Abstract
The penicillin-binding proteins (PBPs) are a set of enzymes that participate in bacterial peptidoglycan assembly. The absolute numbers of each PBP were determined by direct measurement and have been reported for two Staphylococcus aureus strains, RN4220 (methicillin-sensitive S. aureus) and RN450M (methicillin-resistant S. aureus). From the specific activity of the labeled penicillin and the absolute number of disintegrations per minute, and from the number of CFU per milliliter calculated from proteins and optical density, a determination of the number of PBPs per cell was made. These numbers ranged from approximately 150 to 825 PBPs/cell and represent the first direct determination of absolute numbers of PBPs in S. aureus.
The penicillin-binding proteins (PBPs) are a set of enzymes that catalyze the terminal reactions of bacterial peptidoglycan biosynthesis. The transpeptidation and carboxypeptidase functions are inhibited when these proteins are acylated by a β-lactam antibiotic, with the β-lactam ring opening and forming a covalent bond with an active-site serine residue on the PBP (10, 11). Much of the understanding we have about the roles of the individual PBPs comes from work done primarily with the gram-negative rod-shaped organism Escherichia coli and the gram-positive rod-shaped organism Bacillus subtilis (3, 13, 22). Working with E. coli, Spratt published an indirect estimate of the numbers of individual PBPs in an average E. coli cell (18). Nearly 2 decades later, Dougherty et al. directly calculated the numbers of molecules of individual PBPs per cell by measuring both the cell numbers present and the radioactivity of the individual PBPs in a precise quantity of cells (9).
In this work, we measure the numbers of the individual PBPs of the gram-positive coccus Staphylococcus aureus using a similar direct method. Methicillin-resistant S. aureus (MRSA) strains mediate resistance by the acquisition of exogenous DNAs that contains an additional PBP, PBP 2a, encoded by the mecA gene (1, 4, 16, 19). The functioning of this PBP in MRSA strains allows for sufficient cell wall biosynthesis in the presence of β-lactam antibiotics such as methicillin at concentrations that would result in the killing of methicillin-sensitive S. aureus (MSSA) strains (2, 7). Similar to what was done in the previous E. coli study (9), the individual PBPs were saturated with [3H]penicillin and a sufficiently large number of samples were taken in separate experiments to allow statistical assessment of the data sets. The numbers of PBPs were quantitated and compared between genetically related MSSA and MRSA strains, and the number of PBP 2a molecules in the resistant strain was determined.
In vivo labeling and quantitation of PBPs in rapidly growing S. aureus cells.
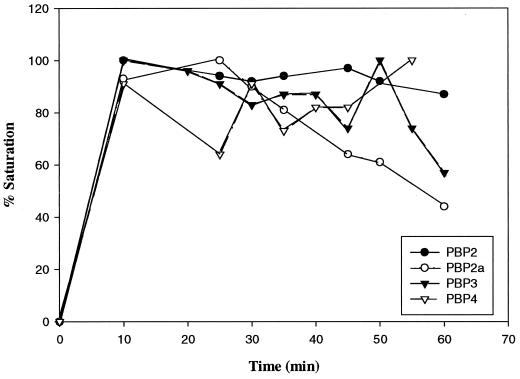
Because of variable losses that occur during the membrane preparation process, and in order to accurately assess the number of PBPs per cell, it was necessary to be able to label cells with [3H]penicillin directly rather than use prepared cell membranes. Previous results indicated that there are no significant differences between the PBP patterns of isolated membranes and in vivo-labeled cells (M. Pucci, unpublished data). Prior to the quantitation experiments, it was necessary to examine binding to the PBPs over time to ensure that the proteins were fully saturated with radiolabeled benzylpenicillin at the time binding was stopped. This was particularly important for PBP 4, as the penicillin-bound enzyme has been shown to be rapidly deacylated, with a half-life of 4 to 6 min compared to the half-life (>40 min) of relatively stable acyl derivatives for PBPs 1, 2, and 3 (6, 21). Cells were incubated with radiolabeled penicillin, and samples were withdrawn at various times from 10 to 60 min and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In order to determine relative values of PBPs from fluorographs, the X-ray films were prefogged (15) and, after exposure, the film was scanned with an LKB Ultroscan XL laser densitometer (Pharmacia LKB Biotech, Uppsala, Sweden). Areas under the curves are automatically calculated by the instrument. The results for PBPs 2, 2a, 3, and 4 are shown in Fig. 1. Full saturation was achieved for all PBPs within 10 to 20 min at 37°C. Interestingly, while binding remained relatively constant over the course of the experiment for PBPs 2, 3, and 4, PBP 2a exhibited progressively less intense bands after 25 min.
FIG. 1.
Percentages of saturation of PBPs with [3H]benzylpenicillin over time at 37°C. Fluorograph bands (film not shown) were scanned (see the text for details), and the highest areas under the curve were arbitrarily set as 100% for each PBP.
One-milliliter samples of cells from mid-logarithmic-phase cultures of S. aureus RN4220 (restriction-deficient ATCC 8325-1; Bla− [17]) and RN450M (ATCC 8325-1 transformed with the COL mec region; Bla−) grown in brain heart infusion broth were taken. The samples for PBP determinations were immediately centrifuged and resuspended in 50 μl of lysis buffer consisting of 50 mM Tris-HCl (pH 7.0), 10 mM MgCl2, 2.5 μg of DNase/ml, and 2.5 μg of RNase/ml (5). Two micrograms (final concentration, 40 μg/ml) of radiolabeled penicillin ([3H]benzylpenicillin, ethylpiperidinium salt, 26.5 Ci/mmol; DuPont NEN, Boston, Mass.) was immediately added, followed by a 20-min incubation at 37°C. Five microliters of lysostaphin was added to a final concentration of 200 μg/ml, and samples were incubated at 37°C for an additional 5 min to allow cell lysis to occur. Immediately after lysis occurred, SDS-PAGE sample buffer was added, the samples were heated for 5 min at 100°C, and the entire volume was loaded into a sample lane for SDS-PAGE. Following electrophoresis, gels were subjected to fluorography for 5 to 7 days to reveal the positions of the radiolabeled PBPs as described previously (9).
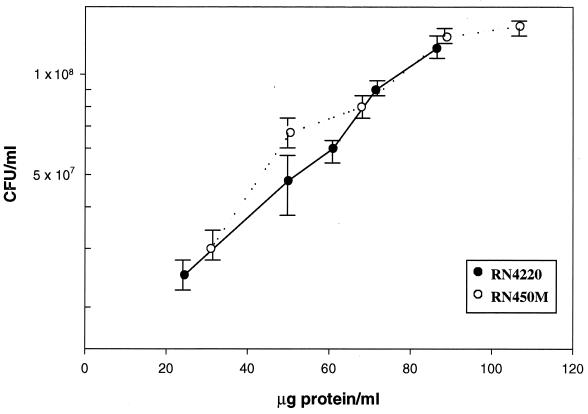
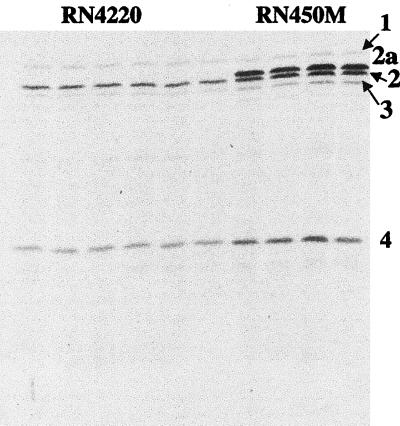
Areas containing the radiolabeled PBPs were cut out from the dried gel, placed in a scintillation vial, rehydrated, and extracted. In order to provide sufficient radioactive counts for counting statistics, two adjacent lanes with identical samples of a given PBP were placed in the same vial to ensure adequate counts per minute per vial. This reduced the probable counting error to below 3% (12). In order to obtain the absolute number of disintegrations per minute from the observed counts per minute, a quench curve was constructed. In all experiments, 12 identical sample lanes were run, thus yielding six determinations for each PBP. Samples for cell number determinations were immediately diluted and plated on brain heart infusion agar for colony enumeration after incubation overnight. Staphylococci undergo cell division in two planes and are often found in clusters. In an attempt to minimize variability in the direct determination of numbers of CFU per milliliter, a conversion from micrograms of protein per milliliter was used. By analysis of multiple fields under phase-contrast microscopy, we found that clusters were predominantly tetrads under the selected growth conditions, and a factor of 4 was used to convert numbers of CFU to numbers of cells (Table 1). Each experiment yielded three values for each individual PBP for each strain, along with the number of CFU per milliliter derived from at least six plate counts of colonies. At the same time, the amount of protein in micrograms per milliliter was also calculated. These data were then used to plot numbers of CFU per milliliter versus micrograms of protein per milliliter (Fig. 2), and it was determined that 106 CFU was equivalent to 1 μg of protein for each strain. Using the total number of micrograms of protein per milliliter, the total number of disintegrations per minute per lane, and the specific activity (26.5 Ci/mmol; determined by high-performance liquid chromatography analysis) of the radioactive penicillin, the number of molecules of radioactive penicillin per microgram of protein could be calculated. By using the above conversion, the number of molecules of radiolabeled penicillin per CFU could be determined. The number of molecules of radiolabeled penicillin per CFU divided by 4 yielded the numbers of individual PBPs per cell. Results from a total of four independent experiments are included in the values in Table 1, and the averages and standard deviations for all 12 data points are presented. A representative PBP profile film is shown in Fig. 3. These results indicate that the numbers of PBPs per cell in S. aureus ranged from about 150 to 200 for PBPs 1 and 3 to about 800 for PBP 2a. This range is similar to what was previously reported for E. coli (9).
TABLE 1.
Numbers of PBPs in MSSA and MRSA strains
| S. aureus strain | PBPa (molecular mass [kDa]) | No. of molecules of [3H]penicillin/μg of proteinb | Avg no. of molecules of [3H]penicillin/CFU ± SD | Avg no. of molecules of [3H]penicillin/cellc ± SD | % of total |
|---|---|---|---|---|---|
| RN4220 | 1 (80) | 7.5 × 108 | 737 ± 32 | 184 ± 8 | 17.1 |
| 2 (73) | 1.8 × 109 | 1,844 ± 243 | 461 ± 61 | 42.8 | |
| 3 (70) | 5.9 × 108 | 593 ± 86 | 148 ± 21 | 13.7 | |
| 4 (46) | 1.1 × 109 | 1,139 ± 117 | 285 ± 29 | 26.4 | |
| RN450M | 1 (80) | 6.7 × 108 | 665 ± 139 | 166 ± 35 | 8.6 |
| 2a (74) | 3.3 × 109 | 3,292 ± 1,519 | 824 ± 379 | 42.6 | |
| 2 (73) | 1.8 × 109 | 1,798 ± 388 | 449 ± 97 | 23.2 | |
| 3 (70) | 7.9 × 108 | 787 ± 241 | 197 ± 60 | 10.2 | |
| 4 (46) | 1.2 × 109 | 1,196 ± 243 | 299 ± 61 | 15.4 |
PBPs are numbered according to standard nomenclature (20).
RN4220, 36 μg of protein/ml; RN450M, 45 μg of protein/ml.
Approximately four cells per CFU.
FIG. 2.
CFU per millilter versus micrograms of protein per milliliter of staphylococcal culture.
FIG. 3.
A representative fluorograph of the PBPs of S. aureus RN4220 and RN450M. The PBPs are numbered on the right according to standard nomenclature. All cell samples received saturating amounts of [3H]benzylpenicillin. First through 6th lanes, RN4220; 7th through 10th lanes, RN450M.
Comparison of direct quantitation and laser densitometry.
The percentages shown in Table 2 are the PBP ratios obtained using the generated absolute values of PBPs by excision and scintillation counting compared with the relative values obtained by laser densitometry. The comparison illustrates that laser densitometry yields relative ratios of PBPs similar to those obtained by direct counting determinations of individual PBPs. This relationship was also observed previously with E. coli PBPs (9). A recalculation of the percentages for RN450M PBPs 1 through 4 after PBP 2a values were subtracted (Table 3) yielded numbers similar to those seen for the MSSA strain RN4220, which lacks PBP 2a, particularly when the direct determination numbers were compared.
TABLE 2.
Relative percentages calculated by densitometric scanning of PBPs compared with those obtained by direct determination
| S. aureus strain | PBP | % of total PBP value
|
|
|---|---|---|---|
| Direct determination | Scanning | ||
| RN4220 | 1 | 17.1 | 13.2 |
| 2 | 42.8 | 46 | |
| 3 | 13.7 | 9.8 | |
| 4 | 26.4 | 31 | |
| RN450M | 1 | 8.6 | 4.8 |
| 2a | 42.6 | 39 | |
| 2 | 23.2 | 26 | |
| 3 | 10.2 | 8.7 | |
| 4 | 15.4 | 21.5 | |
TABLE 3.
Relative percentages calculated by densitometric scanning of PBPs compared with those obtained by direct determination of PBP values minus PBP 2a values for S. aureus RN450M
| PBP | % of total PBP value − PBP 2a valuea
|
|
|---|---|---|
| Direct determination | Scanning | |
| 1 | 14.9 | 7.9 |
| 2a | NA | NA |
| 2 | 40.4 | 42.4 |
| 3 | 17.7 | 14.4 |
| 4 | 26.9 | 35.3 |
NA, not applicable.
To our knowledge, the numbers of PBPs in a gram-positive coccus have not been directly calculated to date. It was of interest to compare the numbers of PBPs with those of the gram-negative rod E. coli using essentially the same experimental technique. The numbers of each individual staphylococcal PBP ranged from 120 to approximately 800 in each cell. Whereas E. coli was found to have about 2,700 PBPs per cell (9), S. aureus RN4220 had 1,078 PBPs and S. aureus RN450M had 1,935 PBPs (1,111 minus values for PBP 2a) (Table 1). An important difference is that E. coli has approximately 1,880 PBPs per cell with carboxypeptidase activity (PBPs 4, 5, 6, 7, and 8). PBP 5 alone has about 800 copies per cell. Staphylococci appear to lack such high numbers of low-molecular-weight PBPs. Therefore, these data suggest that, although staphylococci have thicker cell walls and more layers of peptidoglycan, the actual numbers of PBPs may be less than the total for E. coli. The larger size and, therefore, larger surface area of the E. coli cell can only partially account for the differences in the numbers.
Labischinski et al. found that E. coli appeared to have a single layer of peptidoglycan covering some 70 to 75% of the cell and a triple layer covering the remaining 25 to 30% (14). These findings contrast with the thicker peptidoglycan of gram-positive organisms, estimated to be as much as 20 to 25 layers thick. Studies by Young and colleagues of E. coli, in which multiple PBP deletions had been constructed, indicate that the majority of PBPs in this organism are dispensable (8, 22). Thus, cell wall synthesis in E. coli depends primarily on a numerically small subset of essential PBPs, and other PBPs, while making up the vast majority of the molecules, appear to have fine-tuning functions on the peptidoglycan structure. Thus, the seeming disparity between the amount of peptidoglycan and the number of PBPs in E. coli compared with the relationship seen in S. aureus may not be as great as the raw numbers indicate. In addition, a fraction (∼20%) of the gram-positive cell wall is composed of teichoic acids, a polymer absent from the gram-negative peptidoglycan layer. Finally, another contributing factor may be that E. coli, by virtue of having a larger genome, may simply have more built-in redundancies than gram-positive cocci.
REFERENCES
- 1.Archer, G. L., and D. M. Niemeyer. 1994. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 2:343–347. [DOI] [PubMed] [Google Scholar]
- 2.Berger-Bachi, B. 1999. Genetic basis of methicillin resistance in Staphylococcus aureus. Cell Mol. Life Sci. 56:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers, H. F. 1999. Penicillin-binding protein-mediated resistance in pneumococci and staphylococci. J. Infect. Dis. 179:S353–S359. [DOI] [PubMed] [Google Scholar]
- 4.Chambers, H. F., B. Hartman, and A. Tomasz. 1985. Increasing amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J. Clin. Investig. 76:325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers, H. F., and C. Miick. 1992. Characterization of penicillin-binding protein 2 of Staphylococcus aureus: deacylation reaction and identification of two penicillin-binding peptides. Antimicrob. Agents Chemother. 36:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, H. F., M. J. Sachdeva, and C. J. Hackbarth. 1994. Kinetics of penicillin binding to penicillin-binding proteins of Staphylococcus aureus. Biochem. J. 301:139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248–11254. [PubMed] [Google Scholar]
- 8.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougherty, T. J., K. Kennedy, R. E. Kessler, and M. J. Pucci. 1996. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J. Bacteriol. 178:6110–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghuysen, J.-M. 1991. Serine β-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 45:37–67. [DOI] [PubMed] [Google Scholar]
- 11.Granier, B., M. Jamin, M. Adam, M. Galleni, B. Lakaye, W. Zorzi, J. Grandchamps, J.-M. Wilkin, C. Fraipont, B. Joris, C. Duez, M. Nguyen-Disteche, J. Coyette, M. Leyh-Bouille, J. Dusart, L. Christiaens, J.-M. Frere, and J.-M. Ghuysen. 1994. Serine-type d-ala-d-ala peptidases and penicillin-binding proteins. Methods Enzymol. 244:249–266. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, Y., and D. V. Maudsley. 1974. Biological applications of liquid scintillation counting, p.184. Academic Press, Inc., New York, N.Y.
- 13.Koch, A. L. 2000. Penicillin binding proteins, beta-lactams, and lactamases: offensives, attacks, and defensive countermeasures. Crit. Rev. Microbiol. 26:205–220. [DOI] [PubMed] [Google Scholar]
- 14.Labischinski, H., E. W. Goodell, A. Goodell, and M. L. Hochberg. 1991. Direct proof of a “more-than-single-layered” peptidoglycan architecture of Escherichia coli W7: a neutron small-angle scattering study. J. Bacteriol. 173:751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laskey, R. A., and A. D. Mills. 1975. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur. J. Biochem. 56:335–341. [DOI] [PubMed] [Google Scholar]
- 16.Matsuhashi, M., M. D. Song, F. Ishino, M. Wachi, M. Doi, M. Inoue, K. Ubukata, N. Yamashita, and M. Konno. 1986. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to β-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 167:975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novick, R. N. 1991. Genetic systems in Staphylococci. Methods Enzymol. 204:587–636. [DOI] [PubMed] [Google Scholar]
- 18.Spratt, B. G. 1977. Properties of the penicillin-binding proteins of Escherichia coli K-12. Eur. J. Biochem. 72:341–352. [DOI] [PubMed] [Google Scholar]
- 19.Ubukata, K., R. Nonoguchi, M. Matsuhashi, and M. Konno. 1989. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J. Bacteriol. 171:2882–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ubukata, K., N. Yamashita, and M. Konno. 1985. Occurrence of a β-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 27:851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waxman, D. J., and J. L. Strominger. 1979. Cephalosporin-sensitive penicillin-binding proteins of Staphylococcus aureus and Bacillus subtilis active in the conversion of [14C]penicillin G to [14C]phenylacetylglycine. J. Biol. Chem. 254:10256–10261. [PubMed] [Google Scholar]
- 22.Young, K. D. 2001. Approaching the physiological functions of penicillin-binding proteins in Escherichia coli. Biochimie 83:99–102. [DOI] [PubMed] [Google Scholar]