Abstract
By using a panel of 603 commensal and pathogenic Escherichia coli and Shigella isolates, we showed that mutation rates of strains vary considerably among different ecotypes. Uropathogenic strains had the highest frequency of mutators, while strains from patients with bacteremia had the lowest mutation rates. No correlation between the mutation rates and antibiotic resistance was observed among the studied strains.
Bacterial populations with a high level of genetic variability have a higher probability of survival in constantly changing environments (18). Since genetic variability is generated mostly by mutagenesis, bacterial strains with high mutation rates are expected to have higher capacities for adaptation. Such strains are favored by selection when the advantage of beneficial mutations is greater than the cost of being a mutator due to the overproduction of lethal and deleterious mutations (6, 17, 19). Mutator strains, having a defective mismatch repair system, have indeed been observed in natural populations of Escherichia coli, Salmonella enterica, Neisseria meningitidis, and Pseudomonas aeruginosa (5, 7, 10, 12). Because most of these isolates are pathogens, it has been hypothesized that mutator and hyperrecombination phenotypes may accelerate the evolution of pathogenic strains by, e.g., increasing the variation of surface antigens, as well as by facilitating the acquisition of pathogenic determinants and antibiotic resistance. Indeed, it has been observed that the levels of resistance to antibiotics were significantly higher in mutator than in nonmutator pathogenic P. aeruginosa isolates (10) and that mismatch repair-deficient N. meningitidis strains displayed high phase variation rates (12).
However, from the available data, it is not clear whether high mutation rates are particularly important for the evolution of pathogens in general or for the evolution of only some pathogenic groups (5, 7, 10, 11). Furthermore, a mutator phenotype may not be specific to pathogens, since mutators have also been observed in commensal populations (7). In order to examine the link between a particular bacterial lifestyle, mutation rate, and antibiotic resistance, we used a collection of 603 human E. coli (including Shigella) isolates, either commensal isolates or ones involved in various pathologies, such as enteroinvasive and enterohemorrhagic diseases, urinary tract infection (UTI), bacteremia, pus production from miscellaneous infections, and newborn meningitis (NBM). A detailed list of strains is given in Table 1.
TABLE 1.
Strains studied
| Strain ecotype or pathology | Total no. of strains | Origin(s)a (no. of strains) | Reference(s) or source |
|---|---|---|---|
| Bacteremia | 93 | Paris (three university hospitals) (88), Avignon (one general hospital) (5) | 4a, 11a; unpublished data |
| Commensal organisms | 217 | Croatia (61), Mali (57), France (70), ECOR collection (Sweden and United States) (29) | 2a, 4a, 9a |
| Enteroinvasive disease | 86 | Mainly France: Shigella (73), EIEC (13) | 13a |
| Enterohemorrhagic disease | 26 | France | Unpublished data |
| NBM | 60 | Mainly France | 1 |
| Pus from miscellaneous infections | 30 | Paris (two university hospitals) (24), Avignon (one general hospital) (5), Paris (outpatient) (1) | 11a; unpublished data |
| UTI | 91 | Paris (one university hospital) (49), Avignon (one general hospital) (21), Paris (outpatient) (10), ECOR collection (Sweden) (11) | 9a, 11a; unpublished data |
ECOR, E. coli reference; ECEI, enteroinvasive E. coli.
Variations of mutation rates.
The mutation rates of the studied strains were estimated by monitoring the strains’ capacities to generate mutations conferring resistance to rifampin in at least six independent cultures for each strain (Fig. 1 and 2), as described previously (16). Between 102 and 103 cells from an overnight culture were inoculated onto nitrocellulose filters (NC45; Schleicher and Schuell) laid on plates containing fresh 869 medium (NaCl, 5 g/liter; Bacto Tryptone, 10 g/liter; yeast extract, 5 g/liter; agar, 15 g/liter). The plates were incubated at 37°C for 24 h. The cells were resuspended in 1 ml of 869 medium and incubated for 1 h at 37°C to allow for rifampin resistance expression. Appropriate dilutions were then spread on 869 medium plates with rifampin (100 μg/ml; Sigma) or without. The rifampin-resistant mutants were counted after 24 h at 37°C.
FIG. 1.
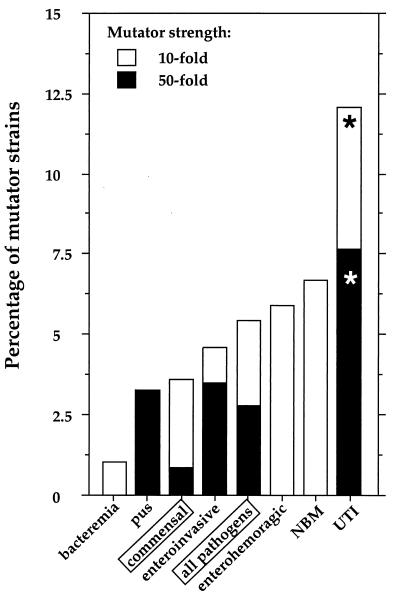
Mutator strains belonging to different human E. coli and Shigella commensal and pathogenic groups. Bacterial strains are grouped according to their origins and the different pathologies in which they have been involved, including bacteremia strains (n = 93), strains isolated from pus (n = 30), commensal strains (n = 217), enteroinvasive strains (n = 86), enterohemorrhagic strains (n = 26), NBM strains (n = 60), and UTI strains (n = 91). In addition, all pathogenic strains are presented as one group in order to facilitate a comparison of that group with commensal strains. Strains were considered mutators when they exhibited frequencies of mutations conferring resistance to rifampin (100 μg/ml) that were 10-fold higher than the median value of mutagenesis (5.04 × 10−9) observed for all studied strains (n = 603) (10-fold mutators). Strains that displayed a >50-fold increase in mutagenesis were considered strong mutators (50-fold mutators). Percentages of mutator strains were calculated for every group of studied strains. UTI strains had significantly higher (according to the χ2 test) fractions (*) of 10- and 50-fold mutators than commensal strains (P = 0.005 and P = 0.001 for 10- and 50-fold mutators, respectively), as well as other pathogens (P = 0.001 and P = 0.001 for 10- and 50-fold mutators, respectively).
FIG. 2.
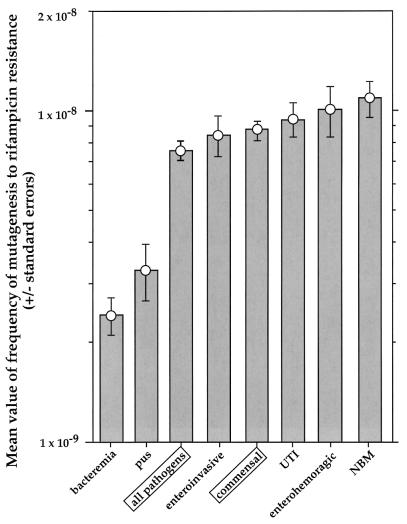
Variability of mutation rates of human E. coli and Shigella isolates after elimination of mutators. The mean value (± standard error) of mutation frequency, after removal of those for 10- and 50-fold mutators (see legend to Fig. 1), is shown for each group.
We found mutators among commensals and pathogens, but the frequencies of mutators in these two groups of strains were not significantly different (Fig. 1 and 2). However, when pathogenic strains were analyzed as members of different ecotypes, mutator strains were found to occur significantly more frequently among UTI strains than they did among commensals and also more frequently among UTI strains than among all other pathogenic strain groups. It is interesting to note that bacteremia strains, which include urosepsis isolates, had the smallest fraction of mutator strains but also, significantly (according to the t test), the lowest mutation rates of all strains (Fig. 1 and 2). Strains isolated from pus also have significantly lower mutation rates than all other strains (except bacteremia strains) (Fig. 1 and 2).
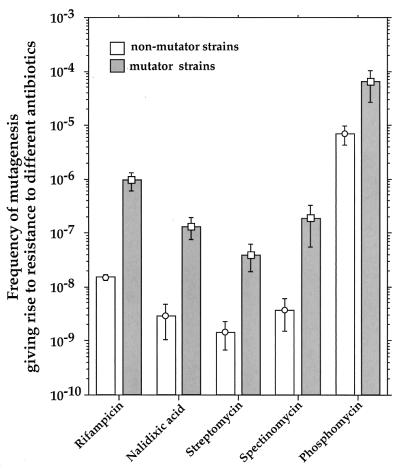
We confirmed that strains generating rifampin-resistant mutants at a high rate correspond to generalized mutators by measuring the frequencies of the mutations that confer resistance to the following four additional antibiotics (at indicated concentrations) in at least six independent cultures for each strain: nalidixic acid (40 μg/ml), phosphomycin (30 μg/ml), spectinomycin (100 μg/ml), and streptomycin (100 μg/ml) (all from Sigma) (Fig. 3).
FIG. 3.
Capacities of mutator strains to generate mutations conferring resistance to different antibiotics. The results are presented as mean values (± standard errors) for mutator (>10-fold increase in mutagenesis; n = 21) and nonmutator (n = 47) strains.
Why do UTI strains have the highest frequency of mutator strains?
One possible explanation for the high frequency of mutators in UTI strains is that mutator strains belong to one clone which has increased in frequency in populations of UTI strains due to the action of positive selection. However, we found that strong UTI mutator strains belong to different E. coli phylogenetic groups: A, B2, and D. Furthermore, by sequencing metabolic genes (trpA, trpB, putP, and papB), we have also confirmed that the group B2 UTI mutator strains (most abundant among UTI strains) did not belong to the same clone (data not shown). Therefore, our data suggest that there is no correlation between mutation rate and phylogenetic group.
The possibility that UTI mutators are better adapted to growth in urine, due to the acquisition of adaptive mutations or to a pleiotropic effect linked to a modified DNA repair ability, seems to be marginal at best, since both mutator and nonmutator UTI strains grow easily in fresh urine (with no significant difference between them), reaching concentrations of about 108 CFU/ml (data not shown).
Another possibility is that mutators are less frequently counterselected in the urinary tract than in other body compartments. It has been demonstrated that mutators can suffer a reduction of fitness due to the accumulation of deleterious mutations (3, 4). One of the measurable phenotypes of fitness reduction is loss of the capacity to grow on minimal synthetic medium. This handicap might be less important, at least in the short run, in urine, as suggested by a higher incidence of auxotrophs (25%) among UTI strains than among strains from fecal samples (5.8%) (13). However, we did not observe more auxotrophs among UTI mutator strains than among UTI nonmutators (data not shown).
Finally, it is possible that UTI mutators are selected because they generate mutations that increase adaptation to the urinary tract at a higher rate than that generated by nonmutators. For example, it has been shown that point mutations in fimH genes increase binding of the adhesin to monomannose residues, structures that are abundant in the urothelial glycoproteins, conferring increased virulence in a UTI mouse model (15) as well as an increased capacity for biofilm formation (14). However, this hypothesis must be confirmed by in vivo and in vitro reconstruction experiments.
Antibiotic resistance.
It is possible that antibiotic treatments contribute to selection of the mutators, as has been demonstrated in in vitro experiments (6). Mutators can be favored under such conditions because they generate antibiotic resistance-conferring mutations at a higher rate than that generated by nonmutators (Fig. 3). In addition, they also generate more mutations that compensate for the fitness reduction associated with antibiotic resistance (1a).
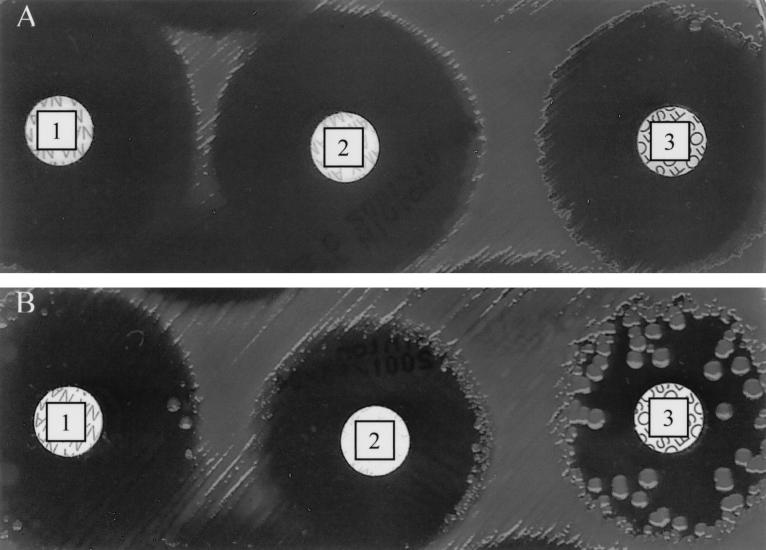
Since most of the antibiotics we used to demonstrate the mutator phenotype of the strains were not of medical relevance, we performed standard antibiogram testing of the activities of amikacin, amoxicillin, amoxicillin-clavulanic acid, ceftazidime, ciprofloxacin, nalidixic acid, trimethoprim-sulfamethoxazole, and phosphomycin (8), as well as a determination of the MICs of ceftazidime, amikacin, and ciprofloxacin (9) for 26 mutator and 42 nonmutator strains. The majority of mutator strains yielded colonies inside the growth inhibition zone (squatter colonies), while no nonmutator strains exhibited that phenotype (Fig. 4). The presence of squatter colonies reflects the high frequency of mutations conferring resistance to antibiotics. The squatter colonies were not observed only when ceftazidime or ciprofloxacin was used. However, mutator strains are not more resistant than nonmutators are, and no mutator strain was resistant to multiple antibiotics (data not shown).
FIG. 4.
Squatter colonies inside growth inhibition zone. Growth inhibitory zones for nalidixic acid (disk 1), amoxicillin (disk 2), and phosphomycin (disk 3) are presented for nonmutator (A) and mutator (B) strains. Note the presence of squatter colonies for the mutator strain only.
Furthermore, additional UTI strains that are resistant to quinolones (n = 9) or that have an overexpressed cephalosporinase (n = 7) (both resistance mechanisms resulting from point mutations) did not show a higher mutation rate than nonmutator strains (data not shown). Therefore, it can be concluded that antibiotics are probably not the major selective pressure that favors mutator strains in natural E. coli populations.
Conclusions.
Although UTI strains have the highest frequency of mutators, the link between high mutation rates and pathogenicity cannot be generalized. Other pathogenic groups do not have more mutators than commensal organisms do. Furthermore, bacteremia and pus isolates have very low mutation rates (Fig. 1 and 2). The reason for the observed high frequency of mutators in populations of UTI strains remains to be determined.
Our finding that mutators are present in almost all studied groups of E. coli ecotypes supports recently published observations which suggest that the majority of E. coli strains repeatedly pass through periods of high mutation rates during their evolutionary history, regardless of whether they are commensal or pathogenic or to which phylogenetic group they belong (2).
Acknowledgments
We thank Jean-Pierre Coutenceau for technical assistance and E. Stewart and O. Tenaillon for critical reading of the manuscript.
This work was supported by grants from the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires—MENRT and the Programme Environnement et Santé—MATE.
REFERENCES
- 1.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642–650. [DOI] [PubMed] [Google Scholar]
- 1a.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479–1482. [DOI] [PubMed] [Google Scholar]
- 2.Denamur, E., G. Lecointre, P. Darlu, O. Tenaillon, C. Acquaviva, C. Sayada, I. Sunjevaric, R. Rothstein, J. Elion, F. Taddei, M. Radman, and I. Matic. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103:711–721. [DOI] [PubMed] [Google Scholar]
- 2a.Duriez, P., O. Clermont, S. Bonacorsi, E. Bingen, A. Chaventre, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology (Reading) 147:1671–1676. [DOI] [PubMed] [Google Scholar]
- 3.Funchain, P., A. Yeung, J. L. Stewart, R. Lin, M. M. Slupska, and J. H. Miller. 2000. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics 154:959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606–2608. [DOI] [PubMed] [Google Scholar]
- 4a.Hilali, F., R. Ruimy, P. Saulnier, C. Barnabé, C. Lebouguénec, M. Tibayrenc, and A. Andremont. 2000. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect. Immun. 68:3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208–1211. [DOI] [PubMed] [Google Scholar]
- 6.Mao, E. F., L. Lane, J. Lee, and J. H. Miller. 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179:417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matic, I., M. Radman, F. Taddei, B. Picard, C. Doit, E. Bingen, E. Denamur, and J. Elion. 1997. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277:1833–1834. [DOI] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 1993. Performance for antimicrobial disk susceptibility tests, 5th ed. Approved standard M2-A5. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 9.National Committee for Clinical Laboratory Standards. 1998. Performance for antimicrobial susceptibility testing, 8th informational supplement. M100-S8. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9a.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. [DOI] [PubMed] [Google Scholar]
- 11.Picard, B., P. Duriez, S. Gouriou, I. Matic, E. Denamur, and F. Taddei. 2001. Mutator natural Escherichia coli isolates have an unusual virulence phenotype. Infect. Immun. 69:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson, A. R., and I. Stojiljkovic. 2001. Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol. 40:645–655. [DOI] [PubMed] [Google Scholar]
- 13.Robeson, J. P., R. M. Goldschmidt, and R. D. Curtiss. 1980. Potential of Escherichia coli isolated from nature to propagate cloning vectors. Nature 283:104–106. [DOI] [PubMed] [Google Scholar]
- 13a.Rolland, K., N. Lambert-Zechovsky, B. Picard, and E. Denamur. 1998. Shigella and enteroinvasive Escherichia coli strains are derived from distinct ancestral strains of E. coli. Microbiology (Reading) 144:2667–2672. [DOI] [PubMed] [Google Scholar]
- 14.Schembri, M. A., and P. Klemm. 2001. Biofilm formation in a hydrodynamic environment by novel FimH variants and ramifications for virulence. Infect. Immun. 69:1322–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokurenko, E. V., V. Chesnokova, D. E. Dykhuizen, I. Ofek, X. R. Wu, K. A. Krogfelt, C. Struve, M. A. Schembri, and D. L. Hasty. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA 95:8922–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taddei, F., I. Matic, and M. Radman. 1995. Cyclic AMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736–11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taddei, F., M. Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon, and B. Godelle. 1997. Role of mutators in adaptive evolution. Nature 387:700–702. [DOI] [PubMed] [Google Scholar]
- 18.Taddei, F., M. Vulic, M. Radman, and I. Matic. 1997. Genetic variability and adaptation to stress, p.271–290. In R. Bijlsma and V. Loeschcke (ed.), Environmental stress, adaptation, and evolution, vol. EXS 83. Birkhäuser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 19.Tenaillon, O., B. Toupance, H. Le Nagard, F. Taddei, and B. Godelle. 1999. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]