm. Plating efficiencies derived from dilutions of exponential- or stationary-phase cultures were similar.
Interestingly, exogenous catalase has been shown to improve the recovery of E. coli cells under various stress conditions (1, 14), probably by degrading oxidative compounds. Reactive oxygen species in the agar medium are thought to be a possible source of toxicity. To test whether the low plating efficiency of the recA mutant could result from a defect in the repair of oxidative damage that occurs upon plating, we measured the ability of this strain to form colonies on catalase-containing plates. For both the wild-type strain and the recA mutant, the capacity to form colonies was similar in the absence and presence of catalase (data not shown). However, colonies appeared in only 4 days instead of 7 days, but without restoring viability to the recA mutant.
Defects in cell and nucleoid morphology of the L. biflexa recA mutant.
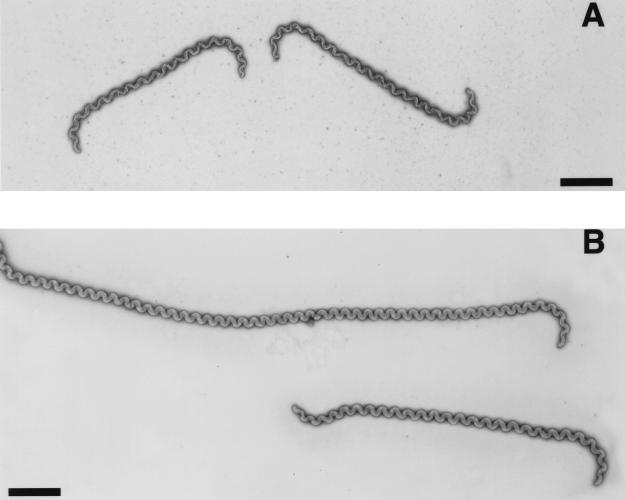
Microscopic observations of exponentially growing cells of the recA mutant showed cells significantly more elongated than those of the wild-type strain. For both strains, cell length becomes longer as cells reach the stationary phase. During entry into stationary phase, cells become smaller and less motile. We measured the length of randomly chosen spirochetes in exponential phase on electron micrographs. The recA mutant cells were significantly longer (average length for 61 cells, 16 μm) and more heterogeneous (cell length, 7 to 22 μm, with 57% of the cells being longer than 14 μm) than those of the wild-type strain (average length for 40 cells, 11 μm; range, 7 to 14 μm). By electron microscopy, we observed no signs of either cell wall defects or septum formation in these elongated cells (Fig. 4). This was also true when the outer membrane was removed under the action of phosphotungstate (data not shown). It has to be noted that cells of the recA mutant retain the characteristic spiral- and/or hook-shaped ends of Leptospira cells (Fig. 4).
FIG. 4.
Microscopic analysis of L. biflexa wild-type strain (A) and recA mutant (B). Electron microscopy of L. biflexa negatively stained with uranyl acetate. Scale bar, 2 μm.
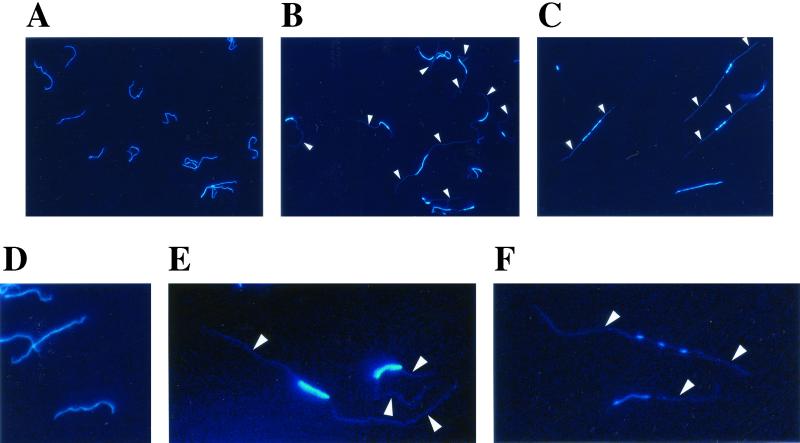
To directly visualize the distribution of DNA inside L. biflexa cells, DNA can be stained with a fluorochrome such as the DNA-specific dye DAPI and examined by fluorescence microscopy. For exponential-phase cultures of the L. biflexa wild-type strain, 100% of the cells were homogeneously stained through most of the cell length; the whole cellular space is occupied by the nucleoid (Fig. 5A and 5D). The fluorescence images of L. biflexa were similar to those of Borrelia spp. (12). In stationary-phase cultures, the proportion of homogeneously stained cells still represented about 99% of the cells (data not shown). For exponential-phase cultures of the L. biflexa recA mutant, only 15 to 50% of the cells possessed homogeneously stained cells, and the remaining cells (50 to 85%) contained small stained spots, mainly at the midcell.
FIG. 5.
Fluorescence microscopy of L. biflexa cells stained with DAPI. Cells of exponential-phase cultures of L. biflexa wild-type strain (A and D) and L. biflexa recA mutant (B, C, E, and F). Arrows point to a faint fluorescence signal in the cellular space of the spirochetes. Pictures from the same groups (top panel: A, B, and C; bottom panel: D, E, and F) are enlarged to the same extent. All images are at the same magnification (original magnification, ×600).
Nucleoids of the recA mutant cells varied in appearance, but most of the time the nucleoid was disorganized in either one or several foci at the midcell and a faint fluorescence signal occupied the entire cell interior (Fig. 5). In stationary phase, 64% of the recA mutant cells exhibited a normal nucleoid-like structure, i.e., DNA was distributed all along the cell length. Other cells had one or several nodes at the midcell (data not shown). Cells of the subpopulation with aberrant nucleoids were significantly more elongated than unaffected cells (Fig. 5), suggesting that the two phenomena (cell elongation and aberrant nucleoid morphology) are linked. Such aberrant nucleoid morphology has never been found in exponentially growing cells of the L. biflexa wild-type strain.
DISCUSSION
RecA is a multifunctional and ubiquitous protein involved in both recombination and DNA repair. Investigations of spirochetal recA so far have been performed with E. coli (23). In this study, we generated an L. biflexa recA mutant. Disruption of the recA gene was confirmed by PCR and Southern blot analysis
DNA repair defects.
The recA gene of L. biflexa was shown to affect responses to DNA damage. This phenotype is similar to that of recA mutants of other bacterial species. Irradiation with UV light is frequently used to compare the effectiveness of DNA repair mechanisms. UV light induces a variety of photoproducts in DNA, such as formation of pyrimidine dimers. In E. coli, UV-induced photoproducts are recognized and repaired by the proteins encoded by the uvr genes (17). Genes for the repair of UV-induced DNA damage (uvrA, uvrB, uvrC, and uvrD) are also present in the genomes of the spirochetes B. burgdorferi and T. pallidum (7, 8). The bifunctional alkylating agent mitomycin C also leads to major DNA lesions, such as cross-linking of DNA strands through reaction with guanine residues. Again, the uvr system recognized the DNA cross-links. In E. coli, repair of DNA cross-links requires RecA-mediated recombination (17).
Our study demonstrates that L. biflexa may also possess a full set of genes involved in DNA repair. DNA repair of the DNA-damaging-agent-induced products may also require the RecA protein for genetic recombination. In contrast, RecA is not involved in protection of Leptospira spp., which are aerophilic bacteria, from oxidative DNA damage as it is in microaerophilic bacteria (5).
DNA segregation defects.
Particularly striking is the slow growth in liquid medium and the poor survival on agar medium of the L. biflexa recA mutant. Growth of the recA mutant and the wild-type strain differed in the length of the lag period and the density of the culture in stationary phase. Bacterial recA mutants usually grow more slowly than isogenic recA+ strains, probably due to DNA degradation. In L. biflexa, the growth defect was accompanied by cell elongation, aberrant nucleoid morphology, and low viability. Viability was assayed by plating cells on solid medium and is therefore referred to as colony-forming ability. Approximately 90% of the recA mutant cells were unable to give rise to colonies. Viability scores for recA mutants of E. coli and Bacillus subtilis were 50 and 12%, respectively (3, 21). Another study has shown that the Streptomyces lividans recA mutant was nonviable (16).
In L. biflexa, the impairment in forming colonies is likely linked to the aberrant nucleoid morphology observed in 50 to 85% of the cells. This can be due to improper DNA segregation and/or cells lacking DNA that cannot grow further. The nonviable but residually dividing cells may contribute to the mass increase of the liquid cultures. Defects in cell and nucleoid morphology were also observed in B. subtilis recA mutants (21). Defects in DNA segregation may result from the inability of a recA mutant to repair stalled DNA replication forks (17), preventing replication fork progression and the following steps such as chromosome partitioning. Micrographs of L. biflexa recA mutant cells may therefore indicate that initiation of replication may take place at the midcell.
During the bacterial cell cycle, a cell goes through different discontinuous processes, such as chromosome replication, nucleoid partition, and cell division. Little is known about all of these processes in spirochetes. A bacterial cell maintains its intracellular DNA in a highly compacted structure to form the nucleoid. For instance, E. coli cells contain a compact and spherical nucleoid (17). Unlike those in E. coli and other bacteria, nucleoids in the spirochete B. burgdorferi appear to be composed of a loose network of strands extending throughout the cell, with no obvious central organizing node (9, 12). In addition, it has been shown that Borrelia cells contain a polyploid structure, containing 8 to 11 copies of its genome per cell (12); each copy of the genome may then be distributed along the entire length of the cell (9, 12).
The pictures observed in Borrelia cells can be extended to Leptospira spp.; DNA in Leptospira cells is distributed all along the cell length, and cells may also contain a polyploid structure. At the exponential phase, chromosome partitioning of nonsynchronized cells of the L. biflexa wild-type strain may form complex fluorescence shapes all along the cell. Observation of a few foci at the midcell in our recA mutant suggests that DNA may not be dispersed randomly throughout the cytoplasm but confined to specific intracellular regions by some unknown active process. Further study of our recA mutant could help in understanding the intracellular distribution of genomic DNA in spirochetes.
Conclusion.
In this study, after knocking out the L. biflexa flaB and metY genes (18), we inactivated the L. biflexa recA gene. The results confirm the efficiency of gene replacement in L. biflexa by treatment of DNA with UV prior to electroporation. However, the number of transformants that we obtained was much lower than those of previously inactivated genes. We can attribute the difficulty in obtaining a recA mutant (compared to flaB and metY mutants) to poor growth of the mutant on solid medium. Our data show that the L. biflexa recA mutant is deficient in recombination repair, suggesting that RecA plays a major role in DNA repair in L. biflexa.
A recombination-deficient strain of L. biflexa would be useful for further genetic studies with this organism. Mutant recA strains are genetically more stable than their recA+ counterparts; they can be used to avoid genetic rearrangements. The expression of foreign antigens may be more stable in a recA mutant. Several reports have demonstrated a role of RecA in virulence of some pathogenic bacteria. It will therefore be interesting to test whether recA mutations affect the virulence of pathogenic Leptospira spp.
Acknowledgments
The Institut Pasteur supported this work.
REFERENCES
- 1.Bredèche, M. F., S. D. Ehrlich, and B. Michel. 2001. Viability of rep recA mutants depends on their capacity to cope with spontaneous oxidative damage and on the DnaK chaperone protein. J. Bacteriol. 183:2165–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenot, A., C. Werts, C. Ottone, N. Sertour, N. W. Charon, D. Postic, G. Baranton, and I. Saint Girons. 2001. First evidence for a restriction-modification system in Leptospira sp. FEMS Microbiol. Lett. 201:139–143. [DOI] [PubMed] [Google Scholar]
- 3.Capaldo, F. N., G. Ramsey, and S. D. Barbour. 1974. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J. Bacteriol. 118:242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens, S. 2000. Borrelia genomes in the year 2000. J. Mol. Microbiol. Biotechnol. 2:401–410. [PubMed] [Google Scholar]
- 5.Duwat, P., S. D. Ehrlich, and A. Gruss. 1995. The recA gene of Lactococcus lactis: characterization and involvement in oxidative and thermal stress. Mol. Microbiol. 17:1121–1131. [DOI] [PubMed] [Google Scholar]
- 6.Ellinghausen, H. C., and W. G. Mc Cullough. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and medium bovine albumin and polysorbate 80. Am. J. Vet. Res. 26:45–51. [PubMed] [Google Scholar]
- 7.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, C. M., S. J. Norris, C. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. Mc Leod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. Mc Donald, P. Artiach, C. Bowman, M. D. Cotton, C. Fujii, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. [DOI] [PubMed] [Google Scholar]
- 9.Hinnebusch, B. J., and A. Bendich. 1997. The bacterial nucleoid visualized by fluorescence microscopy of cells lysed within agarose: comparison of Escherichia coli and spirochetes of the genus Borrelia. J. Bacteriol. 179:2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic leptospires. I. Growth at low temperatures. J. Bacteriol. 94:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlin, S., and L. Brocchieri. 1996. Evolutionary conservation of recA genes in relation to protein structure and function. J. Bacteriol. 178:1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitten, T., and A. G. Barbour. 1992. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics 132:311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar, K. K., R. Srivastava, V. B. Sinha, J. Michalski, J. B. Kaper, and B. S. Srivastava. 1994. recA mutations reduce adherence and colonization by classical and El Tor strains of Vibrio cholerae. Microbiology 140:1217–1222. [DOI] [PubMed] [Google Scholar]
- 14.Mackey, B. M., and D. A. Seymour. 1987. The effect of catalase on recovery of heat-injured DNA-repair mutants of Escherichia coli. J. Gen. Microbiol. 133:1601–1610. [DOI] [PubMed] [Google Scholar]
- 15.Martin, B., P. Garcia, M. P. Castanié, and J. P. Claverys. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15:367–379. [DOI] [PubMed] [Google Scholar]
- 16.Muth, G., D. Frese, A. Kleber, and W. Wohleben. 1997. Mutational analysis of the Streptomyces lividans recA gene suggests that only mutants with residual activity remain viable. Mol. Gen. Genet. 255:420–428. [DOI] [PubMed] [Google Scholar]
- 17.Neidhardt, F. C., R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C.
- 18.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospiraspp.: tivation of L. biflexa flaB results in nonmotile mutants deficient in endoflagella. Mol. Microbiol. 40:189–199. [DOI] [PubMed] [Google Scholar]
- 19.Prod’hom, G., B. Lagier, P. Pelicic, A. J. Hance, B. Gicquel, and C. Guilhot. 1998. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol. Lett. 158:75–81. [DOI] [PubMed] [Google Scholar]
- 20.Saint Girons, I., P. Bourhy, C. Ottone, M. Picardeau, D. Yelton, R. W. Hendrix, P. Glaser, and N. Charon. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa, construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 182:5700–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sciochetti, S. A., G. W. Blakely, and P. J. Piggot. 2001. Growth phase variation in cell and nucleoid morphology in a Bacillus subtilis recA mutant. J. Bacteriol. 183:2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinha, H., A. Pain, and K. Johnstone. 2000. Analysis of the role of recA in phenotypic switching of Pseudomonas tolaasii. J. Bacteriol. 182:6532–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamm, L. V., E. A. Parrish, and F. C. Gherardini. 1991. Cloning of the recA gene from a free-living leptospire and distribution of RecA protein among spirochetes. Appl. Environ. Microbiol. 57:183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′,5′-aminoglycoside phosphotransferase type III. Gene 23:331–341. [DOI] [PubMed] [Google Scholar]
- 25.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]