Abstract
The Clostridium josui aga27A gene encodes the cellulosomal α-galactosidase Aga27A, which comprises a catalytic domain of family 27 of glycoside hydrolases and a dockerin domain responsible for cellulosome assembly. The catalytic domain is highly homologous to those of various α-galactosidases of family 27 of glycoside hydrolases from eukaryotic organisms, especially plants. The recombinant Aga27A α-galactosidase devoid of the dockerin domain preferred highly polymeric galactomannan as a substrate to small saccharides such as melibiose and raffinose.
Multienzyme complexes having high activity against crystalline cellulose, known as cellulosomes, were identified and characterized for some cellulolytic clostridia such as Clostridium josui (14), C. cellulolyticum (3), C. cellulovorans (6), and C. thermocellum (2). A common feature of clostridial cellulosomes is that they consist of a large number of catalytic components arranged around noncatalytic scaffolding proteins. The cellulosomes are devised to degrade plant cell walls but not pure cellulose; i.e., they contain not only celullases but also various hemicellulases as catalytic components. For example, xylanase, mannanase, and chitinase genes were cloned from C. thermocellum, and some gene products were identified in the cellulosome (see reference 2 for citations), although this bacterium is known to utilize only cellulose and cellooligosaccharides as carbon sources. Recently, a pectate lyase has been characterized as a component of the C. cellulovorans cellulosome (25).
α-Galactosidase (EC 3.2.1.22) hydrolyzes α-galactosidic linkages at nonreducing ends in galactose-containing oligosaccharides, galactolipids, and galactomannan (4). Since galactomannan is a hemicellulosic material, the presence of α-galactosidase(s) in the cellulosomes should contribute to the degradation of plant cell walls.
The cipA gene, encoding the scaffolding protein, and some cellulase genes, namely, celB, celD, and celE, were identified in a gene cluster in the C. josui chromosomal DNA (10, 14). In addition, the celA (11) and xynA (7) genes were cloned and characterized along with their translated products. Since we expected that genes encoding cellulases and hemicellulases were clustered in C. josui, we sequenced wide areas around celA and cipA. In this process, we found the aga27A gene, encoding α-galactosidase and classified in family 27 of glycoside hydrolases (reference 13 and information found at the CNRS [Marseille, France] website [http://afmb.cnrs-mrs.fr/CAZY/]), upstream of celA. In this paper, we describe the nucleotide sequence of aga27A and enzyme properties of the recombinant Aga27A α-galactosidase.
Nucleotide sequence of the aga27A gene.
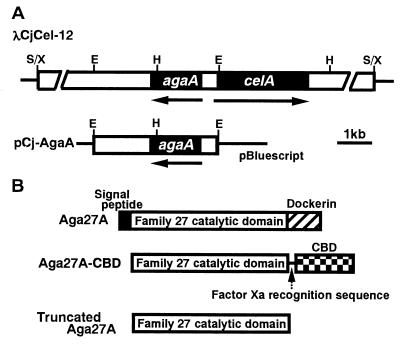
The aga27A gene was identified upstream of celA in λCjCel-12, but the direction of transcription of aga27A was opposite to that of celA (Fig. 1). A 3.5-kb EcoRI fragment of λCjCel-12 containing aga27A was subcloned into the EcoRI site of pBluescript II SK(−), yielding pCj-Aga27A (Fig. 1). The nucleotide sequence of aga27A was determined using an ABI PRISM 310 DNA sequencer system (Perkin-Elmer Applied Biosystems) with a BigDye Terminator sequencing kit (Perkin-Elmer Applied Biosystems) and a series of the subclones. Homology searches in the DDBJ database were carried out with the BLAST program (1).
FIG. 1.
Restriction maps of λCjCel-12 and pCj-AgaA (A) and domains of AgaA protein and its derivatives (B). (A) Thin lines correspond to the vector λGEM-12 or pBluescript II SK(−), and open bars correspond to cloned DNA fragments. The coding region is represented by solid bars. Arrows indicate the direction of the transcription of agaA and celA. E, EcoRI; H, HindIII; S, Sau3AI; X, XhoI.
As shown in Fig. 1, the open reading frame of aga27A consists of 1,434 nucleotides encoding a protein of 478 amino acids with a predicted molecular weight of 52,162. The assigned ATG initiation codon is preceded at a spacing of 8 bp by a potential ribosome-binding sequence (GGAGG), which was homologous with the consensus Shine-Dalgarno sequence (24).
Amino acid sequence of Aga27A.
The deduced N-terminal sequence of 23 amino acids is similar to the signal peptide sequences found in prokaryotic secretory proteins (27). Comparison of the amino acid sequence of Aga27A with those registered in protein databases such as SWISS-PROT and PIR showed that mature Aga27A consists of two distinct functional domains (Fig. 1), i.e., a family 27 catalytic domain of glycosyl hydrolases and a dockerin domain involved in cellulosome assembly (21). As shown in Fig. 2, the family 27 domain of Aga27A exhibited extensive sequence homology with some α-galactosidase catalytic domains in family 27 from various organisms, especially from plants, e.g., Coffea arabica (coffee; 55% sequence identity) (29), Glycine max (soybean; 54%) (DDBJ/EMBL/GenBank accession no. U12926), Phaseolus vulgaris (kidney bean; 54%) (U12927), Cyamopsis tetragonoloba (guar; 54%) (20), Senna occidentalis (senna; 54%) (A63585), Pseudomonas fluorescens subsp. cellulosa (51%) (12), Homo sapiens (human; 45%) (15), Mortierella vinacea (43%) (22, 23), Mus musculus (mouse; 42%) (19), and Saccharomyces carlsbergensis (40%) (26).
FIG. 2.
Alignment of family 27 catalytic domains of α-galactosidases from C. josui (Cj), Caffea arabica (Ca), Cyamopsis tetragonoloba (Ct), G. max (Gm), H. sapiens (Hs), Mus musculus (Mm), Mortierella vinacea (Mv), P. fluorescens subsp. cellulosa (Pf), and Phaseolus vulgaris (Pv). MvI and MvII are α-galactosidase I and α-galactosidase II, respectively, from Mortierella vinacea. Amino acids which are conserved in at least 7 of the 10 sequences are shaded. Dashes indicate gaps left to improve alignment. Numbers refer to amino acid residues at the beginnings of the respective lines; all sequences are numbered from Met-1 of the peptide.
α-Galactosidases are substantially classified into three groups, families 4, 27, and 36 of glycosyl hydrolases, on the basis of amino acid sequence homology (reference 13 and information found at the CNRS [Marseille, France] website [http://afmb.cnrs-mrs.fr/CAZY/]). Almost all of the α-galactosidases from eukaryotic organisms belong to family 27, although an α-galactosidase of Trichoderma reesei (accession no. Z69254) is a member of family 36. On the other hand, most prokaryotic α-galactosidases have been classified into family 4 or 36. Therefore, the C. josui Aga27A α-galactosidase is, along with P. fluorescens subsp. cellulosa Aga27A α-galactosidase (12), the rare exception.
A dockerin domain was found at the C terminus of Aga27A (Fig. 1). Dockerin domains which consist of two duplicated sequences, each of about 22 amino acid residues, are conserved in the components of cellulosomes from C. cellulolyticum, C. cellulovorans, C. josui, and C. thermocellum. Pagès et al. (21) showed that cohesin-dockerin interactions in the C. cellulolyticum and C. thermocellum cellulosomes are species-specific phenomena, and they predicted that four amino acid residues which comprise a repeated pair, AL or AI, in the C. cellulolyticum enzymes and ST in the C. thermocellum enzymes are critical to binding specificity as the recognition code. Recently, Mechaly et al. (17) reported the significance of these residues in the cohesin-dockerin interaction. The dockerin domains of C. josui CelB and CelD were shown to be similar to those of the C. cellulolyticum cellulases; i.e., an AL or AI motif is conserved in the dockerins from C. josui cellulases. A pair of AL motifs was found in the dockerin domain of Aga27A. The presence of a dockerin domain in Aga27A suggested that it is a component of the C. josui cellulosome.
Purification and characterization of the truncated Aga27A.
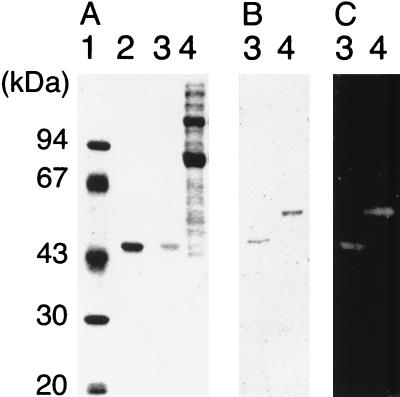
The region encoding the catalytic domain of Aga27A was amplified by PCR using two specific primers, 5′-TTGAATTCGGTGTTTATCTCACCATTTC-3′ and 5′-TTAAGCTTGCTTGTACTTTTAGTACCACT-3′. The PCR product was cloned in pCBD-C for overproduction of the gene product, yielding pCBD-Aga27A. (Fig. 1). Plasmid pCBD-C was obtained from Toyobo (Tokyo, Japan), which provides proteins containing a cellulose-binding domain (CBD) as an affinity tag at their C termini with a factor Xa recognition sequence. Cells of Escherichia coli BL21(pCBD-Aga27A), harvested from 250 ml of an overnight culture in Luria-Bertani broth, were disrupted by ultrasonication, and cell debris was removed by centrifugation. The cell extract was incubated with ball-milled cellulose (BMC) for 10 min on ice to allow Aga27A-CBD to bind to the BMC. The BMC pellet was recovered by centrifugation and washed twice with 0.1 M potassium phosphate buffer (pH 7.0). The resulting BMC pellet was suspended in 3 ml of 5% cellobiose solution in the same buffer (pH 7.0) to elute Aga27A-CBD, and BMC was removed by centrifugation. Aga27A-CBD in the supernatant was treated with trypsin at 4°C overnight to cleave the fusion protein into each moiety, i.e., the catalytic domain of Aga27A, referred to as the truncated Aga27A, and the CBD. The former was separated from the latter by chromatography with a HiLoad Superdex 75pg column (1.6 by 60 cm; Pharmacia Biotech). The final preparation (0.7 mg) gave a single band by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (16), and the molecular weight of the enzyme was estimated to be about 40,000 (Fig. 3A). This value is in good agreement with that deduced from the nucleotide sequence. The molecular weight of Aga27A estimated by gel filtration (about 40,000) suggested that it was a monomeric enzyme. The truncated Aga27A thus obtained was used for the characterization of its enzymatic properties.
FIG. 3.
Expression of AgaA in C. josui and E. coli. The gel was stained with Coomassie brilliant blue (A) or stained for α-galactosidase activity (C). AgaA proteins were detected with a polyclonal mouse antiserum raised against truncated AgaA by Western blot analysis (B). Lane 1, protein mass standards; lane 2, truncated AgaA purified from a recombinant E. coli strain (1 μg); lanes 3, truncated AgaA purified from a recombinant E. coli strain (0.1 μg); lanes 4, cellulosomal proteins of C. josui.
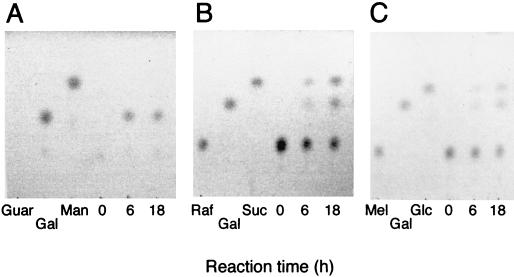
α-Galactosidase activity was measured after a 3-min incubation at 45°C in McIlvaine buffer (a mixture of 0.2 M Na2HPO4 and 0.1 M citric acid [pH 5.5]) in the presence of 10 mM p-nitrophenyl-α-d-galactopyranoside (PNP-Gal; Sigma). One unit of activity was defined as the amount of enzyme releasing 1 μmol of p-nitrophenol from PNP-Gal per min. When melibiose, raffinose, and guar gum (Sigma) were used as the substrates, reducing sugars released from the substrates were measured with the 3,5-dinitrosalicylic acid reagent as described by Miller (18). One unit of activity was defined as the amount of enzyme releasing 1 μmol of galactose from the substrates per min. From Lineweaver-Burk plots, the Km and Vmax values of the enzyme for PNP-Gal were estimated to be 0.81 mM and 92.9 μmol/min/mg, respectively. The optimum pH for activity on PNP-Gal was found to be pH 5.5, and the enzyme was stable in the range of pH 3.0 to 7.0 when it was incubated in buffers of different pHs without the substrate at 30°C for 3 h. The optimum temperature for activity on PNP-Gal was found to be 58°C at pH 5.5. The enzyme was stable at 40°C for 10 min at pH 5.5 in the absence of the substrate. The enzyme displayed similar optimal activities and stabilities for raffinose and guar gum. The truncated Aga27A hydrolyzed the α-1,6-galactoside linkage in guar gum as well as in melibiose and raffinose. The specific activities of the enzyme were 8.3 U/mg for guar gum and 1.7 U/mg for raffinose. The value for guar gum is comparable with that for a T. reesei enzyme, although Aga27A appears to be less active toward raffinose than the fungal enzyme is; i.e., the specific activities of a T. reesei α-galactosidase were 15.3 and 15.2 U/mg for locust bean gum and raffinose, respectively (28). The Km values for Aga27A were determined to be 2.2 and 21.0 mg/ml for guar gum and raffinose, respectively. The release of galactose from guar gum, raffinose, melibiose, and lactose was analyzed by thin-layer chromatography with 1-propanol-nitromethane-water (5:2:3, vol/vol/vol) as the development solvent. The sugars on the plate were visualized with sulfuric acid by heating at 140°C for 5 min. Galactose was detected in the hydrolysates of guar gum, raffinose, and melibiose but not lactose (Fig. 4), showing that the activity of Aga27A is specific for the α-1,6 linkage.
FIG. 4.
Thin-layer chromatography of hydrolysis products from guar gum (A), raffinose (B), and melibiose (C). The reaction mixture was composed of 80 μl of 1% substrate, 80 μl of McIlvaine buffer (pH 5.5), and 40 μl (0.8 U) of enzyme solution. The reaction was done at 35°C, and 20 μl of the reaction mixture was withdrawn at each time indicated. Four microliters of the mixture was used for thin-layer chromatography. Guar, guar gum; Mel, melibiose; Raf, raffinose; Gal, galactose; Glc, glucose; Suc, sucrose; Man, mannanase.
α-Galactosidases can be classified into two groups based on their substrate specificities (5); i.e., one group is specific for small saccharides such as PNP-Gal, melibiose, and raffinose, and the other group can liberate galactose from highly polymerized galactomannans such as guar gum in addition to the small substrates. Some enzymes in family 27 are known to be more active on guar gum than on the small substrates (5). C. josui Aga27A also prefers guar gum to the small substrates. By contrast, bacterial α-galactosidases in family 4 or 36 show high activities toward the small substrates but negligible activities toward guar gum (8, 9). Although P. fluorescens subsp. cellulosa Aga27A, classified in family 27, was capable of hydrolyzing carob galactomannan, its enzyme activity toward this substrate was quite weak, i.e., 0.019 U/mg (12).
Detection of Aga27A in the cellulosomal proteins of C. josui.
Antiserum against the truncated Aga27A was raised in a mouse. The cellulosome was purified from the culture supernatant of C. josui grown on BMC as described previously (14). By Western blot analysis using the antiserum, a major immunoreactive protein with an apparent molecular weight of 52,000 was detected in the cellulosomal proteins (Fig. 3B). The size of this immunoreactive protein was in good agreement with that calculated from the deduced amino acid sequence of Aga27A. Zymogram analysis using 4-methylumbelliferyl-α-d-galactopyranoside as the substrate identified an α-galactosidase with a molecular weight of about 52,000 (Fig. 3C). These results indicate that Aga27A is one of the components of the C. josui cellulosome.
Aga27A was detected in the cellulosome fraction prepared from the culture supernatant of C. josui grown on BMC. Although this bacterium grew quite slowly on guar gum as the carbon source, Western blot analysis indicated that the production of Aga27A was not enhanced by the addition of guar gum (data not shown). These observations suggest that the expression of the aga27A gene is not induced by guar gum but is related to cellulose degradation. It is likely that Aga27A contributes to the degradation of galactomannan present in plant cell walls, allowing the cellulosome access to cellulose chains that are buried in galactomannan and are not accessible unless galactomannan is hydrolyzed and removed.
Nucleotide sequence accession number.
The nucleotide sequence of aga27A reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with accession number AB025362.
Acknowledgments
This work was supported in part by a grant of Rice Genome Project PR-2211, MAFF, Tokyo, Japan.
We thank Yoshikazu Ishida of Toyobo Co., Ltd. for the gift of pCBD-C.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., L. J. W. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes—structure and ultrastructure. J. Struct. Biol. 124:221–234. [DOI] [PubMed] [Google Scholar]
- 3.Bélaich, J.-P., C. Tardif, A. Bélaich, and C. Gaudin. 1997. The cellulolytic system of Clostridium cellulolyticum. J. Biotechnol. 57:3–14. [DOI] [PubMed] [Google Scholar]
- 4.Dey, P. M., and E. D. Campillo. 1984. α-Galactosidase. Adv. Enzymol. Relat. Areas Mol. Biol. 56:141–249. [DOI] [PubMed] [Google Scholar]
- 5.Dey, P. M., S. Patel, and M. D. Brownleader. 1993. Induction of alpha-galactosidase in Penicillium ochrochloron by guar (Cyamopsis tetragonoloba) gum. Biotechnol. Appl. Biochem. 17:361–371. [PubMed] [Google Scholar]
- 6.Doi, R. H., M. Goldstein, S. Hashida, J.-S. Park, and M. Takagi. 1994. The Clostridium cellulovorans cellulosome. Crit. Rev. Microbiol. 20:87–93. [DOI] [PubMed] [Google Scholar]
- 7.Feng, J.-X., S. Karita, E. Fujino, T. Fujino, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Cloning, sequencing and expression of the gene encoding a cell-bound multi-domain xylanase from Clostridium josui and characterization of the translated product. Biosci. Biotechnol. Biochem. 64:2614–2624. [DOI] [PubMed] [Google Scholar]
- 8.Fridjonsson, O., H. Watzlawick, A. Gehweiler, and R. Mattes. 1999. Thermostable α-galactosidase from Bacillus stearothermophilus NUB3621: cloning, sequencing and characterization. FEMS Microbiol. Lett. 176:147–153. [DOI] [PubMed] [Google Scholar]
- 9.Fridjonsson, O., H. Watzlawick, A. Gehweiler, T. Rohrhirsch, and R. Mattes. 1999. Cloning of the gene encoding a novel thermostable α-galactosidase from Thermus brockianus ITI360. Appl. Environ. Microbiol. 65:3955–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujino, T., S. Karita, and K. Ohmiya. 1993. Nucleotide sequences of the celB gene encoding endo-1,4-β-glucanase-2, ORF1 and ORF2 forming a putative cellulase gene cluster of Clostridium josui. J. Ferment. Bioeng. 76:243–250. [Google Scholar]
- 11.Fujino, T., and K. Ohmiya. 1992. Nucleotide sequences of an endo-1,4-β-glucanase gene (celA) from Clostridium josui. J. Ferment. Bioeng. 73:308–313. [Google Scholar]
- 12.Halstead, J. R., M. P. Fransen, R. Y. Eberhart, A. J. Park, H. J. Gilbert, and G. P. Hazlewood. 2000. α-Galactosidase A from Pseudomonas fluorescens subsp. cellulosa: cloning, high level expression and its role in galactomannan hydrolysis. FEMS Microbiol. Lett. 192:197–203. [DOI] [PubMed] [Google Scholar]
- 13.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornreich, R., R. J. Desnick, and D. F. Bishop. 1989. Nucleotide sequence of the human α-galactosidase A gene. Nucleic Acids Res. 17:3301–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 17.Mechaly, A., H. P. Fierobe, A. Bélaich, J. P. Bélaich, R. Lamed, Y. Shoham, and E. A. Bayer. 2001. Cohesin-dockerin interaction in cellulosome assembly: a single hydroxyl group of a dockerin domain distinguishes between nonrecognition and high affinity recognition. J. Biol. Chem. 276:9883–9888. [DOI] [PubMed] [Google Scholar]
- 18.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428. [Google Scholar]
- 19.Ohshima, T., G. J. Murray, J. W. Nagle, J. M. Quirk, M. H. Kraus, N. W. Barton, R. O. Brady, and A. B. Kulkarni. 1995. Structural organization and expression of the mouse gene encoding α-galactosidase A. Gene 166:277–280. [DOI] [PubMed] [Google Scholar]
- 20.Overbeeke, N., A. J. Fellinger, M. Y. Toonen, D. van Wassenaar, and C. T. Verrips. 1989. Cloning and nucleotide sequence of the α-galactosidase cDNA from Cyamopsis tetragonoloba (guar). Plant Mol. Biol. 13:541–550. [DOI] [PubMed] [Google Scholar]
- 21.Pagès, S., A. Bélaich, J.-P. Bélaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517–527. [PubMed] [Google Scholar]
- 22.Shibuya, H., H. Kobayashi, K. Kasamo, and I. Kusakabe. 1995. Nucleotide sequence of α-galactosidase cDNA from Mortierella vinacea. Biosci. Biotechnol. Biochem. 59:1345–1348. [DOI] [PubMed] [Google Scholar]
- 23.Shibuya, H., H. Kobayashi, T. Sato, W. Kim, S. Yoshida, S. Kaneko, K. Kasamo, and I. Kusakabe. 1997. Purification, characterization, and cDNA cloning of a novel α-galactosidase from Mortierella vinacea. Biosci. Biotechnol. Biochem. 61:592–598. [DOI] [PubMed] [Google Scholar]
- 24.Shine, J., and L. Dalgarno. 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254:34–38. [DOI] [PubMed] [Google Scholar]
- 25.Tamaru, Y., and R. H. Doi. 2001. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. USA 98:4125–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turakainen, H., M. Korhola, and S. Aho. 1991. Cloning, sequence, and chromosomal location of a MEL gene from Saccharomyces carlsbergensis NCYC396. Gene 101:97–104. [DOI] [PubMed] [Google Scholar]
- 27.Watson, M. E. E. 1984. Compilation of published signal sequences. Nucleic Acids Res. 12:5145–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeilinger, S., D. Kristufek, I. Arisan-Atac, R. Hodits, and C. P. Kubicek.1 993. Conditions of formation, purification, and characterization of an α-galactosidase of Tricoderma reesei RUT C-30. Appl. Environ. Microbiol. 59:1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, A., and J. Goldstein. 1994. Cloning and functional expression of a cDNA encoding coffee bean α-galactosidase. Gene 140:227–231. [DOI] [PubMed] [Google Scholar]