Abstract
We show that both flanking IS256 elements carried by transposon Tn4001 are capable of generating head-to-tail tandem copies and free circular forms, implying that both are active. Our results suggest that the tandem structures arise from dimeric copies of the donor or vector plasmid present in the population by a mechanism in which an IS256 belonging to one Tn4001 copy attacks an IS256 end carried by the second Tn4001 copy. The resulting structures carry abutted left (inverted left repeat [IRL]) and right (inverted right repeat [IRR]) IS256 ends. Examination of the junction sequence suggested that it may form a relatively good promoter capable of driving transposase synthesis in Escherichia coli. This behavior resembles that of an increasing number of bacterial insertion sequences which generate integrative junctions as part of the transposition cycle. Sequence analysis of the IRL-IRR junctions demonstrated that attack of one end by the other is largely oriented (IRL attacks IRR). Our experiments also defined the functional tips of IS256 as the tips predicted from sequence alignments, confirming that the terminal 4 bp at each end are indeed different. The appearance of these multiple plasmid and transposon forms indicates that care should be exercised when Tn4001 is used in transposition mutagenesis. This is especially true when it is used with naturally transformable hosts, such as Streptococcus pneumoniae, in which reconstitution of the donor plasmid may select for higher-order multimers.
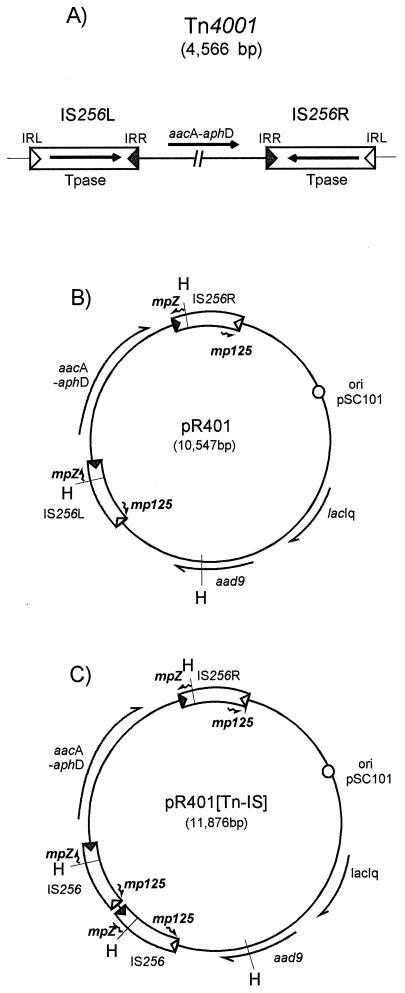
Transposon Tn4001 was originally isolated from Staphylococcus aureus (22) and has proved to be a useful tool for genetic analysis of several gram-positive bacteria, including Streptococcus gordonii (18), and mollicutes, such as the mycoplasmas (7, 10), spiroplasmas (8, 9), and acholeplasmas (41). This compound transposon is composed of the aacA-aphD genes, conferring resistance to gentamicin, kanamycin, and tobramycin, flanked by two copies of IS256 in inverted orientation (Fig. 1A) (22).
FIG. 1.
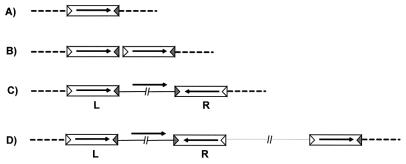
Maps of Tn4001 and vector plasmids. (A) Tn4001. The flanking left (IS256L) and right (IS256R) insertion sequences are indicated by open boxes. IRL and IRR are indicated by open and grey triangles, respectively. The directions of transcription of the IS256 transposase (Tpase) gene and of the antibiotic resistance gene are indicated by arrows. The IS256 copies were designated left and right with respect to the orientation of transcription of the aacA-aphD gene region. This is the inverse of the nomenclature of Byrne et al. (3). (B) Map of pR401. The Tn4001 component insertion sequences are indicated as described above. The short zig-zag arrows indicate oligonucleotides. The positions and directions of transcription of the transposon-associated antibiotic resistance (aacA-aphD), plasmid-associated resistance (aad9), and lacIq genes are indicated by arrows. The plasmid origin of replication derived from pSC101 is indicated by an open circle. H, HindIII restriction sites. (C) Map of pR401[Tn-IS]. The symbols are as described above. This map shows a head-to-tail tandem dimer of IS256 located at the left end of Tn4001. Note that molecules with this configuration (pR401[Tn-IS256L]) and molecules in which the duplicated IS is located at the right end (pR401[IS256R]) have been observed.
Although Tn4001 has been used to mutagenize various gram-positive bacteria, the initial evidence suggested that some of the transposition products might be complex (20). We also observed complex transposition behavior during Tn4001-mediated transposition mutagenesis of Streptococcus pneumoniae. These results prompted us to investigate transpositional recombination of Tn4001 in more detail.
IS256 itself is the founding member of a family of insertion sequences which includes examples from a variety of gram-positive and gram-negative bacterial species (4, 23). It is 1,324 bp long and carries 26-bp terminal imperfect inverted repeats (an inverted left repeat [IRL] and an inverted right repeat [IRR]); in the case of IS256, but not in the majority of family members, these inverted repeats diverge at the terminal dinucleotides. IS256 generally generates 8-bp direct flanking target repeats upon insertion and includes a single long open reading frame encoding the transposase. Very little is known about the transposition mechanism of this family of elements (4, 23).
Like many transposases, that of IS256 exhibits a potential DDE motif assumed to be part of its catalytic site (11, 13). For the elements encoding DDE transposases whose activities have been investigated, the chemical steps catalyzed by the enzymes are quite similar, if not identical; they promote single-strand cleavage (hydrolysis) at the ends of the transposon to generate a 3′ OH group (3′ ends), which attacks the target DNA in a second transesterification reaction, and this results in transfer of the corresponding DNA strands to the target site (26). In spite of these similarities in the reaction mechanisms, transposition pathways can vary quite considerably from element to element. The differences are determined by the strategy adopted for treatment of the second, complementary transposon strand (5′ ends) (45).
Several distinct pathways have been adopted for processing the 5′ transposon end. One major pathway adopted by members of several IS families, including the IS3 family (16, 37) and probably the IS21 and IS30 families (1, 15), involves an initial single-strand cleavage at only one end, the donor end. The free 3′ OH formed is then directed to attack the opposite end on the same DNA strand, the target end, to generate an intermediate in which the two ends are joined by a single-strand bridge. This is then converted into a transposon circle (29; G. Duval-Valentin and M. Chandler, unpublished data). The transposon circle is characterized by a reactive junction in which abutted left and right IS ends (IRL and IRR) are separated by a short spacer whose length is characteristic for the particular IS and which is derived from nucleotides flanking the target end. Each end then undergoes single-strand cleavage to liberate a 3′ OH. The resulting OH groups again act as nucleophiles in a final strand transfer step into the insertion site, which results in integration of the transposon circle. Reactive junctions have been observed for several ISs, including members of the IS1 (39, 44), IS3 (38, 47), IS21 (1, 32), IS30 (27), IS110 (28, 40), and ISL3 families (14). These structures may arise either by circularization of the IS or by tandem duplication (15, 46) and are more active as substrates in transposition than are ISs with distant flanking ends (1, 43).
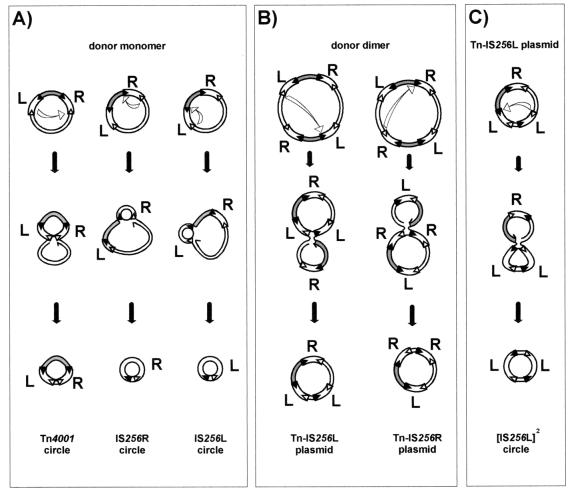
In the case of IS256 (Tn4001), early studies of Staphylococcus aureus (20) revealed structures which resembled tandem IS copies. In this study, we confirmed and extended the initial observations and showed that tandem IS copies are generated from Tn4001 in Escherichia coli. We also observed and characterized circular copies of IS256. Our results suggest that IS256 and presumably other members of the same IS family have a transposition pathway which can use a reactive IRR-IRL junction. As in the case of other ISs which form directly repeated tandem copies, generation of these structures probably involves prior creation of a dimeric donor plasmid molecule (15, 46) (see Fig. 5).
FIG. 5.
Possible intermediate transposition steps leading to integrative IRR-IRL junctions. (A) IRL from one IS256 copy attacks an IRL of the second IS256 copy, leading to circularization of Tn4001; IRL attacks IRR carried by the same IS256 copy, leading to circularization of IS256L or IS256R. (B) IRL of IS256L from one Tn4001 copy attacks IRR of IS256L from the second Tn4001 copy, leading to formation of a pR401[Tn-IS256L] tandem structure; IRL of IS256R from one Tn4001 copy attacks IRR of IS256R from the other Tn4001 copy, leading to formation of a pR401[Tn-IS256R] tandem structure. (C) External IRL of the tandemly repeated IS256L attacks the IRR carried by the second IS256L copy, leading to circularization of the tandemly repeated IS256L.
In studies of S. pneumoniae in which we used a nonreplicative plasmid carrying Tn4001 introduced by natural transformation, we observed that, in a significant proportion of mutants, insertion of Tn4001 was accompanied by insertion of the donor plasmid backbone. This result may be a direct consequence of transposition from plasmid dimers in the population in which the DNA segment which transposes includes one of the two plasmid copies together with the flanking transposable element, as observed for IS1 (2). Alternatively, it may be due to the formation of transposon-IS structures from plasmid dimers, as observed in this study, and integration promoted by a resulting reactive IRL-IRR junction. Although at this time we are not able to distinguish between these possibilities, our results clearly underline a need for caution when Tn4001 is used as a mutagen in S. pneumoniae (and presumably in other natural transformation systems).
MATERIALS AND METHODS
Bacterial strains and media.
The E. coli strains used for plasmid construction and preparation were LN2843 (F− thy leu rpsL lacI::amp), GM2163 [F− ara-14 leuB6 thi-1 fhuA31 lacY1 tsx-78 galT22 supE44 hisG4 rpsL136 xyl-5 mtl-1 dam-13::Tn9 dcm-6 mcrB1 hsdR2(rK− mK+) mcrA], and CT45 [K-12] (F− mcrA thr-1 leuB6 thi-1 lacY1 supE44 tonA21 recA1). Cultures were grown in Luria-Bertani broth (35) supplemented, when necessary, with spectinomycin (50 μg/ml). Plasmids were introduced by electroporation by using a Bio-Rad gene pulser as recommended by the manufacturer. E. coli recombinants were selected on L plates with appropriate antibiotics (obtained from Sigma).
Wild-type nonencapsulated S. pneumoniae strain R800 was used to detect Tn4001 transposition. In all experiments, S. pneumoniae was grown in Casamino Acids-tryptone medium (30), and precompetent and competent cells were prepared as described previously (24). The precompetent cells were activated with synthetic competence-stimulating peptide 1 at a concentration of 25 ng/ml (12). DNA carrying Tn4001 was added at a concentration of 5 or 60 μg/ml. Transformants carrying transposition products were selected on D blood agar plates with 20 μg of gentamicin per ml. As the frequency of Genr transformants depends on the competent cell batch, pR401[Tn-IS] DNA was used to normalize and estimate the ratio between the different plasmid forms. Typically, we obtained frequencies of Genr colonies between 2 × 10−6 and 4 × 10−6 with dam dcm unmethylated pR401[Tn-IS] DNA. Each transformation experiment was repeated with several different cell batches.
Plasmids.
Plasmid pα (18, 19) containing Tn4001 was kindly provided by R. Lunsford; pR401 was constructed by ligating the 5,016-bp BamHI-KpnI restriction fragment from pR217 containing the pSC101 replication origin (31) to the 5.5-kbp BamHI-KpnI fragment containing Tn4001 from pα. Plasmid pR401[aad]2, carrying a tandem duplication of 1,300 bp of the aad (spectinomycin resistance) gene, was constructed by ligating XbaI-linearized pR401 to the XbaI-digested PCR fragment amplified from pR401 with primers mp125 and mpY.
To determine the events leading to establishment of Genr colonies, S. pneumoniae chromosomal DNA was extracted from clones obtained following transformation with pR401 or pR401[Tn-IS] plasmid DNA preparations. Transposition of IS256 independent of Tn4001 would not have been detected in these experiments. Chromosomal DNA was digested with BglII and separated by electrophoresis. Following staining with SyBr green, the gel was dried and hybridized either with a PCR fragment that exhibited homology to the pSC101-derived vector backbone or with an HindIII DNA fragment from within Tn4001 which carried the entire aacA-aphD gene region together with approximately 300 bp of flanking DNA from the right end of IS256.
DNA procedures.
Standard techniques were used for DNA manipulation and cloning (35). Restriction and DNA-modifying enzymes were purchased from New England Biolabs. DNA molecules were isolated from agarose gels with a QIAquick gel extraction kit (Qiagen). Plasmid DNA was extracted with miniprep or maxiprep kits (Qiagen). PCR products were purified with a QIAquick PCR purification kit (Qiagen). The following oligonucleotides were used in this study: mp125 (5′CCGTAAAAGGACTGTTATATGGC 3′), mpY (5′GCTCTAGA-AGCCTGTTCGGTTCGTAAGC 3′), and mpZ (5′AGATATTAACTTAGCGCGTGAGG 3′). PCR amplification was performed using Hot Tub polymerase (Amersham), with 30 cycles consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min per kb amplified.
Gel electrophoresis and hybridization with radiolabeled probes.
Reaction products were separated on 0.7% agarose gels in TAE buffer at room temperature and were detected either with ethidium bromide or SyBr green or by Southern hybridization with [γ-33P]ATP-radiolabeled probes. DNA hybridization was performed directly in the gel. The gel was dried for 60 min at 60°C, and the DNA was denatured with 0.5 M NaOH–0.15 M NaCl for 20 min and neutralized with 0.5 M Tris (pH 8)–0.15 M NaCl. After prehybridization in a solution containing 6× SSC (0.9 M NaCl plus 0.09 M sodium citrate), 0.5% sodium dodecyl sulfate, 100 mg of calf thymus DNA per ml, and 0.1% nonfat milk for 2 to 4 h, hybridization was carried out with a large excess of radiolabeled oligonucleotides for 12 to 15 h in the same buffer. Both steps were performed at 8°C below the melting temperature of the probe. The gel was then washed three times in 6× SSC–0.1% sodium dodecyl sulfate at room temperature, dried, and exposed either to Kodak Biomax film for autoradiography or in a Fuji X BAS1000 phosphorimaging system coupled to the PCBas software package.
Oligonucleotides were radiolabeled for sequencing. One microliter of a 10 μM oligonucleotide solution was incubated for 45 min at 37°C with 30 μCi of [γ-33P]ATP, 1 μl of kinase buffer, and 5 U of T4 polynucleotide kinase in a 10-μl (final volume) mixture. The kinase was inactivated by incubation for 10 min at 68°C. One microliter of radiolabeled oligonucleotide was used directly for sequencing by using the Amersham thermosequenase cycle sequencing protocol. Radiolabeled oligonucleotide mp125 was used to sequence the IS junctions of the different forms isolated from the agarose gel after electrophoresis. Five microliters of each sequencing reaction mixture was loaded on an 8% sequencing gel and electrophoresed at 40 W for 2 h. After drying, the gel was exposed on Biomax autoradiography film (Kodak).
RESULTS
Experimental context.
The initial goal of this study was to generate a bank of mutants of S. pneumoniae by using the IS256-based transposon Tn4001 (Fig. 1A). Plasmid pα, a derivative of pBR322 that carried Tn4001 and was incapable of replication in S. pneumoniae, had been used previously in S. gordoni to deliver Tn4001 (18). It appeared to be inappropriate for this study because the S. pneumoniae strain that was going to be mutagenized already carried integrated pBR322 DNA sequences. Introduction of pα by natural transformation into this host would have resulted in homologous recombination between the pα sequence and the chromosomal pBR322 copy. To circumvent this, Tn4001 was transferred to a different plasmid backbone. Low-copy-number plasmid pR217 (31), a derivative of pSC101, was chosen for this purpose. Cloning of Tn4001 (Fig. 1A) as a BamHI/KpnI fragment into BamHI/KpnI-cleaved pR217 yielded plasmid pR401 (Fig. 1B). Surprisingly, pR401 DNA proved to be about 10-fold less efficient than pα DNA for transforming a strain of S. pneumoniae lacking pBR322 homologous sequences in its chromosome (data not shown).
Since studies with pα indicated that DNA prepared in the absence of dam and dcm methylation yielded significantly more transformants than methylated plasmid DNA (18), pR401 DNA was prepared from an E. coli dam dcm host (GM2163) carrying a wild-type recA gene. The presence of an active homologous recombination system in this strain might have influenced the plasmid content and led to the differences observed in transformation efficiencies. When DNA of pα and pR401 isolated from the E. coli dam dcm strain were analyzed by gel electrophoresis, a large fraction of the pBR322 derivative, pα, appeared as multimers, whereas the pSC101 derivative, pR401, did not appear as multimers. This probably reflected the absence of a dimer resolution system in pBR322 and the presence of an active system in pSC101 (5). Since installation of multimeric autonomous replicating plasmid DNA in S. pneumoniae during natural transformation of competent cells is more efficient than installation of monomeric forms (36), it seemed possible that the lower apparent transposition efficiency from pR401 was due to a lower efficiency of reconstitution of a complete copy of Tn4001 when it was carried by the largely monomeric pR401 plasmid. The steps involved in transposition of Tn4001 from the nonreplicative pR401 plasmid following transformation of S. pneumoniae are not known, although the pathway is obviously complex (see below). During transformation, exogenous double-stranded DNA is randomly nicked and ingested by the recipient cell. Only one strand enters, with a leading 3′ end, and the complementary strand is degraded (25). Transposase expression requires reconstitution of an intact double-stranded DNA in the cell, and transposition requires establishment of an active complex between the transposase and the transposon DNA. Therefore, in analogy to installation of multimeric replicative plasmids during transformation (36), reconstitution of the nonreplicative plasmid inside the S. pneumoniae cell could be a limiting step for transposition. To determine whether multimeric copies of pR401 could improve transposition frequencies, dimeric forms were generated, and their capacities to generate S. pneumoniae mutants resulting from Tn4001 insertions were compared to those of monomeric plasmids, as described below.
Isolation of multimeric forms of pR401.
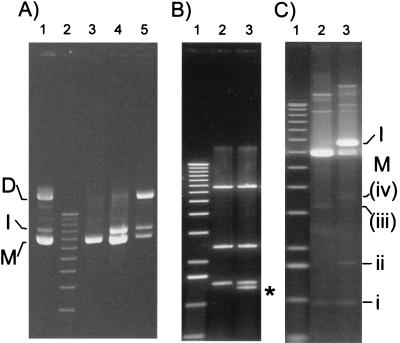
Multimeric forms of pSC101 derivatives were isolated as follows. Plasmid pR401 carries the lacIq gene. When this plasmid is introduced into an E. coli strain (LN2843) with a mutation in the chromosomal lacI gene, colonies carrying pR401 should appear white on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) indicator plates. Plasmidless cells, resulting from segregation of multimeric plasmids, should lose the lacIq gene and thus appear blue (Lac+). Indeed, blue sectors developed in some of the colonies after several days of growth. Analysis of the plasmid contents of the neighboring white regions of such colonies revealed three major species on an agarose gel (Fig. 2). Two of these species migrated as expected for monomeric and dimeric plasmids, while the third migrated at a position slightly above that of monomeric plasmid DNA (intermediate form I in Fig. 2A, lane 1). This form was restricted to colonies with associated blue sectors and, like plasmid dimers, was not detected in the entirely white colonies.
FIG. 2.
Physical nature of pR401 populations. (A) Evolution of different plasmid forms. Lane 1 contained the population of pR401 molecules isolated from LN2843. M, I, and D indicate the relative positions of presumed monomer, intermediate, and dimer plasmid species, respectively. Lanes 3, 4, and 5 contained plasmid populations isolated from E. coli recA which had been transformed with DNA from lane 1. (B) HindIII restriction patterns. Lane 2, monomer species; lane 3, intermediate species. The asterisk indicates the position of the additional DNA fragment in the intermediate species molecules. (C) Low-level species revealed by SyBr green staining. Lane 2, undigested monomer species; lane 3, undigested intermediate species. The major additional bands are indicated by i, ii, (iii), and (iv); parentheses indicate that a species has not been positively characterized yet. The molecular size standard used (panel A, lane 2; panel B, lane 1; panel C, lane 3) was a 1-kb ladder from Gibco-BRL whose largest component was 12 kb long.
To isolate the pR401 dimer for transformation of S. pneumoniae, DNA purified from the white regions of colonies with blue sectors was used at a low concentration to transform a naive, recombination-proficient E. coli dam dcm host. The dam and dcm markers were used initially to obtain more efficient transformable DNA for eventual introduction into S. pneumoniae. Although transformant colonies were obtained in which one of the three species appeared to be the majority form, no clones carrying a single plasmid DNA species were obtained (data not shown). This suggested that there may be interconversion among the three species either by homologous recombination or by transposition. To investigate this further, the same DNA preparations were used to transform an E. coli recA host (CT45). Whereas it was possible to isolate clones carrying only monomeric plasmid DNA (Fig. 2A, lane 3), clones containing a majority of dimeric DNA (Fig. 2A, lane 5) always contained some monomers and form I, and clones containing form I also contained the monomeric plasmid (Fig. 2A, lane 4). This suggested that the plasmid dimer gives rise to both the intermediate and monomeric species, whereas the form I species can generate plasmid monomers via a recA-independent pathway.
Characterization of pR401 derivative form I.
To determine the nature of the novel pR401 species, form I was purified following gel electrophoresis and digested with HindIII, which cleaved once within each IS256 copy and once within the pR401 backbone (Fig. 1B). The results are shown in Fig. 2B. This form gave rise to an additional 1.3-kbp fragment (Fig. 2B, lane 3) compared to the results obtained for the purified monomer (lane 2). The size of this fragment was the size expected for a tandem head-to-tail dimer of the IS (Fig. 1C). Southern hybridization performed with an oligonucleotide specific for the left end of IS256 interior to the terminal inverted repeat (mp125) (Fig. 1B and C) confirmed that the additional fragment carried the predicted portion of IS256 (data not shown). Form I was therefore considered to be a new plasmid species and provisionally designated pR401[Tn-IS].
Although we did not investigate whether the parental plasmid, pα, was able to generate such derivatives, it should be noted that Lyon et al. (20) observed equivalent plasmid forms obtained from a Tn4001-carrying plasmid, pSK1α, the prototype aminoglycoside resistance plasmid isolated from Australian strains of S. aureus (21).
IS circle formation.
The structure of the pR401[Tn-IS] species described above is reminiscent of the structure of transposition intermediates of ISs whose transposition pathway includes formation of an IRR-IRL reactive junction. An additional characteristic of these ISs is their capacity to generate transpositionally active circular forms (11). Such species were not observed in the ethidium bromide-stained gel containing undigested plasmid DNA shown in Fig. 2A. Moreover, since digestion of such circles would be expected to generate a fragment of the same size as that obtained from the transposon-IS junction, they could not have been detected in the digested DNA preparations shown in Fig. 2B. To determine whether IS256 generated such IS circles, more sensitive SyBr green staining was used. Typical results are shown in Fig. 2C. Several high-mobility forms were revealed by this treatment (forms i, ii, iii, and iv). Two of these forms, forms i and ii, migrated at positions compatible with the positions of covalently closed single (monomeric) and double (dimeric) IS circular species, respectively. Form ii was observed only in samples containing pR401[Tn-IS], while form i was observed in all samples. Southern hybridization of this gel with probes specific for different parts of pR401 demonstrated that both form i and form ii carried IS256 DNA but not the plasmid backbone or other regions of Tn4001 (data not shown). We did not analyze the other, minor forms, but their migration was consistent with migration of closed circular pR401 and pR401[Tn-IS] molecules from which the transposon had been deleted (forms iii and iv, respectively). It should be noted that form iv, like form ii, was present only in samples containing pR401[Tn-IS].
Structure and sequence of the transposon-IS and IS circle junction.
Another characteristic of ISs which transpose by using a reactive junction is that the abutted inverted repeats are invariably separated by a short DNA linker derived from nucleotides flanking the target end in the parental insertion. A given IS element generates a linker with a defined length which is not necessarily the same as that of the direct repeats generated by the IS upon insertion (1, 16, 32).
The nature of the junction was determined as follows. Plasmid pR401[Tn-IS] DNA was gel purified from a sample containing a population of molecules (Fig. 2A, lane 1) and used to transform an E. coli recA strain. Total plasmid DNA was prepared from cultures of five isolated colonies and separated by gel electrophoresis. After Sybr green staining of the gel, the IRL-IRR junctions of pR401[Tn-IS] and of the two putative IS circular forms (forms i and ii) were PCR amplified directly by using oligonucleotides mp125 and mpZ (Fig. 1B and C). An identical analysis was performed for form i species obtained from pR401 monomer molecules. Each species yielded a fragment that was approximately 430 bp long, the size predicted for a junction fragment. The nucleotide sequences of five transposon-IS junctions and forms i and ii derived from three of these junctions were determined. In addition, the junction sequences of three form i species derived independently from pR401 monomers were determined.
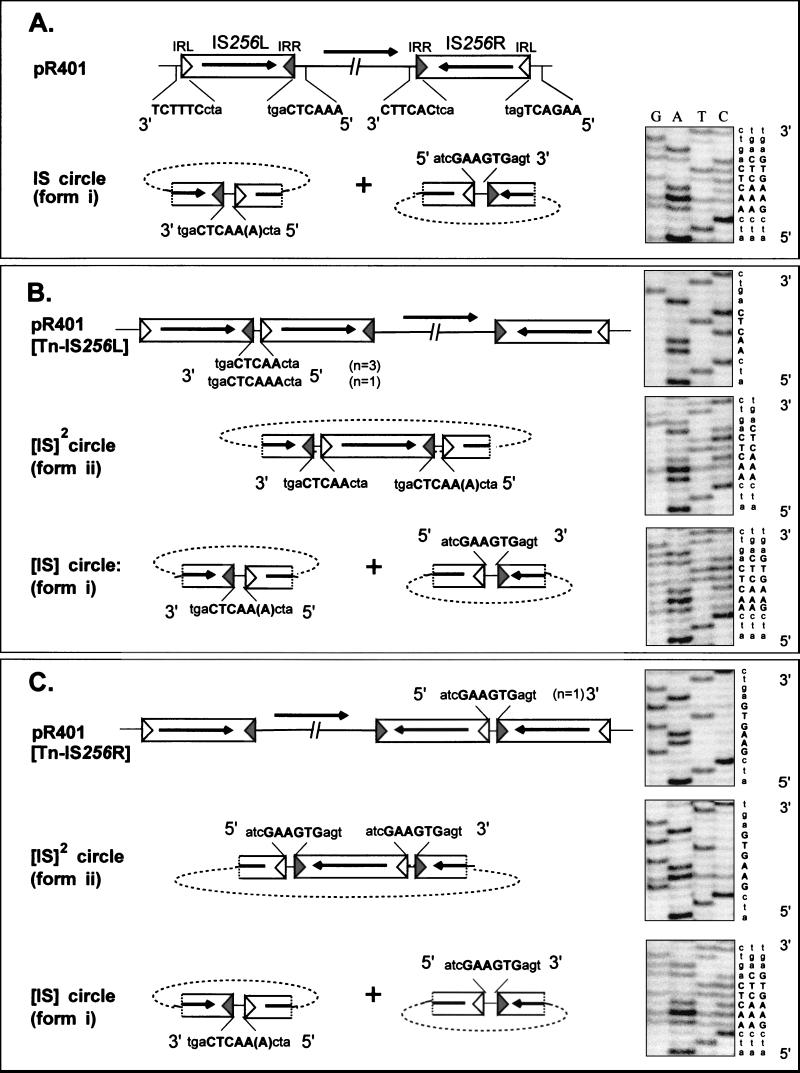
Representative results and a schematic interpretation are shown in Fig. 3. The junction sequence obtained from the population of form i products (monomeric circles) in samples of the pR401 parental plasmid was mixed (Fig. 3A). It included 5- and 6-bp spacers from the right end of IS256L (5′AACTC3′ and AAACTC) and a 6-bp spacer from the right end of IS256R (GAAGTG). This finding is consistent with the presence of IS circles derived from both component IS256 copies and indicates that IRL was the attacking end and IRR was the target end. In addition, it implies that the attack occurred in a majority of cases 6 bp distal to IRR for IS256R (GAAGTG), while in the case of IS256L the attack occurred either at 5 bp (AACTC) or 6 bp (AAACTC) in the population. The results imply that both component copies of IS256 in Tn4001 are active.
FIG. 3.
Nucleotide sequences of different IS junctions and diagrams showing an interpretation of these sequences. The symbols are the same as those described in the legend to Fig. 1. Lowercase letters indicate the sequences of the ends of IS256, showing the difference in the terminal nucleotides of IRL and IRR. Uppercase letters indicate the flanking DNA sequences. Boldface letters indicate the flanking sequences observed to form part of the junction. The parentheses around the final nucleotide in the IRL-associated sequence indicate the appearance of mixed 5- or 6-bp sequences. (A) Monomeric parental plasmid pR401 together with IS circles derived from the left and right IS256 copies giving rise to a mixed junction sequence. (B) Junction sequences obtained from a pR401[Tn-IS] derivative with the IS duplication at the left end of Tn4001. Of four independent isolates of this type, three exhibited a unique 5-bp sequence, while one exhibited a unique 6-bp sequence. Starting with a derivative with a unique 5-bp sequence, form ii DNA was observed to carry a mixed 5-bp–6-bp junction sequence. Form i DNA exhibited the mixed 5-bp–6-bp sequence originally flanking the left IS together with a 6-bp sequence derived from sequences originally flanking the right IS. (C) Junction sequences obtained from a pR401[Tn-IS] derivative with the IS duplication at the right end of Tn4001. In this case, form ii carried a unique 6-bp sequence derived from nucleotides originally flanking the right IS, whereas form i carried the mixed 5-bp–6-bp sequence from the left IS together with the 6-bp sequence derived from the right IS.
Of five independently isolated pR401[Tn-IS] clones, four appeared to carry tandem dimers including IS256L (pR401[Tn-IS256L]) (Fig. 3B), while one carried dimers including IS256R (pR401[Tn-IS256R]) (Fig. 3C). The pR401[Tn-IS256L] dimer junction spacer was derived from the five (AACTC; three individual clones sequenced) or six (AAACTC; one clone sequenced) neighboring IRR-flanking bases, confirming the alternative position of IRL attack. The sequence of the form i junction was again mixed and was similar to the sequence obtained from the form i species of pR401 itself. It included sequences from the pR401[Tn-IS256L] tandem dimer junction, as well as IRR-flanking DNA from IS256L. This conclusion was based on the observation that form i junctions from a pR401[Tn-IS] derivative with only a 5-bp pR401[Tn-IS256L] tandem dimer junction (AACTC) had a mixed sequence including both 5-bp (AACTC) and 6-bp (AAACTC) spacers. It also included a 6-bp sequence from the right end of IS256R (GAAGTG), again indicating that both flanking IS elements contribute to the formation of form i. Form ii, a form observed only in pR401[Tn-IS] preparations, also had mixed 5- and 6-bp spacers derived from the right end of IS256L. No corresponding flanking sequences from IS256R were detected, indicating that this form is derived solely from IS256L. Moreover, form ii DNA from a pR401[Tn-IS] derivative with only a 5-bp pR401[Tn-IS256L] junction (AACTC) also had a mixed sequence including both 5-bp (AACTC) and 6-bp (AAACTC) spacers. This finding is consistent with the notion that form ii is a circular IS dimer ([IS256]2 circle).
Finally, the sequence of the pR401[Tn-IS256R] (Fig. 3C) tandem dimer junction carried by the single clone of pR401[Tn-IS] appeared to be unique and was composed of 6 bp of IRR-flanking DNA (GAAGTG). The form ii species from this preparation contained a similar junction. In contrast, the sequence of the form i junction was mixed, as it was in the other form i derivatives.
These results demonstrate that both IS copies in Tn4001 are active in circle or tandem dimer formation; that attack of one end by the other is largely oriented (IRL attacks IRR); and that events in which IS256L is used generate 5- or 6-bp junction spacers, whereas junctions generated by IS256R carry a majority of 6-bp spacers. Our experiments also defined the functional tips of IS256 as the tips predicted from sequence alignments, confirming that the sequences of the terminal 4 bp at each end are indeed different.
Tn4001-mediated recombination in S. pneumoniae.
Tn4001 transposition in S. pneumoniae depends on a series of relatively complex steps. Transformation of S. pneumoniae involves initial ingestion of single-stranded DNA by the recipient bacteria. To transpose from pR401, Tn4001 requires transposase expression from the resident IS256 copy. For this to occur, a double-stranded plasmid copy must presumably be reconstituted. This chain of events is complicated further by the fact that pR401 does not replicate in S. pneumoniae. Quantitation of Tn4001 transposition is therefore not straightforward. Three observations were made during our attempt to understand the behavior of Tn4001 in S. pneumoniae.
The frequencies of establishment of transposon-specified gentamicin resistance were compared following transformation of S. pneumoniae with pR401 or pR401[Tn-IS] DNA. Plasmid pR401[Tn-IS] gave rise to an approximately 10-fold-higher level of Genr colonies than did pR401. This relatively small difference in frequency could have been due to various factors, such as the presence of a reactive IRL-IRR junction or more efficient reconstitution of pR401[Tn-IS] because of the sequence redundancy resulting from the additional IS256 copy.
The effect of sequence redundancy alone on reconstitution was also investigated. A derivative of pR401, pR401[aad]2, which contained a directly repeated sequence from the aadA gene whose length was similar to that of IS256, was constructed. The frequency of Genr colonies obtained following transformation was fivefold higher than the frequency obtained with pR401 but twofold lower than the frequency obtained with pR401[Tn-IS]. Thus, sequence redundancy improved the efficiency of plasmid reconstitution but may have been less effective than a structure which also included an IRR-IRL junction.
Finally, we assessed the ability of the IRL-IRR junction to promote integration of the entire donor plasmid. Efficient integration has previously been observed with other similar structures (1, 27, 33, 39). The total DNA of 12 independent Genr clones obtained from pR401 and 12 Genr clones obtained from pR401[Tn-IS] were examined following appropriate digestion, electrophoretic separation, and hybridization with probes specific for Tn4001 or for the donor plasmid. Five of the 12 pR401[Tn-IS]-derived clones resulted from integration of the entire plasmid, whereas only 2 of the 12 pR401-derived clones showed this pattern. The potential structures of these integration products are shown in Fig. 4. All other clones carried only Tn4001 (data not shown).
FIG. 4.
Putative transposition products in S. pneumoniae chromosome. The symbols are the same as those described in the legend to Fig. 1. The dashed lines represent chromosomal DNA. (A) IS256 insertion. (B) Double IS256 insertion. The products shown in panels A and B can result from transposition from a pR401 plasmid or from integration via a junction (from an IS256 circle or a [IS256]2 circle), but they cannot be detected in our system since they do not confer resistance to gentamicin. (C) Tn4001 insertion can result from transposition from a pR401 plasmid or from integration via a junction (from a Tn4001 circle). (D) Tn4001, plasmid backbone, and IS256 insertion can result from transposition from dimeric pR401 or from integration via a tandem junction (pR401[Tn-IS256]).
Although further analysis is necessary to determine the role of the IRR-IRL junction in IS256 transposition, our results clearly underline the necessity for prudence when transposons of this type are used as mutagens in S. pneumoniae.
DISCUSSION
Transposition-related recombination of Tn4001.
Transposon Tn4001 is composed of two inverted copies of insertion sequence IS256 flanking an aacA-aphD gene region (20). This transposon has been correlated with the presence of plasmids providing multiple antibiotic resistance to nosocomial strains. Moreover, IS256 itself has been shown to be involved in phase variation of Staphylococcus epidermidis (49). While isolating plasmid pSK1α, the original source of Tn4001, from a clinical isolate of S. aureus, Lyon et al. (20) observed a second plasmid, pSK1β. This plasmid proved to be identical to pSK1α except for the presence of an additional copy of IS256 directly flanking the left copy of IS256 and in the same orientation. We demonstrated here that this type of [Tn-IS] structure is readily generated from a Tn4001-carrying plasmid in E. coli. In our case, an additional IS256 element was found abutting either the left (n = 4) or right (n = 1) copy of IS256 (Fig. 1). We also found that IS256 is capable of forming IS circles. Both types of structure involve a left IS256 end (IRL) and a right IS256 end (IRR) separated by 6 bp (or more rarely 5 bp). Furthermore, interconversion among tandem plasmid, monomeric plasmid, and circles suggest that the IRR-IRL junction is active in promotion of transposition. This behavior resembles that of an increasing number of bacterial insertion sequences which generate integrative junctions as part of the transposition cycle.
For members of the IS3 family of bacterial insertion sequences, transposon circles are generated by intra-IS recombination between the two ends of the element (for a review see reference 33); an initial single-strand cleavage occurs at one end, the donor end, and the free 3′ OH formed is then directed to attack the opposite end on the same DNA strand, the target end, to generate an intermediate in which the two ends are joined by a single-strand bridge (29). If the substrate is circular, the resulting figure-eight form is then converted into a covalently closed circular IS by elimination of the intervening plasmid DNA (46). For IS911, since attack generally occurs precisely 3 bases outside the target end, the ends are separated by a 3-bp linker derived from sequences flanking the target end. Tandem IS dimers have also been observed for members of the IS3 (46), IS21 (32), and IS30 (15) families. Formation of the dimers can occur in two ways (1, 15, 46). One pathway involves prior formation of a plasmid dimer or cointegrate. An initial recombination, similar to that involved in IS circle formation, between the two copies of the IS generates a figure-eight form. Resolution that generates the equivalent transposon circle eliminates the intervening plasmid DNA to generate the tandem structure (46). The observation that these events occurred preferentially in situations in which the donor plasmid was able to multimerize supports this view. Alternatively, an IS circle may be inserted in a sequence-specific manner next to a resident IS (15; C. Loot, C. Turlan, and M. Chandler, unpublished data).
The different types of recombination which we propose occur during transposition of Tn4001 are presented in Fig. 5. We note that the sequence data obtained with E. coli for the different IRL-IRR junctions suggest that in all cases IRL is the attacking end while IRR is the target. This is because the intervening 5- or 6-bp sequence between the two ends is always the sequence which originally flanks IRR (Fig. 3). This may be a reflection of the significant differences in the sequences of the terminal 4 bp of the inverted repeats. This type of asymmetric behavior has been noted previously in the case of IS2 (16, 16a).
There are only two ways in which a copy of IRL can attack a suitably oriented copy of IRR in a monomeric plasmid carrying Tn4001 (Fig. 5A). These ways are both intra-IS recombination events and would generate circular forms of either IS256L or IS256R. While we identified circular copies of IS256L and IS256R in a population of pR401 monomers in E. coli (Fig. 3A), we did not positively identify a circular copy of the entire transposon. As shown in Fig. 5A, formation of such a structure would require attack by one copy of IRL of its homologue from the second IS256. Moreover, this species carries inverted repeats that are the length of IS256, a structure which would be expected to be unstable. For a dimeric plasmid (Fig. 5B), in addition to the intra-IS recombination events, there are four ways in which an IRL copy might attack an IRR copy on a partner IS256 element in an inter-IS recombination event; two of these ways involve IS256L, and two involve IS256R. Resolution would give rise to a plasmid carrying a transposon with either two abutted IS256L copies or two IS256R copies, as we observed (pR401[Tn-IS256L] and pR401[Tn-IS256R]) (Fig. 3B and C). A further round of attack in these [Tn-IS] structures (Fig. 5C) would result in the formation of a circular tandem IS256 dimer ([IS]2 circle) (Fig. 3B and C ). Such dimer circles could be generated only from pR401[Tn-IS] or from dimeric forms of pR401, as we observed. Based on observations made with other IS elements, abutted IS ends are very active in transposition; all species carrying abutted ends would be considered intermediates in transposition and would undergo efficient insertion into a suitable target molecule.
It is interesting that while the number of interstitial base pairs between IRL and IRR at the junction can be identical to the length of the direct flanking target repeat generated upon insertion, as in the case of IS911, this is not a general rule. For example, IS2 generates a 5-bp target duplication upon insertion, but only 2 bp are located between the two ends in the junction (16). For IS1, the transposon circles have an 8-bp interstitial region, while IS1 integration generates a 9-bp target duplication (44). This implies that the geometry of the recombination complex which generates the junction differs from the geometry of that leading to insertion. Insertion of IS256 generally generates an 8-bp direct flanking target repeat (3). However, the number of interstitial base pairs observed at the transposon-IS junction is five or six (Fig. 3). On closer inspection, junctions formed with IS256L appeared to be separated by either 5 or 6 bp, while all of the junctions generated with IS256R appeared to be 6 bp long (Fig. 3). The reason for this spacer difference between [Tn-IS256L] and [Tn-IS256R] is not clear.
Potential promoter in the IRR-IRL junction.
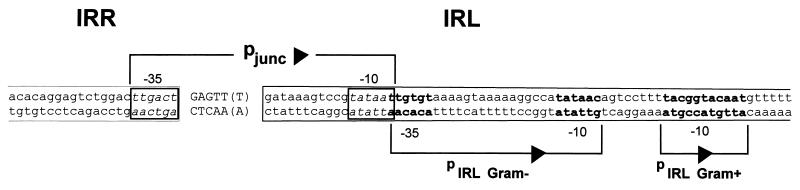
A characteristic of several (1, 6, 28, 42), but not all, elements which undergo tandem dimerization or circularization is that formation of the IRR-IRL junction results in assembly of a promoter, pjunc, capable of driving transposase synthesis. In these cases, IRL carries a −10 promoter element directed inward, while IRR carries a −35 element directed outward. These junction promoters are generally stronger than the resident pIRL promoter, and it is probable that their formation leads to a burst of transposase synthesis that results in efficient integration of the IRR-IRL junction. In the case of IS911, pjunc stimulates transposition about 50-fold (6a).
Analysis of the junction sequences observed here indicated that IS256 may also assemble a pjunc during its transposition cycle. A reasonable −35 element (TTGACT), typical of E. coli ς70 promoters, can be found in IRR (48), while a −10 element which perfectly matches the consensus element (TATAAT) is found in IRL. Formation of the junction with an intervening 6 bp would place these two elements at an optimal distance (pjunc) (Fig. 6). Although no extended −10 sequence (TaTGgTATAAT) typical of promoters from gram-positive bacteria (34) is evident at the IRR-IRL junction, Lyon et al. (20) correlated the presence of IS256 tandem structures with increases in gentamicin, trimethoprim, and kanamycin resistance in S. aureus. Since no transcriptional termination signals between the IS256 IRR-IRL junction and the aacA-aphD gene region were evident (3), the increase in antibiotic resistance might be explained by reconstitution of a strong promoter at the IRR-IRL junction. Figure 6 also shows the elements of a potential resident IS256 promoter which could drive transposase expression (pIRL Gram−), as has been noted previously (3). The 16-bp spacer between these elements suggests that this indigenous promoter may not be very active (17). It is also worth noting that an extended gram-positive −10 element is also present in IS256 and may represent an alternative internal promoter active in this class of bacteria (pIRL Gram+) (Fig. 6). Further studies are required to establish whether these potential promoter elements play a role in IS256 transposition.
FIG. 6.
Sequence of a pR401[Tn-IS256L] junction, showing the −35 and −10 components of a potential junction promoter, pjunc. The IRR and IRL ends are indicated by lowercase letters and are enclosed in boxes. Potential −35 and −10 sequences of pjunc are enclosed in thick black boxes. The intervening sequence is indicated by uppercase letters. The positions of potential gram-positive and gram-negative pIRL promoters are also indicated in bold type.
Tn4001 as a tool for mutagenesis of S. pneumoniae.
The experiments reported here were designed to investigate the reasons for the low apparent efficiency of transposition of Tn4001 from pR401 into the S. pneumoniae genome. Our results are consistent with the idea that this low efficiency is due largely to the low levels of the dimeric plasmid. Analysis with E. coli, as described above, showed that a dimeric pR401 plasmid can give rise to tandem species (pR401[Tn-IS]), as well as monomeric and dimeric IS256 circles, all carrying an IRR-IRL junction. Since this type of structure has been shown to be very active in transposition for a number of other ISs, it seems reasonable to hypothesize that these structures were responsible for at least part of the increased transposition from dimeric pR401.
It should be emphasized, however, that many different steps are involved in generation of the final transposition products in S. pneumoniae and that the type of DNA species which acts as an intracellular transposon donor is not known. Therefore, at present it is not possible to define the contribution of monomeric and dimeric plasmid forms or of plasmids carrying [Tn-IS] to the transposition events. The following considerations must be taken into account in determining the overall transposition frequency. Transformation occurs by entry of single-stranded DNA. Dimeric plasmid DNA is more efficient in transformation than monomeric DNA (36), probably due to the increased probability of regenerating an intact molecule (for example, by pairing between two single-stranded complementary sequences). Reconstitution of a double-stranded plasmid is thought to be a prerequisite for transposition in a first step, for transposase expression, and in a second step, to generate an appropriate DNA substrate. Our results provide some support for this view. A pR401 derivative, pR401[aad]2, carrying a 1.3-kbp duplication, exhibited a 5-fold-higher level of transposition than pR401, while a pR401[Tn-IS] structure exhibited a 10-fold-higher level than pR401, suggesting that the presence of the IRR-IRL junction may also stimulate transposition. Moreover, analysis of chromosomal DNA following transformation by pR401 and pR401[Tn-IS] showed that 2 of 12 and 5 of 12 clones, respectively, carried not only the Tn4001 sequence but also the plasmid backbone sequence. This type of transposition product (Fig. 4D) can result from two transposition pathways. The first involves transposition using the IRL of an IS256L (or IS256R) copy and the IRR of the other IS256L (or IS256R) copy carried by a dimeric plasmid. The second involves integration using the IRR-IRL junction carried by a tandem IS structure. Such complex transposition products make localization of chromosomal insertions with simple PCR-based procedures more difficult, and moreover, the presence of three copies of IS256 would certainly lead to further rearrangements.
In summary, the results presented here demonstrate that IS256 is capable of forming tandem IS dimers and IS circles in E. coli. For other IS elements belonging to different families, these types of structures have been shown to be very active in transposition. Although more detailed studies with a controlled experimental system are necessary to define the IS256 transposition mechanism in gram-positive bacteria, our results highlight the complexity of transposition mutagenesis in naturally transformable bacteria and underline some of the inadequacies of using Tn4001 as a mutagen.
Acknowledgments
M. Prudhomme and C. Turlan contributed equally to this article.
We thank members of the Mobile Genetic Elements Group (B. Ton Hoang, P. Rousseau, Z. Nagy, C. Loot, G. Duval-Valentin, and R. Alazard), B. Martin of the Genetic Transformation Group, D. Lane, and A. J. Carpoussis for discussions and for critically reading the manuscript, B. Marty and C. Granadel for technical assistance, and R. D. Lunsford for providing pα.
This work benefited from grants from the CNRS (UPR9007/UMR5100), the “Programme Physique et Chimie du vivant” (CNRS), and the “Programme microbiologie” (MENRST).
REFERENCES
- 1.Berger, B., and D. Haas. 2001. Transposase and cointegrase: specialized transposition proteins of the bacterial insertion sequence IS21 and related elements. Cell Mol. Life Sci. 58:403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biel, S. W., and D. E. Berg. 1984. Mechanism of IS1 transposition in E. coli: choice between simple insertion and cointegration. Genetics 108:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361–367. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II, p.305–366. ASM Press, Washington, D.C.
- 5.Cornet, F., I. Mortier, J. Patte, and J. M. Louarn. 1994. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol. 176:3188–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalrymple, B. 1987. Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression. Mol. Gen. Genet. 207:413–420. [DOI] [PubMed] [Google Scholar]
- 6a.uval-Valentin, G., C. Normand, V. Khemici, B. Marty, and M. Chandler. 2001. Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J. 20:5802–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dybvig, K., C. T. French, and L. L. Voelker. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J. Bacteriol. 182:4343–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foissac, X., C. Saillard, and J. M. Bove. 1997. Random insertion of transposon Tn4001 in the genome of Spiroplasma citri strain GII3. Plasmid 37:80–86. [DOI] [PubMed] [Google Scholar]
- 9.Gaurivaud, P., F. Laigret, M. Garnier, and J. M. Bove. 2000. Fructose utilization and pathogenicity of Spiroplasma citri: characterization of the fructose operon. Gene 252:61–69. [DOI] [PubMed] [Google Scholar]
- 10.Hahn, T. W., E. A. Mothershed, R. H. Waldo III, and D. C. Krause. 1999. Construction and analysis of a modified Tn4001 conferring chloramphenicol resistance in Mycoplasma pneumoniae. Plasmid 41:120–124 [DOI] [PubMed] [Google Scholar]
- 11.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245–281. [DOI] [PubMed] [Google Scholar]
- 12.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16:6849–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kallastu, A., R. Horak, and M. Kivisaar. 1998. Identification and characterization of IS1411, a new insertion sequence which causes transcriptional activation of the phenol degradation genes in Pseudomonas putida. J. Bacteriol. 180:5306–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiss, J., and F. Olasz. 1999. Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles. Mol. Microbiol. 34:37–52. [DOI] [PubMed] [Google Scholar]
- 16.Lewis, L. A., and N. D. Grindley. 1997. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol. Microbiol. 25:517–529. [DOI] [PubMed] [Google Scholar]
- 16a.Lewis, L. A., M. Greene, R. Saby, N. Gadura, and N. D. F. Grindley. The basis of asymmetry in IS2 transposition. Mol. Microbiol., in press. [DOI] [PubMed]
- 17.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lunsford, R. D. 1995. A Tn4001 delivery system for Streptococcus gordonii (Challis). Plasmid 33:153–157. [DOI] [PubMed] [Google Scholar]
- 19.Lunsford, R. D., and A. G. Roble. 1997. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence-factor-dependent DNA transformation in Streptococcus gordonii. J. Bacteriol. 179:3122–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyon, B. R., M. T. Gillespie, and R. A. Skurray. 1987. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J. Gen. Microbiol. 133:3031–3038. [DOI] [PubMed] [Google Scholar]
- 21.Lyon, B. R., J. W. May, and R. A. Skurray. 1983. Analysis of plasmids in nosocomial strains of multiple-antibiotic-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 23:817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon, B. R., J. W. May, and R. A. Skurray. 1984. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 193:554–556. [DOI] [PubMed] [Google Scholar]
- 23.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, B., P. Garcia, M. P. Castanie, and J. P. Claverys. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15:367–379. [DOI] [PubMed] [Google Scholar]
- 25.Mejean, V., and J. P. Claverys. 1988. Polarity of DNA entry in transformation of Streptococcus pneumoniae. Mol. Gen. Genet. 213:444–448. [DOI] [PubMed] [Google Scholar]
- 26.Mizuuchi, K. 1992. Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem. 61:1011–1051. [DOI] [PubMed] [Google Scholar]
- 27.Olasz, F., R. Stalder, and W. Arber. 1993. Formation of the tandem repeat (IS30)2 and its role in IS30-mediated transpositional DNA rearrangements. Mol. Gen. Genet. 239:177–187. [DOI] [PubMed] [Google Scholar]
- 28.Perkins-Balding, D., G. Duval-Valentin, and A. C. Glasgow. 1999. Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J. Bacteriol. 181:4937–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polard, P., and M. Chandler. 1995. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 9:2846–2858. [DOI] [PubMed] [Google Scholar]
- 30.Porter, R. D., and W. R. Guild. 1976. Characterization of some pneumococcal bacteriophages. J. Virol. 19:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prudhomme, M., V. Mejean, B. Martin, and J. P. Claverys. 1991. Mismatch repair genes of Streptococcus pneumoniae: HexA confers a mutator phenotype in Escherichia coli by negative complementation. J. Bacteriol. 173:7196–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimmann, C., R. Moore, S. Little, A. Savioz, N. S. Willetts, and D. Haas. 1989. Genetic structure, function and regulation of the transposable element IS21. Mol. Gen. Genet. 215:416–424. [DOI] [PubMed] [Google Scholar]
- 33.Rousseau, P., C. Normand, C. Loot, C. Turlan, R. Alazard, G. Duval-Valentin, and M. Chandler. 2002. Transposition of IS911. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II, p.367–383. ASM Press, Washington, D.C.
- 34.Sabelnikov, A. G., B. Greenberg, and S. A. Lacks. 1995. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250:144–155. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Saunders, C. W., and W. R. Guild. 1981. Monomer plasmid DNA transforms Streptococcus pneumoniae. Mol. Gen. Genet. 181:57–62. [DOI] [PubMed] [Google Scholar]
- 37.Sekine, Y., K. Aihara, and E. Ohtsubo. 1999. Linearization and transposition of circular molecules of insertion sequence IS3. J. Mol. Biol. 294:21–34. [DOI] [PubMed] [Google Scholar]
- 38.Sekine, Y., N. Eisaki, and E. Ohtsubo. 1994. Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235:1406–1420. [DOI] [PubMed] [Google Scholar]
- 39.Shiga, Y., Y. Sekine, and E. Ohtsubo. 1999. Transposition of IS1 circles. Genes Cells 4:551–561. [DOI] [PubMed] [Google Scholar]
- 40.Smokvina, T., D. J. Henderson, R. E. Melton, D. F. Brolle, T. Kieser, and D. A. Hopwood. 1994. Transposition of IS117, the 2.5 kb Streptomyces coelicolor A3(2) ‘minicircle’: roles of open reading frames and origin of tandem insertions. Mol. Microbiol. 12:459–468. [DOI] [PubMed] [Google Scholar]
- 41.Tigges, E., and F. C. Minion. 1994. Physical map of the genome of Acholeplasma oculi ISM1499 and construction of a Tn4001 derivative for macrorestriction chromosomal mapping. J. Bacteriol. 176:1180–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ton-Hoang, B., M. Betermier, P. Polard, and M. Chandler. 1997. Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J. 16:3357–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ton-Hoang, B., P. Polard, and M. Chandler. 1998. Efficient transposition of IS911 circles in vitro. EMBO J. 17:1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turlan, C., and M. Chandler. 1995. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 14:5410–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turlan, C., and M. Chandler. 2000. Playing second fiddle: second-strand processing and liberation of transposable elements from donor DNA. Trends Microbiol. 8:268–274. [DOI] [PubMed] [Google Scholar]
- 46.Turlan, C., B. Ton-Hoang, and M. Chandler. 2000. The role of tandem IS dimers in IS911 transposition. Mol. Microbiol. 35:1312–1325. [DOI] [PubMed] [Google Scholar]
- 47.Welz, C. 1993. Funktionelle Analyse des Bakteriellen Insertionelements IS150. Ph.D. thesis. Fakultät für Biologie der Albert-Ludwigs-Universität Freiburg, Freiburg, Germany.
- 48.Wosten, M. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127–150. [DOI] [PubMed] [Google Scholar]
- 49.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345–356. [DOI] [PubMed] [Google Scholar]