Abstract
Intimin is an important virulence factor in two groups of enteric pathogens: enteropathogenic Escherichia coli (EPEC), which is a major cause of infant diarrhea in the developing world, and enterohemorrhagic E. coli (EHEC), which has caused large food-borne outbreaks of hemorrhagic colitis in the United States and other developed countries. Intimin is encoded on a 35-kb pathogenicity island called the locus of enterocyte effacement (LEE). At least five antigenic types have been described for the highly variable gene, and each type is generally characteristic of particular evolutionary lineages. We determined the nucleotide sequences of intimin and other LEE genes in two O111 clones that have not been amenable to typing. The sequences from both O111:H8 and O111:H9 differed from the Int-β that is typical of other clones in the same evolutionary lineage. The sequence from the O111:H8 strains was a mosaic of divergent segments that alternately clustered with Int-α, Int-β, or Int-γ. The sequence from the O111:H9 clone consistently showed a close relationship with that from E2348/69, a distantly related strain that expresses Int-α. The results suggest that there have been multiple acquisitions of the LEE in the EHEC 2/EPEC 2 clonal lineage, with a recent turnover in either O111:H8 or its close relatives. Amino acid substitutions that alter residue charge occurred more frequently than would be expected under random substitution in the extracellular domains of intimin, suggesting that diversifying selection has promoted divergence in this region of the protein. An N-terminal domain that presumably functions in the periplasm may also be under positive selection.
Intimin is an outer membrane protein that functions in the virulence of enteropathogenic Escherichia coli (EPEC), a pathotype that is a leading cause of infant diarrhea in the developing world (24). The protein is encoded by the eae gene, which is part of a pathogenicity island called the locus of enterocyte effacement (LEE) (22). In addition to intimin, the LEE island encodes several secreted proteins, a type III secretion system, and Tir, a protein that is translocated to the host cell membrane, where it serves as the receptor for intimin. The proteins produced by the LEE island mediate a characteristic histopathology called attaching and effacing lesions (8). The mucosal lesions result from the intimate attachment of EPEC to the host intestinal epithelium, effacement of microvilli, and the formation of actin-rich pedestals that elevate individual bacteria above the host cell membrane (17). The attaching and effacing phenotype can also be exhibited by enterohemorrhagic E. coli (EHEC), a pathotype implicated in food- and waterborne outbreaks of hemorrhagic colitis (HC) and hemolytic-uremic syndrome (HUS) in the United States and other developed countries (5).
The acquisition of the LEE in the EPEC and EHEC clonal lineages is considered a key evolutionary event that set the stage for the parallel emergence of pathotypes (29). According to one model (38), LEE inserted into the selC site and the subsequent divergence and acquisition of different mobile virulence elements gave rise to two pathogenic lineages, EPEC 1 and EHEC 1 (40). In another ancestral clone, LEE inserted into the pheU site (32) and the EHEC 2 and EPEC 2 clones diverged through a second series of acquisition events (5). The stepwise evolutionary scenario provides a working hypothesis for the persistence of LEE in clonal lineages and the subsequent in situ divergence of genes through mutation and recombination.
Genetic variation in the intimin gene is essentially concordant with the evolutionary model. At least five antigenic types have been detected, and the allelic types are generally associated with particular clonal lineages (1). Int-α is characteristic of EPEC 1 strains, whereas Int-γ is found in EHEC 1 and closely related O55:H7 strains. The two sister groups, EPEC 2 and EHEC 2, usually carry Int-β. However, there are puzzling exceptions to the clonal framework. For example, the EHEC 2 strains with serotype O111:H8 generally do not react with Int-β antibodies, nor do they amplify with Int-β specific PCR primers (28). This is an enigma because O111:H8 strains typically carry an eae homologue, detectable by either gene hybridization (4, 31) or PCR (28), and these strains can produce mucosal lesions in experimental infections (34). How can this be? One possibility is that the O111 eae sequence is a minor variant, say, of the Int-β allele, that has diverged by point mutations. A second possibility is that it is a novel sequence resulting from intragenic recombination, such as seen in the α, β, and γ intimin variants (23). Another possibility is that the distinct intimin of the O111:H8 clone reflects the acquisition of an entirely different LEE island into an EHEC 2 clonal frame.
The objective of this study was to infer the evolutionary history of the intimin gene in the O111:H8 clone. To accomplish this goal, we sequenced the eae gene in five O111:H8 strains originally recovered from separate cases of disease more than 40 years apart. We also included two isolates of an O111:H9 clone whose intimin type is unknown. The nucleotide sequences were used to construct gene phylogenies for all or parts of the intimin alleles, to detect recombination within the eae gene, and to assess whether natural selection has promoted or constrained the rate of amino acid change in different intimin domains.
MATERIALS AND METHODS
Bacterial strains.
The strains used in the study (Table 1) were originally isolated from patients with diarrheal disease. Strains DEC 8a (CDC 2198-77), DEC 8b (CDC3030A-86), and CL-37 were recovered from cases of HC or HUS in North America (Table 1). Strain 3215-99 (Texas SHD no. F6627) was isolated from a patient in an outbreak in Texas, the first HC outbreak in the United States attributable to a Shiga toxin-producing O111 strain (2). Strain 412/55 was collected in 1955 and is one of the earliest EHEC isolates known (H. Karch, personal communication). Strain 921-B4 is an O111:H9 strain originally isolated from an outbreak of diarrhea in Finland in 1987 that affected more than 700 people, including healthy adults (35).
TABLE 1.
Clinical isolates of E. coli for which the eae gene was sequenced
| Isolate | Serotype | Locality (yr) | Source or reference |
|---|---|---|---|
| DEC 8a | O111:NM | Maryland (1977) | Centers for Disease Control and Prevention |
| DEC 8b | O111:H8 | Idaho (1986) | 3 |
| CL-37 | O111:H8 | Canada (1982) | 16 |
| 3215-99 | O111:H8 | Texas (1999) | Centers for Disease Control and Prevention |
| 412/55 | O111:? | Germany (1955) | H. Karch |
| 921-B4 | O111:H9 | Finland (1987) | 35 |
| 9084-83 | O111:NM | Peru (1983) | Centers for Disease Control and Prevention |
Genomic DNA extraction.
Cultures were grown overnight in Luria-Bertani broth at 37°C. DNA was extracted with a Puregene DNA extraction kit (Gentra Systems, Minneapolis, Minn.) following the manufacturer’s recommendations for gram-negative bacteria.
Enzymatic amplification and nucleotide sequence determination.
The program Primer Designer (version 2.0) was used to design primers in the two genes flanking eae (escD and cesT) (6, 7): escD1143F 5′-CAT TCT GAA AGG AGG CTA TGT C-3′; and cesT242F, 5′-TAT GGT TTG CAG AGA ATG GTG G-3′. The primers were used in the PCR at a final concentration of 0.5 μM with deoxynucleoside triphosphates at 0.2 mM each, 2.5 U of TaqPlus Precision (Stratagene), and 100 ng of DNA template.
The thermal cycle, which was proceeded by a 3-min soak at 94°C, was run for 35 cycles on a Perkin-Elmer 9700 with the following parameters: 92°C, 40 s; 66°C, 1 min; and 72°C, 3.5 min. The resulting 3.4-kb amplicons were purified with Qiaquick PCR purification kits (Qiagen). DNA was electrophoresed in ethidium bromide-stained gels and quantified under UV illumination by comparison to a low-mass DNA ladder (Gibco BRL).
Cycle sequencing reactions were performed with CEQ dye terminator cycle sequencing kits (Beckman) with approximately 50 fmol of template and a final primer concentration of 2 μM. The thermal cycle was run for 30 cycles with the following parameters: 94°C, 20 s; 57°C, 20 s; and 60°C, 4 min. Reactions were purified with Centricep columns (Princeton Separations), and the DNA ladder was detected on a Beckman CEQ2000 capillary sequencer. Sequences were concatenated and aligned with the program SEQMAN in the computer package DNASTAR. Internal primers for sequencing were sequentially designed as data were generated.
Sequence analysis.
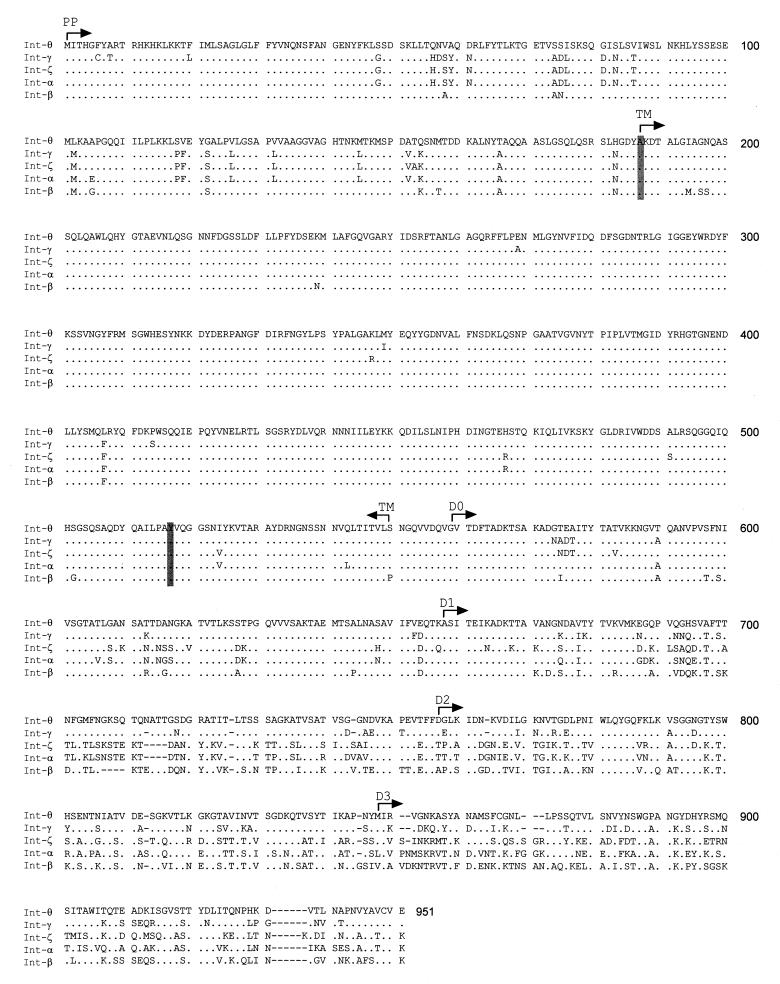
CLUSTAL X (33) was used to produce a multiple alignment of 28 inferred amino acid sequences, which included the seven sequences determined here and 21 sequences retrieved from GenBank. Phylogenetic trees were constructed with the neighbor-joining algorithm (30) with the program MEGA version 2.0 (18). Trees were based on synonymous distance (dIs) calculated by the modified Nei-Gojobori method (15, 25) with a Jukes-Cantor correction applied to account for multiple substitutions at single nucleotide sites. The genes were partitioned into three segments for phylogeny estimation. The first segment specifies the 186 residues at the N terminus, which presumably functions as the periplasmic (PP) domain. The second segment includes the conserved central domain identified by McGraw et al. (23), which spans the region from the alanine residue at position 187 through the lysine residue at position 517 (Fig. 1). The third segment included the residues at the C terminus (residues 518 to 945), which comprises the four extracellular (EC) domains identified by Luo et al. (20).
FIG. 1.
Amino acid alignment of five intimin sequences. Dots indicate identity to Int-θ, and dashes indicate alignment gaps. Consensus gaps indicate amino acid residues present in sequences that are not in those shown here. Shaded residues delineate the conserved central domain identified by McGraw et al. (23). Arrows demarcate the functional domains (20). GenBank accession numbers for previously published sequences are Int-α, AF022236; Int-β, AF081187; and Int-γ, AF081182.
Heterogeneity in sequence divergence resulting from either recombination or natural selection was assessed by the maximum chi-square method of Maynard Smith (21). The computer program, MAXCHI, implements a version of this method to identify the ends of segments of a mosaic allele. The program compares each sequence to a reference sequence and finds the point, kmax, at which the chi-square statistic achieves a maximum. The sequence was then divided into two segments, and a new maximum was found within each segment. This cycle was repeated four times so that 16 maxima were found. The significance of the kmax values was tested by a Monte Carlo procedure (23).
We used two methods to detect the past action of natural selection in intimin evolution. First, the proportion of synonymous differences per synonymous site (pS) and the proportion of nonsynonymous difference per nonsynonymous site (pN) were estimated by the Nei-Gojobori method (25) with MEGA (18). Variation in functional constraint across the eae gene was examined by tabulating the average pS and pN for each of the functional domains delineated by Luo et al. (20) and for 30-codon subsets in a sliding window using a computer program called PSWIN. These quantities have been used to detect adaptive evolution (26), because the rates of evolution per site are expected to be equal for selectively neutral mutations (pS = pN), the synonymous rate exceeds the nonsynonymous rate for purifying (negative) selection (pS > pN), and the nonsynonymous rate exceeds the synonymous rate for diversifying (positive) selection (pN > pS). Second, we used the method of Hughes et al. (13) to assess whether amino acid replacements that change residue property (radical change) occur more often than chance would dictate. The method estimates the rate of radical nonsynonymous change (pNR) versus the rate of conservative nonsynonymous change (pNC). We computed pNC and pNR with radical changes defined as charge or polarity for the transmembrane (TM) and EC domains. These are “per site” measures that are expected to be equal for selectively neutral mutations (13).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been submitted to GenBank under accession numbers AF449414 to AF449420. The eae alignment is available from the authors’ website (http://foodsafe.msu.edu/whittam/).
RESULTS
Nucleotide polymorphism in eae.
The intimin gene was amplified and sequenced from the seven O111 strains listed in Table 1. The sequences fell into two distinct groups. The eae gene of the O111:H8 isolates and nonmotile relatives was 2,808 nucleotides in length; it encodes a protein of 935 amino acids. Only one polymorphic site was observed among the five sequences: strain CL-37 differed from the remaining four by a single base change that predicts an amino acid replacement (G662E). The five sequences were similar to the eae sequence published for isolate 95NR1 (36), a Shiga toxin-producing O111:H− strain implicated in a foodborne outbreak of HUS in Australia (27). Among these six sequences, there were 11 polymorphic sites, seven of which are nonsynonymous substitutions. The second group includes the eae sequences from strains 921-B4 and 9084-83, which were identical and predict a slightly larger protein 938 amino acids in length.
To determine the evolutionary relationships of the eae genes from O111 strains with other known intimin variants of other pathovars, the O111 eae sequences were aligned with 21 homologous genes retrieved from GenBank. A multiple alignment with CLUSTAL X yielded a total of 951 amino acid positions of the combined data set of 28 inferred amino acid sequences. A number of gaps were introduced in the alignment, the majority of which were in the C-terminal region of the gene. Among the 28 sequences, there were 21 unique variants at the nucleotide level. Part of the multiple alignment, including the predicted amino acid sequences for the intimins of the O111:H8 and O111:H9 strains to the α (EPEC O127:H6), β (EPEC O111:H2), and γ (EHEC O157:H7) intimins, is shown in Fig. 1. The alignment reveals that the eae sequence of the O111:H8 clone is divergent from other intimins (Fig. 1). We hereafter refer to O111:H8 intimin as the “Int-θ” allele. Likewise, the sequence in O111:H9 is similar to the allele designated “ζ” (GenBank accession no. AJ298279.1) (J. Jores, K. Zehmke, L. Roumer, and L. Wieler, unpublished data), so we refer to the O111:H9 eae sequence as the Int-ζ allele class.
The level of divergence between intimins varies in different regions of the molecule (Table 2). For example, in the N-terminal region (codons 1 to 550) (Fig. 1), Int-θ is most closely related to Int-β, differing at 3.1% of the amino acid positions. In the carboxyl end of the molecule (codons 551 to 951) (Fig. 1), however, Int-θ is most similar to Int-γ and is more than 35% different from the other intimin alleles. For the Int-ζ of the O111:H9 lineage, the primary structure is nearly identical to Int-α in the N terminus but is much more divergent (24%) in the C-terminal domains.
TABLE 2.
The percentage of inferred amino acid differences between major alleles of intimina
| Intimin allele (strain) | % of differences with:
|
||||
|---|---|---|---|---|---|
| Int-θ | Int-γ | Int-ζ | Int-α | Int-β | |
| Int-θ (3215-99) | 0 | 5.5 | 5.1 | 4.9 | 3.1 |
| Int-γ (EDL-933) | 18.3 | 0 | 2.2 | 2.0 | 5.6 |
| Int-ζ (921-B4) | 37.9 | 36.3 | 0 | 0.9 | 5.3 |
| Int-α (E2348/69) | 35.5 | 36.6 | 24.1 | 0 | 4.9 |
| Int-β (DEC12a) | 36.9 | 33.4 | 34.7 | 33.7 | 0 |
Codon positions with gaps in the multiple alignment (Fig. 1) were excluded from the calculation. The values for the conserved N-terminal region (550 codon positions compared) are given above the diagonal and the values for the EC C-terminal region (377 codon positions compared) are given below the diagonal.
Based on the crystal structure of α-intimin in complex with Tir, Luo and colleagues (20) identified five functional domains: the N-terminal membrane anchor region, which includes the PP and TM domains, and four EC domains, labeled D0 to D3 (Fig. 1). Of these five domains, the PP and TM domains were the most conserved, with 104 (19%) of the 550 amino acid positions being variable. Most of the variable amino acid positions were found in the C terminus: 237 out of 393 sites (60%) were polymorphic across the four EC domains, and D3—the domain that binds to Tir—had the greatest concentration of variable amino acid positions.
At the nucleotide level, the five domains differ dramatically in the proportions of pS and pN nucleotide substitutions (Table 3). The average pS (× 100) equals 12.0 in the N-terminal domains and is more than four times greater in the EC domains. The average pN is also substantially greater in the EC domains than in the N-terminal region, with the greatest value in D3. The ratio of pS to pN, a measure of the strength of natural selection at the molecular level, ranges from the most conserved value of 4.4 in the PP + TM domains to the least conserved of 1.4 in D3 (Table 3). These ratios indicate that on average the N-terminal membrane anchor region has twice the selective constraint of the EC domains of intimin.
TABLE 3.
Variation in the proportion of pS and pN substitutions in five functional domains of intimin of Luo et al. (20)a
| Domain | pS × 100 (SE) | pN × 100 (SE) | Ratio |
|---|---|---|---|
| PP + TM | 12.00 (0.86) | 2.72 (0.30) | 4.4 |
| D0 | 19.68 (2.30) | 6.90 (1.34) | 2.9 |
| D1 | 40.34 (2.61) | 19.80 (2.13) | 2.0 |
| D2 | 43.28 (2.24) | 21.70 (2.36) | 2.0 |
| D3 | 41.66 (2.36) | 30.59 (2.38) | 1.4 |
| Entire gene | 21.51 (0.87) | 9.50 (0.54) | 2.3 |
The average pairwise values are expressed as number of substitutions per 100 sites based on 21 eae nucleotide sequences. D0 through D3 denote the four EC domains.
Phylogenetic analysis.
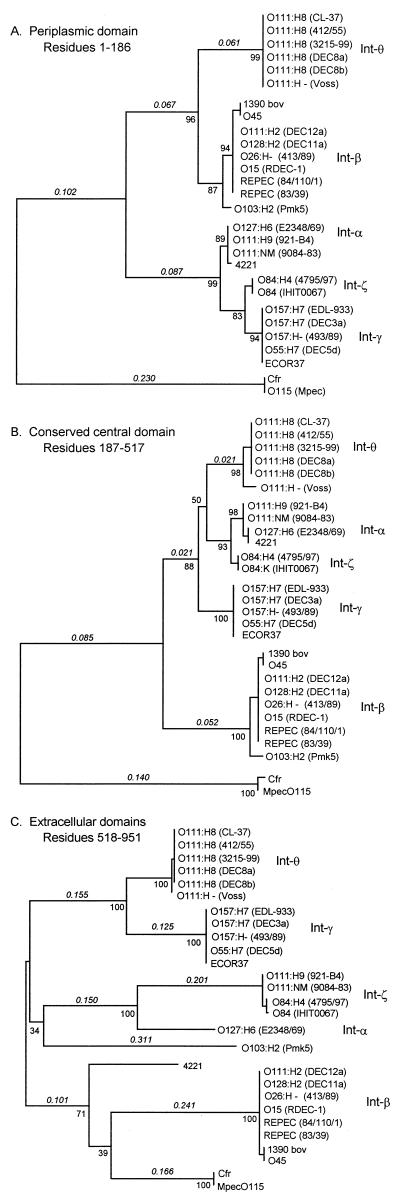
To elucidate the origin of Int-θ and Int-ζ in pathogenic O111 strains, we inferred a phylogeny for the three main regions of the eae gene: the N-terminal 186 residues that comprise the putative PP domain, the 331 residues that comprise the central conserved domain, and the 434 residues of the C-terminal EC domains (Fig. 2). The evolutionary relationship of the O111:H8 Int-θ allele with the α, β, and γ alleles depends on the segment of the gene that was used to infer a phylogeny. The topology for the N-terminal segment places Int-θ with the Int-β allele cluster, a cluster that includes an EPEC O111 strain and an EHEC O26 strain (Fig. 2A). In contrast, Int-θ clusters first with Int-α in the tree constructed from the conserved central domain (Fig. 2B). The tree based on the EC domains indicates a third relationship in which Int-θ is most closely related to the Int-γ alleles characteristic of the EHEC O157:H7 strains (Fig. 2C). The close connection between Int-θ and Int-γ was also obtained when individual gene trees were constructed for each EC domain separately (results not shown).
FIG. 2.
Gene phylogenies inferred for three regions of the intimin gene. Trees were constructed from ds with the neighbor-joining algorithm. Branch lengths are shown in italics above each branch. The numbers at nodes are the percentage of bootstrap replications in which a particular node is supported.
The Int-ζ sequence for O111:H9 strains consistently showed a closer relationship with the Int-α than with either Int-β or Int-γ, regardless of the segment that was used to construct the gene tree. Sequences for two O84 isolates (Int-ζ) also consistently clustered with Int-α and were closest to the O111:H9 eae sequences. However, the Int-α sequence was divergent from the four ζ alleles in the EC domains (Fig. 2C).
Such striking differences in the phylogeny for different parts of a molecule can result from natural selection altering the rate of molecular evolution (and thus distorting the tree) or from past horizontal transfers and recombination which can create mosaic alleles from gene segments with distinct histories. In the next sections, we analyze the mosaic structure of intimin alleles and present evidence for radical amino acid change in the EC domains.
Heterogeneity and mosaic structure.
To determine points of significant heterogeneity in sequence divergence, we applied the maximum chi-square method (21) to pairs of intimin alleles. The comparison of Int-θ to Int-α disclosed seven breakpoints with significant kmax values. The notable segment is the piece between positions 34 and 182, which differs at 12% of the nucleotides and is embedded in a region of 1 to 2% sequence divergence (Fig. 3). At the 3′ end, there is remarkable heterogeneity ranging from 7 to 88% sequence difference in short stretches of 50 to 100 bp of DNA. Int-θ and Int-γ also show differences in the 3′ end of the gene which encodes the PP domain. In this case, the conservation of the central domain extends through to codon 675 and includes D0. The final EC segment is 16.1% divergent in the two sequences, which reflects substantial sequence divergence but is less than half the divergence seen in the EC domains between other intimin allele classes. There were no significant breakpoints detected in the 5′ end of the Int-θ and Int-β comparison. The first breakpoint is at position 490, followed by three points marking significant heterogeneity spaced at ∼100-bp intervals in the 3′ end of the sequences.
FIG. 3.
Plot of the variable nucleotide sites and location of significant breakpoints (kmax) in pairwise comparisons of intimin alleles. Each breakpoint is denoted by a vertical slash with the nucleotide position shown below. Each vertical line marks the location of a nucleotide difference between Int-θ and the eae sequence labeled in the figure. Numbers above are the percentages of differences in the segment. The level of divergence is highlighted with colors: yellow, <5%; orange, 5 to 10%; pink, 10 to 20%. Domain structure is drawn below with the conserved central (CC) region shaded in gray.
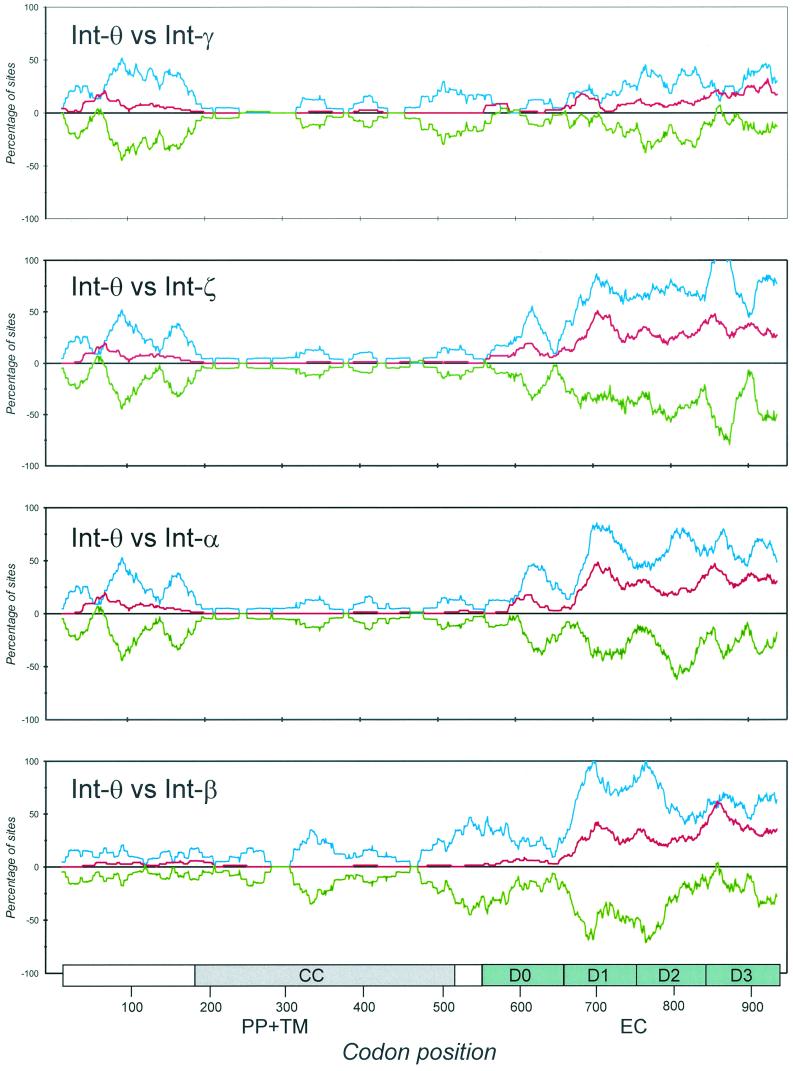
The degree of selective constraint on the amino acid sequence of intimin is shown by the pairwise comparisons of pS and pN substitutions (Fig. 4). In all five intimin domains, pS exceeded pN, over most of each region, indicating that purifying selection predominates in intimin evolution. Both pS and pN show a marked increase in the last three EC domains, with the highest value of pN in D3. The increase in pN indicates that there is less constraint on amino acid replacements in the EC domains than in the TM domain.
FIG. 4.
Plot of pS and pN substitutions per 100 sites in a 30-codon sliding window. Each panel is a comparison of two intimin alleles. Colored lines represent the following values: blue, pS; red, pN; and green, pN − pS. The difference (pN − pS) measures the degree of selective constraint on a region. The boundaries of each functional domain are drawn at the bottom of the diagram.
In two regions, pN > pS, which suggests that selection is favoring amino acid replacements. The first region is centered on residue 60 in the 5′ end of the gene and is seen in comparison of Int-θ with -γ, -β, and -α. This region presumably is involved in the periplasmic functions of intimin. The second region is in D2 and D3 of the C terminus of the protein. In this case, pN > pS only in the comparison of Int-θ with Int-γ and Int-β.
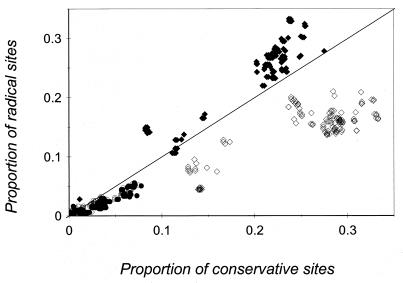
The comparison of pNC versus pNR shows that a greater proportion of amino acid replacements in the TM domain involves conservative substitutions. In contrast, amino acid replacements that involve charge changes occur more frequently in the EC domains than expected under random substitution (Fig. 5). However, radical changes that involve polarity occur less frequently. The results suggest that amino acid replacements that alter charge are selectively favored in the external domains of intimin.
FIG. 5.
Proportion of pNR plotted against the proportion of pNC for all pairwise comparisons of intimin TM and combined EC domains. Diamonds, EC domains; and circles, TM domain. Solid symbols indicate comparisons where radical substitutions involve residue charge change; open symbols indicate comparisons where radical substitutions involve change in polarity.
DISCUSSION
An interesting finding of this study is the extensive divergence of eae in O111:H8 relative to other strains in the EHEC 2 lineage. The results are particularly surprising because of the close relationship between O111:H8 and the O26:H11 clone. The two EHEC 2 strains are virtually indistinguishable on the basis of multilocus enzyme electrophoresis (39) and multilocus sequencing (29), so they would be predicted to have nearly identical intimin sequences as well. The similarity of the eae gene in O111:H9 to that in E2348/69 was also unexpected. While the relationship of the O111:H9 clone to other strains in the EHEC 2 and EPEC 2 cluster has not been fully resolved, its placement in the group is strongly supported by multilocus sequencing (29) and restriction fragment data (10).
Why is the intimin in the O111 strains uncharacteristic of that in the EHEC 2/EPEC 2 evolutionary lineage? One hypothesis is that recombination could have altered intimin in the two O111 clones. Alternatively, it is possible that the O111 strains have independently acquired a different copy of the LEE. To better resolve the history of the O111 sequences, we sequenced additional LEE genes from two isolates of O111:H8 (CL-37 and 3215-99) and one of O111:H9 (921-B4). We selected two genes that are known to be highly polymorphic: tir, which is in the same operon as eae, and sepZ, which is located ∼10 kb upstream. The tir and sepZ sequences from the O111:H8 lineage clustered with those from EPEC 1 strain E2348/69 (results not shown), which raises the possibility that the backbone of the LEE in O111:H8 is most closely related to the Int-α-associated LEE from EPEC 1. Sequences for O111:H9 also clustered with sequences from E2348/69. The results from additional LEE genes suggest a third hypothesis: that α-LEE is ancestral and that the O111 clones have retained the ancestral copy while related strains have lost α-LEE and gained a divergent copy of the island that carries Int-β.
Sperandio et al. (32) found that two O111:H9 strains shared with EPEC 1 the selC insertion site for the LEE. Thus, it is clear that the LEE in O111:H9 has been acquired independently of other strains in EHEC 2 and EPEC 2, in which the LEE is typically in the pheU site. It is plausible that O111:H9 has retained an ancestral copy of the LEE in the selC site; however, on the basis of a phylogenetic reconstruction of pathogenic E. coli (5), at least six parallel losses of the LEE would be required if this hypothesis were true. A greater number could be required, depending on the exact placement of the O111:H9 clone in a complete phylogeny for EHEC 2 and EPEC 2. If O111:H9 separated prior to the diversification of the rest of the group, then a single loss of α-LEE and gain of β-LEE are required in the clone that gave rise to the rest of the group. However, if O111:H9 diverged long after the diversification of the group, then many more independent losses and gains would have to be inferred. The most parsimonious explanation (and hence the likeliest) minimizes the number of evolutionary events that are required under a given hypothesis. It is more parsimonious to assume that the O111:H9 clone has lost (or never had) the β-LEE backbone and independently acquired an α-LEE.
Retention of an ancestral α-LEE is not a plausible explanation for O111:H8 because the selC site is not occupied and because the LEE island is presumably in the pheU site (32). Intimin in O111:H8 is a mosaic of segments that have different evolutionary histories, with sequence from one region (the PP domain) clustering with those from other EHEC 2 strains. Has recombination simply modified an Int-β in O111:H8? This seems unlikely because the divergence of Int-θ from Int-β at synonymous sites (dS = 0.095) in the PP domain is 30 times greater than the average divergence among Int-β alleles (dS = 0.003); the distance between the two-allele classes is considerable, as it represents about half the divergence among intimin allele classes in E. coli (Fig. 2A). Moreover, not only is the intimin in O111:H8 different from other EHEC 2 clones, but the sequences for tir and sepZ are actually more similar to those from the EPEC 1 strain that harbors Int-α. Thus, the LEE in O111:H8 is a novel mosaic made up of divergent segments that differ from those found in other E. coli strains. One implication of these results is that the LEE has turned over recently either in O111:H8 or in its sister O26 clone. It is interesting that O111:H8 has a Tir-binding domain that is γ-like, whereas Tir itself is α-like. It remains to be investigated how modified LEE genes can function in a divergent LEE backbone, how such divergence influences pathogenesis, and how each LEE backbone is regulated in different genetic backgrounds.
Evidence for selection on intimin domains.
Bacterial genes encoding proteins that are secreted or exposed on the cell surface characteristically show a high level of sequence polymorphism (19, 37). One hypothesis to explain the variation in exposed proteins is that diversifying selection accelerates the rate of amino acid substitutions, thereby generating new protein variants that are not recognized by the host immune system. The observation that the outer domains of intimin are highly immunogenic and highly polymorphic raises the possibility that diversifying selection promotes evolutionary change in these domains (1, 23). Alternatively, the constraints on amino acid substitution may be relaxed in the intimin EC domains relative to the TM domain (where hydrophobicity must be maintained in order to anchor the protein in the cell membrane) so that amino acid changes accumulate more rapidly in the EC domains under neutral evolution.
If the external domains of intimin are evolving under diversifying selection, then the rate of nonsynonymous substitutions should be higher than the rate of synonymous substitutions (pN − pS > 0). In this case, amino acid substitutions are favored by selection so that replacement substitutions accumulate at a higher rate than synonymous substitutions. Alternatively, if the domains evolve under neutrality, then the rates of pN and pS should be similar (pN − pS ≈ 0). The pattern of substitution for eae (Fig. 4) suggests that purifying selection predominates in intimin evolution over time, as synonymous changes outnumber nonsynonymous changes (pN − pS < 0), even over most of the EC domain. The rate of nonsynonymous substitution is higher in the external and PP domains than in the TM domain, indicating that amino acid changes are less constrained in the two end regions. Despite the prevalence of purifying selection, some evidence for positive selection over restricted regions was apparent both in the PP and EC domains (Fig. 4).
Although we predicted that diversifying selection would be apparent in the EC domains, we did not anticipate finding evidence that amino acid changes are favored in the PP region. The boundary of the domain was not clearly defined based on crystallography data (20), nor has its function been well characterized. Here the PP domain comprises the 186 amino acids at the N terminus of the conserved central domain; the central domain was defined by McGraw et al. (23) based on amino acid conservation between intimin and invasin and was delineated by the outermost two conserved amino acid residues that did not encompass any alignment gaps. Because the function of the domain is not clear, it is difficult to speculate why amino acid substitutions would be favored. It is plausible that it must evolve to interact with different or divergent proteins in the various E. coli strains that harbor the LEE island.
Although we found evidence of diversifying selection in the EC domains, the effect was not as great as we expected. A complicating factor in detecting diversifying selection in highly divergent genes such as intimin is that sites that are free to vary may have become saturated (have undergone multiple substitutions), so that further changes are obscured. Among the intimin sequences, most pairwise comparisons involve either very closely related sequences, where there are too few changes for statistical inference, or very distantly related sequences, where sites are likely saturated. To provide additional evidence for diversifying selection, we used another test that compares two classes of nonsynonymous substitutions: those that are conservative with respect to amino acid properties and those that are radical, where the substitution alters a residue characteristic such as charge or polarity. Natural selection may act on the amino acid replacements so that substitutions are not random with respect to a residue property. When pNC > pNR, substitutions are occurring in such a way to maintain amino acid characteristics, whereas when pNR > pNC, substitutions that change residue property are occurring more frequently than expected by chance; the implication is that natural selection resists or favors changes in residue property, respectively.
The comparison of pNR and pNC provides not only evidence for selection but can also reveal the residue property that is under selection. Residue charge is important, as it is a major determinant in protein binding. Comparison of pNR and pNC suggests that natural selection may favor residue charge changes in surface proteins of pathogens as well as in host defense molecules. For example, some comparisons of the peptide binding region of major histocompatibility complex class I molecules suggest that natural selection favors replacements that alter residue charge (12): in the products of two class I loci, HLA-A and HLA-B, pNR significantly exceeded pNC. In addition, the pattern of charge changes differed between the two loci, and there was a correspondence between charge variation in each HLA protein and the peptides that it binds (11). Residue charge changes also appear to be favored in defensins (14), which are small, antimicrobial peptides that are secreted into the gut lumen in response to microbial invasion (9). Some surface pathogen proteins also show a propensity towards charge changes. In Plasmodium falciparum, amino acid replacements that altered charge occurred more frequently than conservative changes for four of five surface proteins (but not in four nonsurface proteins) (11). The implication from these studies is that charge changes in pathogen surface proteins are favored by selection because the changes allow the pathogen to escape host defenses.
The comparison of radical and conservative amino acid replacements in intimin suggests that the EC domains of intimin are under diversifying selection (Fig. 5). If the EC domains were evolving under neutrality, then amino acid replacements would be random with respect to charge and polarity. However, changes are not random with respect to either property. When radical amino acid replacements are defined as those that alter residue charge, radical changes outnumber conservative changes (pNR > pNC), suggesting that such replacements are selectively favored. However, when radical changes involve polarity differences, pNC exceeds pNR, so alterations in this residue property are not favored (Fig. 5). The propensity for charge-changing amino acid substitutions supports the hypothesis that diversification of intimin extracellular domains is driven by natural selection, perhaps as a means to escape immune surveillance in vertebrate hosts.
Acknowledgments
We thank Jia Sohn and Sheila Plock for technical assistance. We also thank Nancy Strockbine, Centers for Disease Control and Prevention, and Helge Karch for kindly supplying strains.
The research was supported by grants from the National Institutes of Health.
REFERENCES
- 1.Adu-Bobie, J., L. R. Trabulsi, M. M. Carneiro-Sampaio, G. Dougan, and G. Frankel. 1998. Identification of immunodominant regions within the C-terminal cell binding domain of intimin α and intimin β from enteropathogenic Escherichia coli. Infect. Immun. 66:5643–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2000. From the Centers for Disease Control and Prevention. Escherichia coli O111:H8 outbreak among teenage campers—Texas, 1999. JAMA 283:2517–2518. [PubMed] [Google Scholar]
- 3.Bopp, C. A., K. D. Greene, F. P. Downes, E. G. Sowers, J. G. Wells, and I. K. Wachsmuth. 1987. Unusual verotoxin-producing Escherichia coli associated with hemorrhagic colitis. J. Clin. Microbiol. 25:1486–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos, L. C., T. S. Whittam, T. A. T. Gomes, J. R. C. Andrade, and L. R. Trabulsi. 1994. Escherichia coli serogroup O111 includes several clones of diarrheagenic strains with different virulence properties. Infect. Immun. 62:3282–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 107:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliot, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. Deng, L.-C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176–1189. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, S. J., J. Yu, and J. B. Kaper. 1999. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67:4260–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganz, T. 1999. Defensins and host defense. Science 286:420–421. [DOI] [PubMed] [Google Scholar]
- 10.Herbelin, C. J., S. C. Chirillo, K. A. Melnick, and T. S. Whittam. 2000. Gene conservation and loss in the mutS-rpoS genomic region of pathogenic Escherichia coli. J. Bacteriol. 182:5381–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes, A. L. 1999. Adaptive evolution of genes and genomes. Oxford University Press, New York, N.Y.
- 12.Hughes, A. L., and M. K. Hughes. 1995. Natural selection on the peptide-binding regions of major histocompatibility complex molecules. Immunogenetics 42:233–243. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, A. L., T. Ota, and M. Nei. 1990. Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol. Biol. Evol. 7:515–524. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, A. L., and M. Yeager. 1997. Molecular evolution of the vertebrate immune system. Bioessays 19:777–786. [DOI] [PubMed] [Google Scholar]
- 15.Ina, Y. 1995. New methods for estimating the numbers of synonymous and nonsynonymous substitutions. J. Mol. Evol. 40:190–226. [DOI] [PubMed] [Google Scholar]
- 16.Karmali, M. A., B. T. Steele, M. Petric, and C. Lim. 1983. Sporadic cases of haemolytic-uremic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619–620. [DOI] [PubMed] [Google Scholar]
- 17.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. Jakobsen, and M. Nei. 2000. MEGA 2: Molecular Evolutionary Genetics Analysis Program. Version 2.0. Pennsylvania State University, University Park.
- 19.Li, J., H. Ochman, E. A. Groisman, E. F. Boyd, F. Solomon, K. Nelson, and R. K. Selander. 1995. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc. Natl. Acad. Sci. USA 92:7252–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073–1077. [DOI] [PubMed] [Google Scholar]
- 21.Maynard Smith, J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129. [DOI] [PubMed] [Google Scholar]
- 22.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12–22. [DOI] [PubMed] [Google Scholar]
- 24.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426. [DOI] [PubMed] [Google Scholar]
- 26.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, N.Y.
- 27.Paton, A. W., R. M. Ratcliff, R. M. Doyle, J. Seymour-Murray, D. Davos, J. A. Lanser, and J. C. Paton. 1996. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J. Clin. Microbiol. 34:1622–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid, S. D., D. J. Betting, and T. S. Whittam. 1999. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J. Clin. Microbiol. 37:2719–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64–67. [DOI] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- 31.Scotland, S. M., G. A. Willshaw, H. R. Smith, B. Said, N. Stokes, and B. Rowe. 1993. Virulence properties of Escherichia coli strains belonging to serogroups O26, O55, O111, and O128 isolated in the United Kingdom in 1991 from patients with diarrhoea. Epidemiol. Infect. 111:429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperandio, V., J. B. Kaper, M. R. Bortolini, B. C. Neves, R. Keller, and L. R. Trabulsi. 1998. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol. Lett. 164:133–139. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzipori, S., R. Gibson, and J. Montanaro. 1989. Nature and distribution of mucosal lesions associated with enteropathogenic and enterohemorrhagic Escherichia coli in piglets and the role of plasmid-mediated factors. Infect. Immun. 57:1142–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viljanen, M. K., T. Peltola, S. Y. T. Junnila, L. Olkkonen, H. Järvinen, M. Kuistila, and P. Huovinen. 1990. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet 336:831–834. [DOI] [PubMed] [Google Scholar]
- 36.Voss, E., A. W. Paton, P. A. Manning, and J. C. Paton. 1998. Molecular analysis of Shiga toxigenic Escherichia coli O111:H− proteins which react with sera from patients with hemolytic-uremic syndrome. Infect. Immun. 66:1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittam, T. S. 1995. Genetic population structure and pathogenicity in enteric bacteria, p. 217–245. In S. Baumberg, J. P. W. Young, E. M. H. Wellington, and J. R. Saunders (ed.), Population genetics of bacteria. Cambridge University Press, Cambridge, England.
- 38.Whittam, T. S., and E. A. McGraw. 1996. Clonal analysis of EPEC serogroups. Rev. Microbiol. 27(Suppl. 1):7–16. [Google Scholar]
- 39.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Ørskov, I. Ørskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieler, L. H., T. K. McDaniel, T. S. Whittam, and J. B. Kaper. 1997. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of strains. FEMS Microbiol. Lett. 156:49–53. [DOI] [PubMed] [Google Scholar]