Abstract
In many yeast species, including Kluyveromyces lactis, growth on certain sugars (such as galactose, raffinose, and maltose) occurs only under respiratory conditions. If respiration is blocked by inhibitors, mutation, or anaerobiosis, growth does not take place. This apparent dependence on respiration for the utilization of certain sugars has often been suspected to be associated with the mechanism of the sugar uptake step. We hypothesized that in many yeast species, the permease activities for these sugars are not sufficient to ensure the high substrate flow that is necessary for fermentative growth. By introducing additional sugar permease genes, we have obtained K. lactis strains that were capable of growing on galactose and raffinose in the absence of respiration. High dosages of both the permease and maltase genes were indeed necessary for K. lactis cells to grow on maltose in the absence of respiration. These results strongly suggest that the sugar uptake step is the major bottleneck in the fermentative assimilation of certain sugars in K. lactis and probably in many other yeasts.
Kluyveromyces lactis and many other yeast species can grow on galactose and certain oligosaccharides (such as raffinose and maltose) aerobically, but they cannot grow on these sugars anaerobically or in the absence of respiration (15, 18, 19, 35). Assimilation of these carbon sources occurs only under respiring conditions. The phenomenon has been known by the classical name of the Kluyver effect. The kind of sugars involved varies depending on the species and sometimes on the strains within a species. Although the reason for this apparent dependence on respiration for the assimilation of certain sugars is not clear, the phenomenon does appear to be brought about by the interplay of several factors involving lowered rate of transport and metabolism of certain sugars (4). Saccharomyces cerevisiae generally does not show this phenomenon (Kluyver effect negative), although K. lactis and S. cerevisiae seem to use similar pathways to metabolize galactose, raffinose, and maltose.
In the present work we show that, by introducing additional Saccharomyces sugar permease genes, K. lactis cells can be released from their dependence on respiration for the assimilation of galactose and raffinose. High dosages of both permease and maltase genes were indeed necessary for K. lactis cells to grow on maltose in the absence of respiration.
MATERIALS AND METHODS
Strains, media, and growth conditions.
K. lactis strains used in this study were PM4-4B (MATα ade1 ade2 uraA), PM1-11B (MATa metA uraA lys arg rag2), P11-1D (uraA lys his2-2 lac12-230), P11-1A (lys his2-2 LAC12), 11D304 (MATα his2-2 lac12-230 [32]), JBD100 (MATα trp1 ura3-100 lac4-1), JBD100/M3 (MATα trp1 ura3-100 lac4-1 cyt1 [16]), and JA6 (MATα ade1 ade2 trp1 uraA [9]). The P11-1D lac12 mutant was obtained by sporulation of the P11 diploid constructed by crossing the original mutant 11D304 lac12 (32) with a ura3 LAC12 strain, PM1-11B. As described for the 11D304 lac12 strain (32), the lac12 mutant P11-1D is able to grow on galactose (data not shown).
Genetic procedures for crossing and sporulation have been described previously (18, 40). Rich medium was 1% (wt/vol) Bacto-yeast extract and 2% Bacto-peptone (YP). Mineral medium contained 6.7 g of yeast nitrogen base (Difco) per liter without amino acids (YNB) supplemented with the appropriate amino acids and bases. Various carbon sources were added at 2%. Media were solidified with 20 g of Bacto agar per liter. Incubations were done at 28°C. For respiration-dependent growth, the cultures were grown on a reciprocating shaker at 110 rpm.
The Escherichia coli strains JM83 [ara Δ(lac-proAB) rpsL (=strA) φ80 lacZΔM15] was used for plasmid amplification by a standard procedure (33).
Fermentation test.
As a rapid test to determine the presence or absence of fermentation, fermentation basal medium test (44) was used with minor modifications: production of carbon dioxide leads to a change in color of the indicator dye bromothymol blue (BTB). Precultures, grown for 2 days in liquid minimal medium supplemented with 2% glucose and appropriate auxotrophic requirements, were harvested and diluted at 2 × 106 cells/ml in YP plus galactose (YPGal) liquid medium containing 6 ml of BTB (stock solution: 400 mg of BTB, 1.5 g of NaOH, and H2O to a final volume of 100 ml, brought to pH 6.5). The cell suspensions were maintained at 28°C for 2 days. The pH indicator dye BTB in the medium turns green to yellow when active fermentation occurs, producing CO2.
Plasmids and genomic library.
The gene vectors and plasmids are described in Table 1. All plasmids contained a URA3 marker and a replication origin for K. lactis. The K. lactis-S. cerevisiae shuttle library of S. cerevisiae genomic DNA (30) contained Sau3AI partial digests of S. cerevisiae DNA inserted into the yeast shuttle vector pSK1 (11), which can replicate in both yeasts. K. lactis transformation was carried out according to Bianchi et al. (5).
TABLE 1.
List of vectors and plasmids
| Plasmid | Structure, properties, and origin (reference) | Reference |
|---|---|---|
| pUK-S11 | Multicopy vector derived from plasmid pKD1 | 12 |
| pUK-GAL2 | pUK-S11 carrying GAL2 gene (2.6-kb HindIII-EcoRI fragment) (14) | This study |
| pUK-MAL22 | pUK-S11 carrying K. lactis maltase gene MAL22 (5-kb HindIII-PstI fragment from pEFHP) (obtained from A. Dominguez, University of Salamanca; unpublished) | This study |
| KCp491 | Centromeric vector (KEp6 + KlCEN2), low copy number (<3) | 42 |
| KCp-GAL2 | KCp491 carrying GAL2 gene (3.5-kb HindIII fragment) (14) | This study |
| pAF1 | pUK-S11 carrying HXT4 gene (3.8-kb XbaI-BglII fragment from clone F) | This study |
| pKS1 | pUK-S11 carrying HXT1/DDSEI segment (4.0-kb HindIII fragment from clone F) | This study |
| pKK1 | pUK-S11 carrying MAL6T gene (3.7-kb HindIII-BglII fragment) (13) | This study |
| pKL1 | KCp491 carrying the same MAL6T as above | This study |
| pMM1 | pUK-S11 carrying MAL6T-MAL6S segment (7.5-kb HindIII fragment) (13) | This study |
| pKM1 | KCp491 carrying the same MAL6T-MAL6S as above | This study |
DNA sequencing and sequence analysis.
Sequencing was performed by the dideoxy chain termination method (34) with Sequenase version 2 (USB). Sequence analysis was performed with the BlastP program (2).
[14C]galactose uptake.
The method of sugar uptake was essentially that described by Bisson and Fraenkel (7). Cells grown in minimal medium supplemented with 2% galactose were harvested at the exponential phase of growth (A600, 1 to 2), washed twice with ice-cold 0.1 M potassium phosphate buffer (pH 6.5), and resuspended in the same buffer at an A600 of 90. A 60-μl aliquot of cell suspension (about 3.5 mg, dry weight) was used for each measurement. Uptake was initiated by addition of 60 μl of radiolabeled 10 mM galactose (148 Bq/μmol). Aliquots of 27 μl were taken at different time intervals, and uptake was terminated by addition of 5 ml of cold water. Cells were collected and washed on a filter paper before radioactivity was measured with a Tricarb liquid scintillation counter (Packard). d-[U-14C]galactose was obtained from Amersham Pharmacia Biotech. For uptake measurements in the presence of 2.5 μM antimycin A, the cell suspension was preincubated for 5 min with the inhibitor.
RESULTS AND DISCUSSION
Galactose utilization is respiration dependent in K. lactis.
Galactose utilization in K. lactis, as in S. cerevisiae, is a galactose-inducible activity. Most of the galactose regulon genes are equivalent in the two species (10, 39, 45). Galactose is first taken up by a permease, which in S. cerevisiae is encoded by GAL2 (29, 36). In K. lactis, the equivalent galactose permease is the product of LAC12, which is known as the lactose permease gene (32). Some S. cerevisiae strains or variants cannot grow on galactose when respiration is blocked by antimycin A or by a mutation causing respiratory deficiency. The reason for this deficiency has been found to be the presence of an inactive gal2 allele (= imp1) in these strains (1, 14). gal2 mutants still grow slowly on galactose, but this growth is completely abolished when antimycin A is added or when the strain is transformed into a petite strain. Normal K. lactis strains show a behavior reminiscent of gal2 strains of S. cerevisiae. K. lactis cannot grow on galactose in the presence of antimycin A, as if the LAC12 product were deficient. It is, however, not deficient, since K. lactis grows well on galactose when respiration is allowed.
GAL2 gene releases galactose utilization from dependence on respiration in K. lactis.
We asked whether replacement of LAC12 by GAL2 could render K. lactis cells capable of fermentative growth on galactose. We transformed both LAC12 (PM4-4B) and lac12 (P11-1D) strains of K. lactis with GAL2 (carried either by multicopy or low-copy-number shuttle vectors), and the transformed strains were replica plated onto galactose complete medium (YPGal) containing 5 μM antimycin A. All the transformants gained the ability to grow on galactose plus antimycin A plates (data not shown); that is, galactose utilization was no longer respiration dependent. The introduction of the GAL2 gene also seems to confer fermentative activity (see Materials and Methods): the lac12 mutant, which is incapable of fermenting galactose, has become, after transformation, fermentation positive (data not shown).
These observations suggest that the inability of the lac12 mutant to ferment galactose and the inability to grow on galactose plus antimycin A are both due to an insufficient level of galactose permease activity. Moreover, the fact that a single dose of the GAL2 gene was sufficient to allow this conversion even in the lac12 mutant may suggest that S. cerevisiae GAL2 permease is more expressed or more efficient in galactose transport than the LAC12 product, which was originally identified as a lactose transporter.
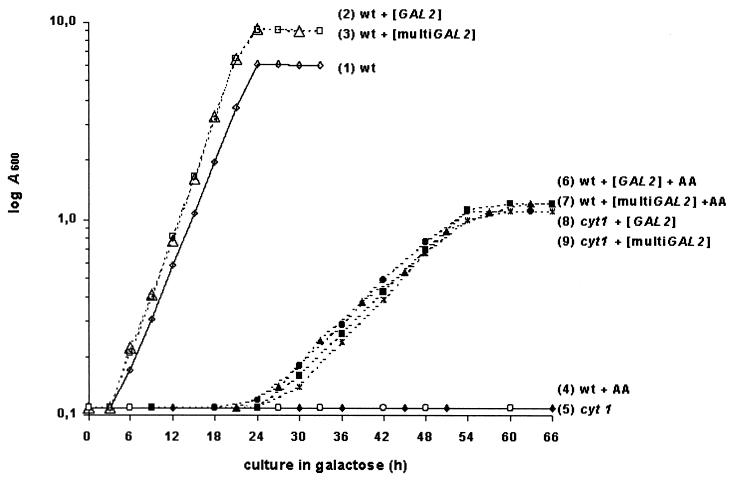
If galactose utilization is dependent on respiration, a K. lactis mutant defective in respiration should be incapable of growing on galactose. Indeed, the cyt1 mutant (respirationless due to a mutation of cytochrome c1 [16]) could not grow on galactose (Fig. 1). Again, transformation of this mutant with the GAL2 gene could restore its growth on galactose. Figure 1 presents quantitative aspects of growth on galactose as a function of respiration. The wild type in the presence of antimycin A as well as the cyt1 mutant did not grow at all on galactose. The doubling time of the wild-type strain was significantly reduced (3.5 to 3 h) by the introduction of GAL2 plasmids. The wild type with GAL2 plasmids and antimycin A as well as the cyt1 mutant with GAL2 plasmids (all respirationless growth) showed similar doubling times of about 10 h. Both the presence of the cyt1 mutation and the addition of antimycin A determined a cell mass yield reduced to the same level, as expected for a typical fermentative growth.
FIG. 1.
Growth curves of wild-type and cyt1 mutant strains in galactose. The wild-type strain JBD100 (wt) and its isogenic cyt1 mutant were transformed with either a multicopy GAL2 plasmid (pUK-GAL2) or a monocopy GAL2 plasmid (KCp-GAL2). The transformants were grown in glucose minimal medium to mid-exponential phase and then transferred to galactose complete medium (YPGal) with or without addition of 5 mM antimycin A (AA). The cultures were grown on a reciprocating shaker at 110 rpm. Growth was followed for 66 h by A600 measurements. Growth curves of nontransformed strains are included for comparison.
Galactose transport activity is reduced by antimycin A and is increased by the addition of the GAL2 gene.
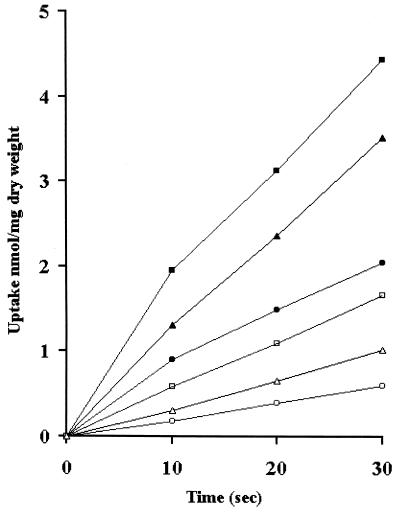
In K. lactis the utilization of respiratory inhibitors is the only way to analyze the effect of respiratory deficiency on galactose transport. Respiratory mutants are, in fact, unable to grow on galactose and cannot grow even with the addition of low glucose concentrations. Higher concentrations of glucose are not convenient because of the repression of the GAL system. An analysis of the rate of d-[1-14C]galactose transport as a function of respiration was performed in wild-type cells induced by including 2% galactose in the medium and then shifted to buffer containing labeled galactose, in the presence or absence of antimycin A (Fig. 2). The drug partially inhibited galactose uptake, leaving about one-third of the activity. In the strain transformed with a low-copy-number GAL2 plasmid, galactose uptake activity was more than doubled, indicating that the GAL2 product was actually contributing to galactose uptake in K. lactis cells. The addition of GAL2 on a high-copy-number plasmid resulted in an increase in galactose uptake activity but at a lower extent than the addition of GAL2 in low copy number. This result suggests that an excess of protein could perturb the equilibrium of the plasma membrane.
FIG. 2.
Effect of antimycin A on galactose transport. K. lactis cells (PM4-4B), transformed or not with GAL2-carrying monocopy plasmid KCp-GAL2 and with GAL2-carrying multicopy plasmid pUK-GAL2, were grown on galactose, washed, and suspended in phosphate buffer. Uptake radiolabeled galactose was determined in the presence and absence of antimycin A as detailed in Materials and Methods. Time zero values were subtracted from all measurements. Symbols: wild-type PM4-4B in the absence (bull;) and presence (○) of antimycin A; PM4-4B/KCp-GAL2 in the absence (▪) and presence (□) of antimycin A; PM4-4B/pUK-GAL2 in the absence (▴) and presence (▵) of antimycin A. The values are means of three independent experiments. In no case was the variation higher than 15%.
The ensemble of results from the galactose experiments supported the idea that in K. lactis galactose uptake is the major limiting step for fermentative growth on galactose.
Additional doses of HXT4 gene release raffinose utilization from dependence on respiration in K. lactis.
Raffinose (α-galactosyl-α-glucosyl-β-fructose) is hydrolyzed by the periplasmic invertase to fructose and melibiose (3). The enzyme is encoded in K. lactis by KlINV1, a homologue of SUC2 of S. cerevisiae (17). Fructose is then transported into the cells by glucose permeases in both species (8, 24, 31, 41). Melibiose is assimilated in S. cerevisiae by the product of several MEL genes encoding an α-galactosidase (26, 27, 28), whereas this sugar is not assimilated by K. lactis (23).
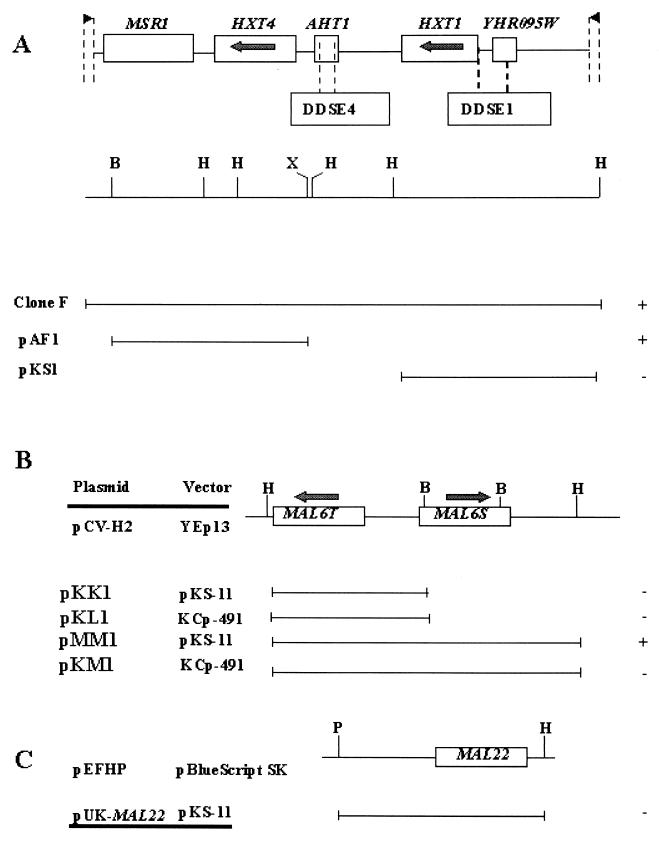
We looked for S. cerevisiae genes which might be able to release K. lactis from its dependence on respiration for raffinose utilization. We transformed the PM4-4B strain with a K. lactis-S. cerevisiae shuttle library of S. cerevisiae genomic DNA and identified five clones that contained a plasmid with an identical DNA insert of 10 kb. One of them (clone F) was sequenced. The cloned DNA was found to be a segment of chromosome VIII of S. cerevisiae (Fig. 3A).
FIG. 3.
(A) Physical map of the HXT4 gene region of S. cerevisiae chromosome VIII. The region cloned into the K. lactis transformant clone F (see text) is shown. The flanking sequences used as primers for sequencing are indicated with arrows and broken lines. pAF1 and pKS1 are subclones. (B and C) Physical map and subcloning of the MAL regions of S. cerevisiae and K. lactis, respectively. Plasmid pCV-H2 contains S. cerevisiae Mal6T (maltose permease) and MAL6S (maltase), and plasmid pEFPH contains the K. lactis MAL22 gene (maltase) (A. Dominguez, unpublished data). pKK1, pKL1, pMM1, pKM1, and pUK-MAL22 are subclones. Open reading frames and DDSE sequences are shown by boxes. DNA inserts are represented by thin lines. Restriction sites: B, BglII; H, HindIII; P, PstI; X, XbaI. The + and − signs stand for the presence and absence, respectively, of transformed clones on raffinose plus antimycin A medium (A) and on maltose plus antimycin A medium (B and C).
This segment contained the genes MSR1, HXT4, and HXT1 and two unknown open reading frames (ORFs), AHT1 and YHR095W. HXT4 and HXT1 are two members of the hexose transporter family. The segment also contained, in the promoter region of the HXT genes, DDSE1 and DDSE4 (sequences involved in glucose sensor Snf3p control [37]). Focusing our attention on the HXT4 and HXT1 genes and DDSE1 sequence, we subcloned the HXT4 gene and a DNA fragment including HXT1 and the DDSE1 region into the K. lactis multicopy vector pUK-S11. We named these plasmids pAF1 and pKS1, respectively (Table 1). They were transformed into PM4-4B, and the transformants were tested for growth on raffinose plus antimycin A plates. As reported in Fig. 3A, only the HXT4 gene was able to support growth in this medium.
The transformation experiment with pAF1 and pKS1 was also performed in the cytochrome c1 mutant JBD100/M3. This respiration-deficient mutant is unable to grow on raffinose. Its growth on raffinose was restored by transformation with the HXT4-carrying plasmid (data not shown). This is consistent with the results of the antimycin A experiments. As observed for growth on galactose, the growth curve of the HXT4-transformed cyt1 mutant displayed a reduced cell yield on raffinose (optical density at 600 nm [OD600] of 0.8) with respect to the wild type (OD600 of 4.2), as expected for fermentative growth.
As the HXT4 gene codes for a hexose transporter, its effect on raffinose utilization was attributed to its supposed permease activity for fructose (a product of raffinose hydrolysis; the other product, melibiose, is not assimilated in this yeast). The increased uptake of fructose might have an additional positive consequence by inducing glycolytic genes like pyruvate decarboxylase (6).
The HXT4 (=LGT1) gene of S. cerevisiae had been identified as a suppressor of the rag1 mutation of K. lactis (30). The rag1 mutant, affected in the low-affinity hexose permease gene RAG1, cannot grow on glucose plus antimycin A (21), that is, the mutant is Kluyver effect positive on glucose. HXT4 complemented this deficiency. We asked if the RAG1 gene has the same effect as HXT4 on raffinose utilization by K. lactis. We found that a multicopy RAG1 plasmid (41) could not restore the growth of PM4-4B on raffinose plus antimycin A medium. This suggests that a specific type of hexose transporter is necessary for restoration of fermentative growth on raffinose. An equivalent of such protein in K. lactis, if it exists, remains to be identified.
Release of maltose utilization from respiration.
Maltose (α-glucosyl-α-glucose) is hydrolyzed by maltase, an α-glucosidase. The enzyme is thought to be located in the inner side of the cytoplasmic membrane, and therefore maltose has to be transported into the cell by a permease. In S. cerevisiae the maltase and the permease are encoded by five unlinked MAL loci (38). In K. lactis, the MAL22 locus, encoding a maltase, and the MAL21 locus, encoding a maltose permease, have been identified recently by A. Dominguez and collaborators at the University of Salamanca (personal communication). These loci are structurally very close to the S. cerevisiae genes. Maltase synthesis in S. cerevisiae is inducible, while in K. lactis induction is strain dependent. In both yeasts, maltase synthesis is repressed by glucose (20, 38).
Assimilation of maltose by K. lactis is also respiration dependent. In order to know whether this dependence is due to a limited maltose permease activity, we examined the effect of addition of the S. cerevisiae MAL6T gene, encoding a maltose permease (13). The study was performed in two strains, PM4-4B and JA6, in which the maltose utilization system is regulated differently, being inducible and constitutive, respectively. We obtained similar results with both strains.
The MAL6T gene carried by a multicopy (pKK1) or a centromeric (pKL1) vector (Table 1) was transformed into K. lactis strains. The transformants were tested for growth on 2% maltose plus antimycin A plates. As shown in Fig. 3B, MAL6T alone did not allow K. lactis to grow on this medium. We therefore tested a multicopy plasmid containing not only MAL6T but also MAL6S, which encodes maltase. When this plasmid, pMM1, was introduced in both strains, growth was fully restored on maltose plus antimycin A (Fig. 3B). However, the combined genes carried by centromeric plasmid pKM1 could not restore growth (Fig. 3B). Therefore, either the maltase or both the maltase and the permease are required at a high level to restore growth. The homologous K. lactis maltase gene, MAL22, on a multicopy vector (plasmid pUK-MAL22, Table 1) could not by itself restore growth on maltose plus antimycin A (Fig. 3B). Thus, we are led to conclude that both the permease and maltase genes are required at high dosage to establish respiration-independent growth on maltose.
The analysis was also performed with the respiration-deficient cyt1 mutant, which cannot grow on maltose. Again, only the MAL6T/MAL6S combined plasmid could restore growth on maltose to this strain (data not shown). The restored growth of the cyt1 mutant gave, however, a reduced cell yield (A600 = 0.7), about one-fifth of that of the wild-type strain JBD100 (A600 = 3.6).
Altogether, these results suggest that, in K. lactis, the available level of maltose-metabolizing enzymes (permease and maltase) is too low to sustain fermentative growth, which demands a high flow of substrate, while this level is sufficient for respiratory growth.
To conclude, we here demonstrated that additional doses of S. cerevisiae sugar tranporter genes can release K. lactis cells from their dependence on respiration for the utilization of galactose, maltose, and raffinose. For galactose and raffinose utilization, the limiting step, in the absence of respiration, is clearly the uptake of the substrate. In the case of maltose, uptake of the substrate is not the only step that limits fermentative growth. In addition to an insufficient permease level, the maltase activity is also too low to ensure fermentative growth.
The Kluyver effect is a widely observed phenomenon in the utilization of various nonglucose sugars by many yeast species. The genetic basis of the phenomenon was examined here on the model of K. lactis with several sugars. The question is of practical importance, since glucose is generally not the main sugar substrate in industrial processes.
One may ask whether lactose utilization by K. lactis is respiration dependent (this yeast and Kluyveromyces marxianus are used for industrial hydrolysis of lactose). A survey on laboratory strains of K. lactis shows that the ability to grow on lactose plus antimycin A is a strain-dependent property (P. Goffrini, unpublished data), suggesting that the lactose permease activity varies in different strains. The present study suggests that the fermentation capacity on lactose may be increased by specifically amplifying the LAC12 gene. The well-known Kluyver effect for maltose utilization in Candida utilis (22, 43) might also involve a limited maltose transport activity of this yeast.
In the genome of S. cerevisiae, a species which has a preference for fermentative life, the sugar transporter genes are amplified to the extreme, and this yeast is considered Kluyver effect negative, but this is not true for all sugars. For example, it is Kluyver effect positive for trehalose, probably because of an insufficient level of the trehalose transporter (AGT1 product), as suggested by Malluta et al. (25).
Acknowledgments
We thank H. Fukuhara for helpful suggestions and for critical comments on the manuscript. We also thank A. Dominguez (Department of Microbiology and Genetics, University of Salamanca, Spain) for the pEFHP plasmid; R. Dickson (Department of Biochemistry, University of Kentucky College of Medicine, Lexington, Ky.) for the Kllac-12 mutant 11D304; J. Subik (Department of Microbiology and Virology, Faculty of Natural Sciences, Comenius University, Bratislava, Slovak Republic) for the cytochrome c1 mutant JBD 100/M3; M. Vanoni (Dipartimento di Biotecnologie e Bioscienze, Università degli Studi di Milano-Bicocca, Milan, Italy) for the pCV-H2 plasmid; and M. Wésolowski-Louvel (Centre de Génétique Moleculaire et Cellulaire, CNRS UMR 5534, Université Claude Bernard, Villeurbanne, France) for the S. cerevisiae genomic library.
This work was supported by grants from the Ministero Università e Ricerca Scientifica e Tecnologica-Università di Parma Cofin 1999 and Cofin 2000.
REFERENCES
- 1.Algeri, A. A., L. Bianchi, A. M. Viola, P. P. Puglisi, and N. Marmiroli. 1981. IMP1/imp1: a gene involved in the nucleo-mitochondrial control of galactose fermentation in Saccharomyces cerevisiae. Genetics 97:27–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, J. A. 1976. The utilization of sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 32:125–234. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, J. A. 1992. Some controls on oligosaccharide utilization by yeasts: the physiological basis of the Kluyver effect. FEMS Microbiol. Lett. 79:371–378. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi, M. M., C. Falcone, X. J. Chen, M. Wésolowski-Louvel, L. Frontali, and H. Fukuhara. 1987. Transformation of the yeast Kluyveromyces lactis by new vectors derived from the 1.6 μm circular plasmid pkD1. Curr. Genet. 12:185–192. [Google Scholar]
- 6.Bianchi, M. M., L. Tizzani, M. Destruelle, L. Frontali, and M. Wesolowski-Louvel. 1996. The ‘;petite-negative’ yeast Kluyveromyces lactis has a single gene expressing pyruvate decarboxylase activity. Mol. Microbiol. 19:27–36. [DOI] [PubMed] [Google Scholar]
- 7.Bisson, L. F., and D. G. Fraenkel. 1983. Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80:730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boles, E., and C. P. Hollenberg. 1997. The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21:85–111. [DOI] [PubMed] [Google Scholar]
- 9.Breunig, K. D. 1989. Glucose repression of LAC gene expression in yeast is mediated by the transcriptional activator LAC9. Mol. Gen. Genet. 216:422–427. [DOI] [PubMed] [Google Scholar]
- 10.Breunig, K. D., M. Bolotin-Fukuhara, M. M. Bianchi, D. Bourgarel, C. Falcone, I. Ferrero, L. Frontali, P. Goffrini, J. J. Krijger, C. Mazzoni, C. Milkowski, H. Y. Steensma, M. Wesolowski-Louvel, and A. M. Zeeman. 2000. Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb. Technol. 26:771–780. [DOI] [PubMed] [Google Scholar]
- 11.Chen, X. J. 1987. Etude du plasmide pKD1 et développement de systèmes d’expression chez la levure. Ph.D. thesis. Université Paris XI, Paris, France.
- 12.Chen, X. J., and H. Fukuhara. 1988. A gene fusion system using the aminoglycoside-3′-phosphotransferase gene of the kanamycin-resistance transposon Tn903: use in the yeast Kluyveromyces lactis and Saccharomyces cerevisiae. Gene 69:181–192. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, J. D., M. J. Goldenthal, T. Chow, B. Buchferer, and J. Marmur. 1985. Organization of the MAL loci of Saccharomyces: physical identification and functional characterization of three genes at the MAL6 locus. Mol. Gen. Genet. 200:1–8. [DOI] [PubMed] [Google Scholar]
- 14.Donnini, C., T. Lodi, I. Ferrero. A. A. Algeri, and P. P. Puglisi. 1992. Allelism of IMP1 and GAL2 genes of Saccharomyces cerevisiae. J. Bacteriol. 174:3411–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Entian, K. D., and J. A. Barnett. 1983. Some genetical and biochemical attempts to elucidate the energetics of sugar uptake and explain the Kluyver effect in the yeast Kluyveromyces lactis. Curr. Genet. 7:323–325. [DOI] [PubMed] [Google Scholar]
- 16.Gbelska, Y., K. Horvathova, Q. J. van der Aart, B. J. Zonneveld, H. Y. Steensma, and J. Subik. 1996. Isolation and molecular analysis of the gene for cytochrome c1 from Kluyveromyces lactis. Curr. Genet. 30:145–150. [DOI] [PubMed] [Google Scholar]
- 17.Georis, I., J. P. Cassart, K. D. Breunig, and J. Vandenhaute. 1999. Glucose repression of the Kluyveromyces lactis invertase gene KlINV1 does not require Mig1p. Mol. Gen. Genet. 261:862–870. [DOI] [PubMed] [Google Scholar]
- 18.Goffrini, P., A. A. Algeri, C. Donnini, M. Wesolowski-Louvel, and I. Ferrero. 1989. RAG1 and RAG2: nuclear genes involved in the dependence/independence on mitochondrial respiratory function for growth on sugars. Yeast 5:99–106. [DOI] [PubMed] [Google Scholar]
- 19.Goffrini, P., A. Ficarelli, C. Donnini, T. Lodi, P. P. Puglisi, and I. Ferrero. 1996. FOG1 and FOG2 genes, required for the transcriptional activation of glucose-repressible genes of Kluyveromyces lactis are homologous to GAL83 and SNF1 of Saccharomyces cerevisiae. Curr. Genet. 29:316–326. [PubMed] [Google Scholar]
- 20.Goffrini, P., A. Ficarelli, and I. Ferrero. 1995. Hexokinase activity is affected in mutants of Kluyveromyces lactis resistant to glucose repression. Microbiology 141:441–447. [Google Scholar]
- 21.Goffrini, P., M. Wesolowski-Louvel, I. Ferrero, and H. Fukuhara. 1990. RAG1 gene of the yeast Kluyveromyces lactis codes for a sugar transporter. Nucleic Acids Res. 18:5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaliterna, J., R. A. Weusthuis, J. I. Castrillo, J. P. Van Dijken, and J. T. Pronk. 1995. Transient responses of Candida utilis to oxygen limitation: regulation of the Kluyver effect for maltose. Yeast 11:317–325. [DOI] [PubMed] [Google Scholar]
- 23.Kurtzman, C. P., and J. W. Fell (ed.). 1998. The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 24.Lagunas, R. 1993. Sugar transport in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 10:229–242. [DOI] [PubMed] [Google Scholar]
- 25.Malluta, E. F., P. Decker, and B. U. Stambuk. 2000. The Kluyver effect for trehalose in Saccharomyces cerevisiae. J. Basic Microbiol. 40:199–205. [DOI] [PubMed] [Google Scholar]
- 26.Naumov, G., H. Turakainen, E. Naumova, S. Aho, and M. Korhola. 1990. A new family of polymorphic genes in Saccharomyces cerevisiae: alpha-galactosidase genes MEL1–MEL7. Mol. Gen. Genet. 224:119–128. [DOI] [PubMed] [Google Scholar]
- 27.Naumov, G., E. Naumova, H. Turakainen, P. Suominem, and M. Korhola. 1991. Polymeric genes MEL8, MEL9 and MEL10—new members of alpha-galactosidase gene family in Saccharomyces cerevisiae. Curr. Genet. 20:269–276. [DOI] [PubMed] [Google Scholar]
- 28.Naumov, G., E. Naumova, H. Turakainen, and M. Korhola. 1996. Identification of the alpha-galactosidase MEL genes in some populations of Saccharomyces cerevisiae: a new gene MEL11. Genet. Res. 67:101–108. [DOI] [PubMed] [Google Scholar]
- 29.Nehlin, J. O., M. Carlberg, and H. Ronne. 1989. Yeast galactose permease is related to yeast and mammalian glucose transporters. Gene 85:313–319. [DOI] [PubMed] [Google Scholar]
- 30.Prior, C., H. Fukuhara, J. Blaisonneau, and M. Wesolowski-Louvel. 1993. Low-affinity glucose carrier gene LGT1 of Saccharomyces cerevisiae, a homologue of the Kluyveromyces lactis RAG1 gene. Yeast 9:1373–1377. [DOI] [PubMed] [Google Scholar]
- 31.Reifenberger, E., K. Freidel, and M. Ciriacy. 1995. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol. 16:157–167. [DOI] [PubMed] [Google Scholar]
- 32.Riley, M. I., K. Sreekrishna, S. Bhairi, and R. C. Dickson. 1987. Isolation and characterization of mutants of Kluyveromyces lactis defective in lactose transport. Mol. Gen. Genet. 208:145–151. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sims, A. P., and J. A. Barnett. 1978. The requirement of oxygen for the utilization of maltose, cellobiose and d-galactose by certain anaerobically fermenting yeast. J. Gen. Microbiol. 166:277–288. [Google Scholar]
- 36.Szkutnicka, K., J. F. Tschopp, L. Andrews, and V. P. Cirillo. 1989. Sequence and structure of the yeast galactose transporter. J. Bacteriol. 171:4486–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theodoris, G., N. M. Fong, D. M. Coons, and L. F. Bisson. 1994. High-copy suppression of glucose transport defects by HXT4 and regulatory elements in the promoters of the HXT genes in Saccharomyces cerevisiae. Genetics 137:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanoni, M., P. Sollitti, M. Goldenthal, and J. Marmur. 1989. Structure and regulation of the multigene family controlling maltose fermentation in budding yeast. Prog. Nucleic Acid Res. Mol. Biol. 37:281–322. [DOI] [PubMed] [Google Scholar]
- 39.Webster, T. D., and R. C. Dickson. 1988. The organization and transcription of the galactose gene cluster of Kluyveromyces lactis. Nucleic Acids Res. 16:8011–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wésolowski, M., A. A. Algeri, P. Goffrini, and H. Fukuhara. 1982. Killer DNA plasmids of the yeast Kluyveromyces lactis. 1. Mutations affecting the killer phenotype. Curr. Genet. 5:191–197. [DOI] [PubMed] [Google Scholar]
- 41.Wesolowski-Louvel, M., P. Goffrini, I. Ferrero, and H. Fukuhara. 1992. Glucose transport in the yeast Kluyveromyces lactis. I. Properties of an inducible low-affinity glucose transporter gene. Mol. Gen. Genet. 233:89–96. [DOI] [PubMed] [Google Scholar]
- 42.Wesoloswki-Louvel, M., K. D. Breunig, and H. Fukuhara. 1996. Kluyveromyces lactis, p.139–201. In K. Wolf (ed.), Nonconventional yeast in biotechnology. Springer-Verlag, Berlin, Germany.
- 43.Weusthuis, R. A., M. A. Luttik, W. A. Scheffers, J. P. van Dijken, and J. T. Pronk. 1994. Is the Kluyver effect in yeasts caused by product inhibition? Microbiology 140:1723–1729. [DOI] [PubMed] [Google Scholar]
- 44.Wickerham, L. J. 1951. Taxonomy of yeast. Tech. Bull. 1029. U.S. Department of Agriculture, Washington, D.C.
- 45.Zachariae, W., P. Kuger, and K. D. Breunig. 1993. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9 (KlGAL4). Nucleic Acids Res. 21:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]