Abstract
The terminal step of triacylglycerol (TAG) formation in the yeast Saccharomyces cerevisiae is catalyzed by the enzyme acyl-CoA:diacylglycerol acyltransferase (DAGAT). In this study we demonstrate that the gene product of YOR245c, Dga1p, catalyzes a major yeast DAGAT activity which is localized to lipid particles. Enzyme measurements employing a newly established assay containing radioactively labeled diacylglycerol (DAG) as a substrate and unlabeled palmitoyl-CoA as a cosubstrate revealed a 70- to 90-fold enrichment of DAGAT in lipid particles over the homogenate but also a 2- to 3-fold enrichment in endoplasmic reticulum fractions. In a dga1 deletion strain, the DAGAT activity in lipid particles is dramatically reduced, whereas the activity in microsomes is affected only to a minor extent. Thus, we propose the existence of DAGAT isoenzymes in the microsomal fraction. Furthermore, we unveiled an acyl-CoA-independent TAG synthase activity in lipid particles which is distinct from Dga1p and the phosphatidylcholine:DAGAT Lro1p. This acyl-CoA-independent TAG synthase utilizes DAG as an acceptor and free fatty acids as cosubstrates and occurs independently of the acyl-CoA synthases Faa1p to Faa4p. Based on lipid analysis of the respective deletion strains, Lro1p and Dga1p are the major contributors to total cellular TAG synthesis, whereas other TAG synthesizing systems appear to be of minor importance. In conclusion, at least three different pathways are involved in the formation of storage TAG in the yeast.
Triacylglycerols (TAGs) are the most important storage form for energy and fatty acids required for membrane biosynthesis in eukaryotic cells. Within the last decade, the formation of TAG in various organisms has been investigated. The most prominent enzyme of this biosynthetic pathway is acyl-coenzyme A (CoA):diacylglycerol acyltransferase (DAGAT; EC 2.3.1.20), which was shown to form TAG from diacylglycerol (DAG) and acyl-CoA as substrates (24). Genes encoding DAGATs homologous to acyl-CoA:cholesterol acyltransferase (ACAT) were identified in plants (14, 30, 42) and in mice (4). In addition, a DAGAT without sequence similarity to ACATs was recently discovered in the oleaginous fungus Mortierella ramanniana (21). DAGAT activity of different species was found mainly in microsomes (16, 31, 33, 42) but also in so-called lipid bodies (17, 18, 19, 29), a compartment where neutral lipids such as TAG and steryl esters are stored. A second type of reaction leading to TAG is catalyzed by a phosphatidylcholine:DAGAT. This pathway was identified in plants (sunflower, Ricinus communis, and Crepis palaestina) (6) and in the yeast Saccharomyces cerevisiae (6, 28). Finally, an acyl-CoA-independent TAG formation different from phosphatidylcholine:DAGAT was found in rat intestine (25) and mouse (32).
TAG synthesis in yeast and fungi has been only poorly studied. DAGAT activity in lipid particles in baker’s yeast (S. cerevisiae) was described by Christiansen (5), but the enzyme catalyzing this reaction was not characterized at the molecular level. In the oleaginous yeast Rhodotorula glutinis, a cytosolic TAG biosynthetic multienzyme complex containing a DAGAT was unveiled (11), suggesting that both the membrane fraction and the soluble fraction are involved in TAG formation. Most recently, a DAGAT from M. ramanniana was identified by Lardizabal et al. (21). While computational searches failed to identify proteins of S. cerevisiae with homology to ACATs and DAGATs previously found in mammals and plants, the amino acid sequence of the DAGAT from M. ramanniana showed distinct similarity to that of the yeast open reading frame (ORF) YOR245c, a gene of as-yet-unknown function. Heterologous expression of YOR245c in insect cells demonstrated a high increase of DAGAT activity in the host, suggesting that YOR245c is indeed involved in TAG synthesis (21).
Another TAG synthase from S. cerevisiae is the phosphatidylcholine:DAGAT Lro1p, which is located in microsomes (6, 28). An lro1 deletion strain showed a 65% decreased incorporation of oleate into TAG, a 75% decreased glycerol incorporation into TAG, and a significantly reduced amount of TAG (28). These results are in line with previous findings from our laboratory (8) but are different from data presented by Dahlqvist et al. (6), who showed that the amount of TAG was only slightly decreased in the lro1 strain compared to the wild type. Nevertheless, these investigations demonstrated that the phosphatidylcholine-dependent TAG synthase Lro1p contributes to TAG formation in yeast. These results also suggested the existence of redundant systems of yeast TAG synthesis.
The study presented here focused on the identification of TAG-synthesizing enzymes in lipid particles of S. cerevisiae. This compartment consists of a highly hydrophobic core of TAGs and steryl esters (approximately 50% each) surrounded by a phospholipid monolayer. A small amount and a limited number of proteins, which are mostly enzymes involved in lipid biogenesis, are embedded in this membrane (for a review see reference 43). TAGs stored in yeast lipid particles can be utilized for membrane biogenesis, especially during depletion of fatty acids (7, 22). We present evidence that DAGAT activity in lipid particles of the yeast can be attributed to the gene product encoded by YOR245c (DGA1). Furthermore, we show that redundant systems of acyl-CoA-dependent and -independent TAG biosynthesis, which are different from Lro1p and Dga1p, exist in this yeast. These systems appear to be distributed between lipid particles and the endoplasmic reticulum.
MATERIALS AND METHODS
Strains and culture conditions.
The wild-type S. cerevisiae strains X2180-1A (MATa SUC2 mal gal2 CUP1), FY1679 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200), BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), and YB332 (MATa NMT1 ura3-52 his3Δ200 ade2-101 lys2-801 leu2-3,112) were used throughout this study. The dga1 mutant (yor245c::KanMX4; isogenic to BY4741) and the lro1 mutant (ynr008w::KanMX4; isogenic to FY1679) were obtained from the strain collection Euroscarf, Frankfurt, Germany. YB526 (MATa NMT1 ura3-52 his3Δ200 ade2-101 lys2-801 leu2-3,112 faa1Δ1.9::HIS3 faa2Δ0.5::LEU2 faa3Δ0.8::LEU2 faa4Δ0.3::LYS2) was kindly provided by J. Gordon, St. Louis, Mo.
Cells were grown aerobically in 2-liter Erlenmeyer flasks to the late logarithmic growth phase (optical density at 600 nm [OD600] ≈ 11.0) at 30°C in YPD medium (1% yeast extract [Oxoid], 2% peptone [Oxoid], 2% glucose [Merck]). Medium (500 ml) was inoculated with 0.5 ml of preculture grown aerobically for 48 h in YPD medium at 30°C.
Strain construction.
The marker module His3MX6, including the Schizosaccharomyces pombe his5+ gene (26, 34), on the vector pFA6a (35) was used to replace the ORF YOR245c in S. cerevisiae FY1679 by the short flanking homology method. Deletion cassettes containing both the start and stop codons of YOR245c and the entire his5+ gene were constructed with the primers (5′)TAAGGAAACGCAGAGGCATACAGTTTGAACAGTCACATAACGGATCCCCGGGTTAATTAA(3′) (forward) and (5′)TTTATTCTAACATATTTTGTGTTTTCCAATGAATTCATTAGAATTCGAGCTCGTTTAAAC(3′) (reverse) (26). The underlined sequences are homologous to YOR245c.
A 1.40-kb PCR fragment was generated with ExTaq DNA polymerase (Takara, Otsu, Japan) by using approximately 150 ng of plasmid as a template in a standard PCR mixture containing PCR buffer (20 mM Mg2+), 0.2 mM concentrations of each deoxynucleoside triphosphate, and 1 μM concentrations of the primers in a total volume of 100 μl. After a denaturation step of 2 min at 94°C, the enzyme was added and fragments were amplified for 10 cycles of 15 s at 94°C, 30 s at 55°C, and 60 s at 72°C and for 30 cycles of 15 s at 94°C, 30 s at 68°C, and 90 s at 72°C, followed by a final elongation step of 12 min at 72°C.
Overnight cultures (OD600 ∼ 0.7) of the wild-type strain FY1679 and the lro1 mutant were used for transformation by the high-efficiency lithium acetate transformation protocol (12). Transformants were grown on plates lacking histidine for 2 to 3 days at 30°C. Only clones that yielded large colonies were considered positive transformants and further checked for correct integration of the deletion cassette. To verify the correct replacement of YOR245c by the His3MX6 module, one forward primer binding outside the target locus and two reverse primers binding within the target locus and within the marker module were designed (35, 36), and analytical PCRs with whole-yeast-cell extracts (15) and AmpliTaq DNA polymerase were performed.
Isolation and characterization of subcellular fractions.
The lipid particle fraction was obtained at high purity from cells grown to the late logarithmic phase as described by Leber et al. (23). The isolation of other subcellular fractions used in this study has been described by Zinser et al. (41).
Protein analysis.
Protein was quantified by the method of Lowry et al. (27) with bovine serum albumin as a standard. Proteins were precipitated with trichloroacetic acid at a final concentration of 10%. The protein pellet was solubilized in 0.1% sodium dodecyl sulfate-0.1% NaOH. Prior to protein analysis, samples of the lipid particle fraction were delipidated. Nonpolar lipids were extracted with 2 volumes of diethyl ether, the organic phase was withdrawn, residual diethyl ether was removed under a stream of nitrogen, and proteins were precipitated as described above.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was carried out by the method of Laemmli (20). Samples were dissociated at 37°C to avoid hydrolysis of polypeptides, which may occur at higher temperatures. Western blot analysis was performed as described by Haid and Suissa (13). Proteins were detected by an enzyme-linked immunosorbent assay with rabbit antiserum as the first antibody and peroxidase-conjugated goat anti-rabbit immunoglobulin G as the second antibody. Antibodies used in this study were directed against the microsomal marker protein BIP/Kar2p and against Erg6p, a marker protein of lipid particles. Relative enrichment and cross-contamination of organelle fractions were assessed as described previously (40).
Measurement of TAG synthase activity.
The DAGAT assay was performed in a final volume of 400 μl containing [14C]DAG (17 μg; 20,000 dpm), 80 nmol of palmitoyl-CoA, 500 μg of bovine serum albumin, 0.25% Triton X-100, 150 mM Tris Cl (pH 7.0), 15 mM KCl, and 15 mM MgCl2. Before addition of the other assay components the 14C-labeled substrate dissolved in chloroform-methanol was brought to dryness under a stream of nitrogen. After vortexing of the assay mixture for 1 min and sonication for 7 min, the enzyme source was added. Homogenate (0.4 to 1 mg of protein), lipid particles (20 to 60 μg of protein), microsomes (200 to 600 μg of protein), plasma membrane (300 to 330 μg of protein), vacuoles (320 to 350 μg of protein), mitochondria (400 to 500 μg of protein), and cytosol (210 to 240 μg of protein) were used as enzyme sources. Incubations were carried out for 30 min at 30°C and were terminated by the addition of 3 ml of chloroform-methanol (1:2, vol/vol) and 1 ml of distilled water. Lipids were extracted for 1 h by vortexing the samples frequently. After removal of the aqueous phase the extract was washed twice with 1 ml of distilled water each. Then the organic phase was taken to dryness under a stream of nitrogen. Lipids were dissolved in 100 μl of chloroform-methanol (2:1, vol/vol), the extracts were applied to high-performance thin-layer chromatography (TLC) plates (silica gel 60; Merck, Darmstadt, Germany), and chromatograms were developed in an ascending manner with the solvent system light petroleum-diethyl ether-acetic acid (35:15:1; per vol). After chromatographic separation, the radioactively labeled lipids were detected by TLC scanning with a Tracemaster 20 automatic TLC linear analyzer (Berthold). In addition, lipids were visualized on TLC plates by staining with iodine vapor, bands were scraped off, and radioactivity was measured by liquid scintillation counting with LSC Safety (Baker, Deventer, The Netherlands) and 5% water as a scintillation mixture.
The acyl-CoA-independent TAG synthase assay was the same as that for DAGAT described above but without the addition of palmitoyl-CoA. Instead, 50 nmol of CoA, 50 nmol of palmitic acid, and 150 nmol of ATP or combinations of these reagents, as described in Results, were added to the assay mixture. Furthermore, assays with [14C]palmitic acid and DAG as substrates were performed with the ingredients described above.
Lipid substrates.
Radioactively labeled 1,2-sn-DAG was synthesized by incubation of the wild-type yeast strain X2180-1A with 0.5 mCi of [1-14C]palmitic acid for 24 h at 30°C in 500 ml of YPD medium. Cells were harvested, washed with distilled water, mixed with distilled water and glass beads (0.30 mm diameter) at a 1:1:1 ratio (wt/vol/wt), and disrupted for 3 min in a Braun-Melsungen homogenizer under CO2 cooling. Then lipids were extracted as described by Folch et al. (10) and separated by TLC with the solvent system light petroleum-diethyl ether-acetic acid (35:15:1, vol/vol/vol). Lipids were visualized by staining with iodine vapor, and phospholipid bands were scraped off. After extraction with chloroform-methanol (1:4, vol/vol) for 3 h, the organic phase was collected and the remaining silica gel was extracted twice with chloroform-methanol (1:4, vol/vol) for 20 min each. The organic phases were combined and taken to dryness, and lipids were dissolved in 750 μl of chloroform-methanol (2:1, vol/vol). To convert phospholipids to 1,2-sn-DAG, 5 μl of phospholipase C (1 U/μl) and 500 μl of a buffer (pH 7.2) containing 20 mM Tris Cl, 0.12 NaCl, 5 mM CaCl2, and 2 mM ZnSO4 was added. This mixture was incubated at 37°C for 6 h, and the reaction was stopped by adding 3 ml of chloroform-methanol (1:2, vol/vol) and 1 ml of distilled water. Lipid extraction was performed for 1 h with frequent vortexing. After removal of the aqueous phase, the organic phase was washed twice with 1 ml of distilled water each and then applied to high-performance TLC plates (silica gel 60; Merck). Chromatograms were developed in an ascending manner with the solvent system light petroleum-diethyl ether-acetic acid (35:15:1; per vol). The DAG band visualized by staining with iodine vapor was scraped off and extracted with chloroform-methanol (1:4, vol/vol) as described above. The substrate was quantified as described with 1,2-diolein as a standard, as described below.
Other substrates used were 1-stearoyl-2-[1-14C]arachidonyl-sn-glycerol (20,000 dpm/nmol) (Amersham Pharmacia Biotech) and [1-14C]palmitic acid (30,000 dpm/nmol) (NEN).
Lipid analysis.
Lipids were extracted from whole yeast cells grown to the late logarithmic phase (OD600 ∼ 11.0) or from subcellular fractions by the procedure described in the previous section. For the quantification of neutral lipids, extracts were applied to silica gel 60 plates with a sample applicator (Linomat IV; CAMAG, Muttenz, Switzerland), and chromatograms were developed in an ascending manner by using the solvent system light petroleum-diethyl ether-acetic acid (25:25:1, vol/vol/vol) for the first third of the distance. Then plates were dried briefly and further developed to the top of the plate with the solvent system light petroleum-diethyl ether (49:1, vol/vol). Quantification of ergosterol and ergosteryl esters was carried out by densitometric scanning at 275 nm using a Shimadzu CS-930 dual-wavelength chromatoscanner with ergosterol as a standard. TAGs were visualized by postchromatographic staining with a chromatogram immersion device (CAMAG). Plates were dipped for 6 s into a developing reagent consisting of 0.63 g of MnCl2 · 4H2O, 60 ml of water, 60 ml of methanol, and 4 ml of concentrated sulfuric acid, briefly dried, and heated at 100°C for 30 min. Quantification of TAG was carried out by densitometric scanning at 400 nm with triolein (NuCheck, Inc., Elysian, Minn.) as a standard. The procedure for quantification of DAG was the same as for TAG but with 1,2-diolein instead of triolein as a standard.
Computational analysis.
Molecular data for proteins were obtained from the Yeast Protein Data Base (www.proteome.com/databases/YPD), the Saccharomyces Genome Data Base (genome-www.stanford.edu/Saccharomyces), SwissProt (www.expasy.ch), and The European Patent Office (http://ep.espacenet.com). Homology searches were performed with BLAST Search (http://dove.embl-heidelberg.de/Blast2/).
RESULTS
DAGAT assay.
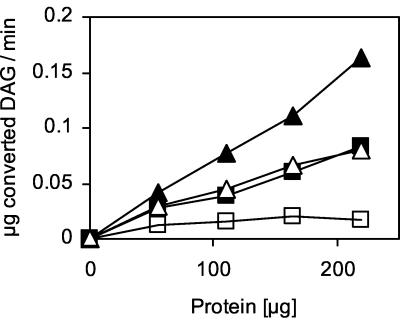
To test DAGAT activity in the yeast, we established an assay containing 14C-labeled DAG as the substrate and acyl-CoA as the cosubstrate. To increase the solubility of the substrates in the aqueous environment, the assay mix was sonicated and the detergent Triton X-100 was added. The pH optimum of acyl-CoA-dependent TAG synthase was between 7.0 and 7.5, and the temperature optimum was 30°C (data not shown). TAG synthase activity showed a clear dependence on the presence of specific ions. Both K+ and Mg2+ stimulated TAG synthase activity in a protein (Fig. 1)- and time (data not shown)-dependent manner. The highest activity was achieved when a combination of K+ and Mg2+ was used in the assay, whereas other ions, such as Ca2+ and Na+, at various concentrations did not show any significant enhancement of enzyme activity (data not shown).
FIG. 1.
Dependence of DAGAT activity on a specific ionic environment. DAGAT assays without ions (open squares) and in the presence of K+ (open triangles), Mg2+ (filled squares), or both ions (filled triangles) were performed with microsomes or lipid particles of the wild-type strain X2180-1A as enzyme sources, as described in Materials and Methods. Means of three independent experiments are shown.
DAGAT is located in lipid particles.
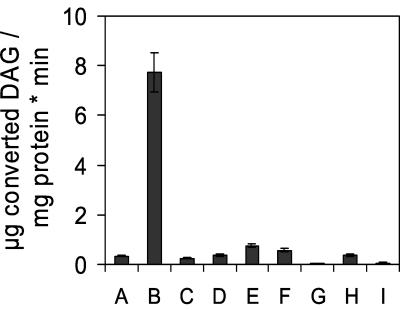
To localize the acyl-CoA-dependent TAG synthase activity, the assay described above was performed with highly purified yeast subcellular fractions. These studies unveiled a 70- to 90-fold enrichment of the specific activity of DAGAT in lipid particles over the homogenate (Fig. 2), a 2- to 3-fold enrichment in 30,000 × g microsomes, and a 1.5- to 2-fold enrichment in 40,000 × g microsomes. The specific activity of DAGAT achieved with lipid particles was 7.72 μg of DAG converted to TAG per mg of protein per min. Other subcellular fractions including the cytosol were practically devoid of DAGAT activity.
FIG. 2.
Subcellular localization of DAGAT. Homogenate (A), lipid particles (B), plasma membrane (C), vacuoles (D), 30,000 × g microsomes (E), 40,000 × g microsomes (F), 100,000 × g microsomes (G), mitochondria (H), and cytosol (I) of the wild-type strain X2180-1A were tested for DAGAT activity as described in Materials and Methods. Values are means from three independent experiments.
YOR245c encodes DAGAT of lipid particles.
To identify the gene encoding the DAGAT localized in lipid particles, we searched databases for yeast homologues to DAGATs from other organisms. As an appropriate candidate we found the yeast ORF YOR245c (DGA1), which showed an overall identity of 44% with the DAGAT of M. ramanniana described recently by Lardizabal et al. (21). Computational analysis of YOR245c revealed a molecular mass of the gene product of approximately 47.7 kDa and the presence of one hydrophobic domain and two potential transmembrane domains. The latter property is only partially in line with our understanding of the possible localization of this polypeptides to lipid particles, because previous studies in our laboratory had shown that lipid particle proteins typically lack transmembrane spanning regions (3).
The dga1 deletion strain showed the same growth phenotype as the wild type (data not shown) and was able to form regular lipid particles. To investigate whether YOR245c encodes the DAGAT of lipid particles and microsomes, we performed enzymatic assays with the respective subcellular fractions of the dga1 deletion strain (Table 1). The specific activity of DAGAT in microsomes from dga1 was only slightly reduced compared to the wild type. In contrast, lipid particles of the mutant showed an activity reduced to only 5% of the wild-type control. These data indicate that Dga1p catalyzes acyl-CoA-dependent TAG synthesis located mainly in lipid particles of S. cerevisiae.
TABLE 1.
DAGAT activity in dga1 and lro1 mutant strainsa
| Strain | DAGAT sp act (μg of converted DAG/mg of protein/min)
|
||
|---|---|---|---|
| Homogenate | Lipid particles | Microsomes | |
| BY4741 (wild type) | 0.080 | 8.51 | 0.17 |
| dga1 strain | 0.073 | 0.19 | 0.13 |
| FY1679 (wild type) | 0.043 | 6.42 | 0.12 |
| lro1 strain | 0.056 | 5.39 | 0.15 |
Homogenate, lipid particles, and microsomes of the dga1 and lro1 mutant strains and the corresponding wild-type strains BY4741 and FY1679, respectively, were prepared and DAGAT activity was measured as described in Materials and Methods. Values are means (standard deviation, ± 10%) from three independent experiments.
To distinguish clearly between acyl-CoA-dependent TAG synthase activity catalyzed by Dga1p and the previously described Lro1p (6, 28) we also analyzed microsomes and lipid particles of lro1. The lro1 mutant strain, however, did not show any significant difference in DAGAT activity compared to the corresponding wild-type strain, FY1679 (Table 1).
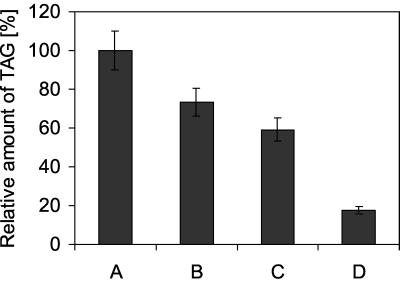
To determine the contribution of Dga1p to total TAG formation in yeast, we analyzed lipid extracts of both wild-type and the dga1 strain grown to the late logarithmic phase (OD ∼ 11.0). The strains contain similar amounts of ergosterol and ergosteryl esters (data not shown), whereas the amount of TAG in dga1 was decreased to 70% of the wild-type amount (Fig. 3). As a consequence, the ratio of ergosteryl ester to TAG in highly purified lipid particles was 35% higher in dga1 than in the wild type.
FIG. 3.
TAG concentration in dga1 and lro1 strains. Neutral lipids of the wild-type strain FY1679 (A) and dga1 (B), lro1 (C), and dga1 lro1 (D) strains were isolated from cells grown to the late logarithmic phase. TAGs were quantified as described in Materials and Methods, and the value for FY1679 was set at 100%. Values are means from three independent experiments.
Similar to dga1, the amount of TAG in lro1 is reduced to 60% of the wild-type value (Fig. 3). In the dga1 lro1 double mutant, the amount of TAG decreased to 20% of the wild-type level. This result indicates that Dga1p and Lro1p are not the only gene products involved in TAG formation. Like the dga1 and lro1 strains, the dga1 lro1 double mutant grows like the wild type (data not shown).
Lipid particles harbor an acyl-CoA-independent TAG synthase.
DAG acylation experiments performed with CoA and palmitic acid as the only cosubstrates showed a moderate but significant TAG synthase activity with wild-type lipid particles as an enzyme source (Table 2). To unveil a possible mechanism for this reaction we carried out control experiments with dga1 and lro1 strains and a quadruple mutant with mutations in faa1 through faa4 (faa1–faa4 mutant) which is defective in the activation of free fatty acids. When palmitoyl-CoA was used as a cosubstrate for TAG synthase, the acyl-CoA-dependent DAGAT activity of lipid particles from dga1 was dramatically reduced, whereas that of the faa1–faa4 mutant was almost unaffected. When CoA and palmitic acid or palmitic acid only was used as a cosubstrate, TAG formation in the wild type was strongly reduced, but it was not further decreased by deletion of DGA1 or FAA1 through FAA4. Addition of ATP to the assay mixture containing CoA and palmitic acid resulted in additive activities of acyl-CoA-dependent and -independent TAG synthase in the wild type because of the ATP dependence of Faa1p to Faa4p. As expected, in the dga1 mutant, the activity remained at the level without ATP, indicating that the DAGAT reaction catalyzed by Dga1p strictly depends on acyl-CoA as a cosubstrate. In the faa1–faa4 quadruple mutant, the acyl-CoA-dependent TAG synthase did not contribute to total TAG synthesis due to the blocked acyl-CoA formation in this strain. It is noteworthy that lipid particles contain Faa1p and Faa4p (3), which can convert free fatty acids to acyl-CoA. Taken together, these results indicate that incorporation of fatty acids into TAG can occur to some extent independently of Dga1p and also independently of acyl-CoA formation. This finding was also confirmed by using unlabeled DAG and [14C]palmitic acid as assay substrates (data not shown). In an lro1 mutant, acyl-CoA-dependent and -independent TAG synthase activities were the same as those in the wild type under all assay conditions, confirming the view that these reactions occur independently of Lro1p (our unpublished data).
TABLE 2.
Acyl-CoA-independent TAG synthase activitya
| Strain | TAG synthase sp act (μg of converted DAG/mg of protein/min)
|
|||
|---|---|---|---|---|
| Palmitoyl-CoA | Palmitic acid + CoA | Palmitic acid + CoA + ATP | Palmitic acid | |
| BY4741 (wild type) | 8.51 | 1.65 | 11.36 | 1.11 |
| dga1 strain | 0.19 | 1.56 | 1.46 | 1.32 |
| YB332 (wild type) | 6.42 | 1.25 | 8.63 | 1.03 |
| faa1–faa4 strain | 5.27 | 1.52 | 1.85 | 1.30 |
TAG synthase activity in lipid particles of the dga1 mutant and the faa1–faa4 quadruple mutant and the corresponding wild types, BY4741 and YB332, was measured as described in Materials and Methods using palmitoyl-CoA, CoA and palmitic acid, CoA, palmitic acid, and ATP, or palmitic acid as a cosubstrate. Values are means (standard deviation, ± 10%) from three independent experiments.
DISCUSSION
The present study showed that YOR245c (DGA1) encodes an acyl-CoA-dependent DAGAT located in lipid particles of S. cerevisiae. Previous work of Lardizabal et al. (21) had shown that heterologous expression of Dga1p in insect cells caused a high increase of DAGAT activity in the host, confirming the view that the gene product is not only an activator of TAG synthesis. The combination of molecular biological, cell biological, and biochemical methods employed in the present study, however, enabled us to obtain more insight into the complex systems of TAG formation in the yeast. Using these methods, we were able to dissect the TAG biosynthetic pathways which we consider relevant by means of subcellular fractionation, the use of mutants with deletions in steps involved, and enzymatic analysis.
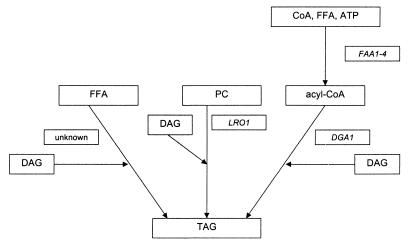
Which pathways contribute to the biosynthesis of TAG in S. cerevisiae? According to our results and those of others (6, 28), three pathways appear to lead to the formation of TAG in the yeast (Fig. 4). (i) Dga1p catalyzes TAG formation by an acyl-CoA-dependent esterification of DAG; activation of free fatty acids used as cosubstrates for this reaction requires activity of Faa1p to Faa4p. (ii) Lro1p was shown to form TAG by a phosphatidylcholine:DAGAT reaction (6, 28). (iii) Finally, free fatty acids can be incorporated into DAG by a reaction that is independent of Dga1p and Lro1p. While Dga1p and Lro1p pathways seem to contribute the majority of TAG synthesis in yeast, the acyl-CoA-independent mechanism is responsible for only a small percentage of the total cellular TAG formation. Incorporation of radioactively labeled free fatty acids into complex lipids of S. cerevisiae has been shown by Wagner and Paltauf (37). Free fatty acids were preferentially incorporated into phospholipids in vivo, suggesting a phospholipase A2-dependent deacylation-reacylation mechanism. In addition, free fatty acids were incorporated into TAG, which led us to speculate that a similar mechanism might be involved in the formation of TAG in yeast. However, the genes and gene products catalyzing such a reaction are still unknown.
FIG. 4.
Pathways for TAG synthesis in S. cerevisiae. FFA, free fatty acid; PC, phosphatidylcholine.
Which organelles contribute to TAG formation in S. cerevisiae? It has been shown before that Lro1p is a microsomal TAG synthase (6, 28). In addition to microsomes, lipid particles contribute to cellular TAG synthesis through the action of Dga1p (Table 1), thus confirming previous results by Christiansen (5). However, Dga1p-independent DAGAT activity was also detected in microsomes, suggesting the existence of DAGAT isoenzymes in the yeast. Finally, acyl-CoA-independent DAG acylation was shown in lipid particles, but this type of reaction may also occur in other subcellular compartments. Preliminary investigations point to a similar mechanism of TAG synthesis in microsomes (our unpublished results). Contrary to the oleaginous yeast R. glutinis, which contains a cytosolic TAG biosynthetic multienzyme complex with DAGAT activity (11), cytosolic TAG formation was not detected in S. cerevisiae. Thus, in baker’s yeast, lipid particles and microsomes (endoplasmic reticulum) appear to be the only or at least the major sites of TAG synthesis. It has been hypothesized that lipid particles and the endoplasmic reticulum are related organelles (for a review see reference 43). Their interaction may be relevant for concerted formation of TAG and for the exclusive storage of this component in lipid particles.
Is synthesis of TAG by Dga1p important for the yeast? It has become clear from the work of Lardizabal et al. (21) and from data presented here that Dga1p is dispensable for the yeast and is responsible for the formation of only 30% of cellular TAG. Obviously, a dga1 mutant strain can form a sufficient amount of TAG through other pathways and, as a consequence, form lipid particles like the wild type. An interesting observation, however, is the fact that lipid particles, which are the depot of TAG, are also able to synthesize a certain portion of this lipid themselves through the action of Dga1p. The substrates required for this reaction appear to be formed in lipid particles in situ. Lipid particles contain the complete set of enzymes needed for the biosynthesis of phosphatidic acid (1, 2). Phosphatidate phosphohydrolase, whose activity was in part detected in the cytosol (9), may have access to the surface of lipid particles and convert phosphatidic acid to DAG. Faa1p and Faa4p, which catalyze the formation of acyl-CoAs, the cosubstrates for the acylation of DAG through Dga1p, were also identified as components of lipid particles (3). Thus, lipid particles appear to be at least in part autonomous for TAG production. TAG newly formed by enzymes bound to the surface monolayer of lipid particles, however, has to be incorporated into the hydrophobic core of this compartment. How this is accomplished is not known at present. Similarly, the remobilization process of TAG from lipid particles is only poorly understood. Recent investigations in our laboratory identified a TAG lipase in lipid particles (unpublished results). We hypothesize that the presence of enzymes catalyzing synthesis of TAG on the one hand and lipolysis on the other hand in lipid particles may regulate accumulation and mobilization of TAG, respectively. The requirement for fatty acids during membrane proliferation or the formation of excess fatty acids may favor one side of the reaction or the other. Although there is at present no proof for such a regulatory mechanism in yeast, investigations with other model systems unveiled the existence of a TAG lipolysis-re-esterification cycle (38, 39), supporting this theory.
Acknowledgments
This work was financially supported by the Fonds zur Förderung der Wissenschaftlichen Forschung in Österreich (projects 13669 and 15141) and the Austrian Ministry of Education, Science and Culture (project AUSTROFAN).
Footnotes
We dedicate this study to Fritz Paltauf on the occasion of his retirement.
REFERENCES
- 1.Athenstaedt, K., and G. Daum. 1997. Biosynthesis of phosphatidic acid in lipid particles and the endoplasmic reticulum of the yeast Saccharomyces cerevisiae. J. Bacteriol. 179:7611–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Athenstaedt, K., S. Weys, F. Paltauf, and G. Daum. 1999. Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athenstaedt, K., D. Zweytick, A. Jandrositz, S. D. Kohlwein, and G. Daum. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cases, S., S. J. Smith, Y. Zheng, H. M. Myers, S. R. Lear, E. Sande, S. Novak, C. Collins, C. B. Welch, A. J. Lusis, S. K. Erickson, and R. V. Farese, Jr. 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 95:13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christiansen, K. 1978. Triacylglycerol synthesis in lipid particles from baker’s yeast (Saccharomyces cerevisiae). Biochim. Biophys. Acta 530:78–90. [DOI] [PubMed] [Google Scholar]
- 6.Dahlqvist, A., U. Stahl, M. Lenman, A. Banas, M. Lee, L. Sandager, H. Ronne, and S. Stymne. 2000. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 97:6487–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daum, G., and F. Paltauf. 1980. Triacylglycerols as fatty acid donors for membrane phospholipid biosynthesis in yeast. Monatsh. Chem. 111:355–363. [Google Scholar]
- 8.Daum, G., G. Tuller, T. Nemec, C. Hrastnik, G. Balliano, L. Cattel, P. Milla, F. Rocco, A. Conzelmann, C. Vionnet, D. E. Kelly, S. Kelly, E. Schweizer, H.-J. Schüller, U. Hojad, E. Greiner, and K. Finger. 1999. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast 15:601–614. [DOI] [PubMed] [Google Scholar]
- 9.Faulkner, A., X. Chen, J. Rush, B. Horazdovsky, C. J. Waechter, G. M. Carman, and P. C. Sternweis. 1999. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 274:14831–14837. [DOI] [PubMed] [Google Scholar]
- 10.Folch, J., M. Lees, and G. H. Sloane-Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497–509. [PubMed] [Google Scholar]
- 11.Gangar, A., A. A. Karande, and R. Rajasekharan. 2001. Isolation and localization of a cytosolic 10S triacylglycerol biosynthetic multienzyme complex from oleaginous yeast. J. Biol. Chem. 276:10290–10298. [DOI] [PubMed] [Google Scholar]
- 12.Gietz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5:255–269. [Google Scholar]
- 13.Haid, A., and M. Suissa. 1983. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96:192–205. [DOI] [PubMed] [Google Scholar]
- 14.Hobbs, D. H., C. Lu, and M. J. Hills. 1999. Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett. 452:145–149. [DOI] [PubMed] [Google Scholar]
- 15.Huxley, C., E. D. Green, and I. Dunham. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6:236. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, F. M., L. Michaelson, T. C. M. Fraser, A. K. Stobart, and G. Griffiths. 1998. Biosynthesis of triacylglycerol in the filamentous fungus Mucor circinelloides. Microbiology 144:2639–2645. [DOI] [PubMed] [Google Scholar]
- 17.Kamisaka, Y., S. Mishra, and T. Nakahara. 1997. Purification and characterization of diacylglycerol acyltransferase from the lipid body fraction of an oleaginous fungus. J. Biochem. (Tokyo) 121:1107–1114. [DOI] [PubMed] [Google Scholar]
- 18.Kamisaka, Y., and T. Nakahara. 1994. Characterization of the diacylglycerol acyltransferase activity in the lipid body fraction from an oleaginous fungus. J. Biochem. 116:1295–1301. [DOI] [PubMed] [Google Scholar]
- 19.Kamisaka, Y., N. Noda, T. Sakai, and K. Kawasaki. 1999. Lipid bodies and lipid body formation in an oleaginous fungus, Mortierella ramanniana var. angulispora. Biochim. Biophys. Acta 1438:185–198. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 21.Lardizabal, K. D., D. Hawkins, and G. A. Thompson. January 2000. Diacylglycerol acyltransferase proteins. U.S. patent WO0001713.
- 22.Leber, R., E. Zinser, C. Hrastnik, F. Paltauf, and G. Daum. 1995. Export of steryl esters from lipid particles and release of free sterols in the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1234:119–126. [DOI] [PubMed] [Google Scholar]
- 23.Leber, R., E. Zinser, G. Zellnig, F. Paltauf, and G. Daum. 1994. Characterization of lipid particles of the yeast Saccharomyces cerevisiae. Yeast 10:1421–1428. [DOI] [PubMed] [Google Scholar]
- 24.Lehner, R., and A. Kuksis. 1996. Biosynthesis of triacylglycerols. Prog. Lipid Res. 35:169–201. [DOI] [PubMed] [Google Scholar]
- 25.Lehner, R., and A. Kuksis. 1993. Triacylglycerol synthesis by an sn-1,2(2,3)-diacylglycerol transacylase from rat intestinal microsomes. J. Biol. Chem. 268:8781–8786. [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. [DOI] [PubMed] [Google Scholar]
- 27.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- 28.Oelkers, P., A. Tinkelenberg, N. Erdeniz, D. Cromley, J. T. Billheimer, and S. L. Sturley. 2000. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275:15609–15612. [DOI] [PubMed] [Google Scholar]
- 29.Pillai, M. G., M. Certik, T. Nakahara, and Y. Kamisaka. 1998. Characterization of triacylglycerol biosynthesis in subcellular fractions of an oleaginous fungus, Mortierella ramanniana var. angulispora. Biochim. Biophys. Acta 1393:128–136. [DOI] [PubMed] [Google Scholar]
- 30.Routaboul, J., C. Benning, N. Bechtold, M. Caboche, and L. Lepiniec. 1999. The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol. Biochem. 37:831–840. [DOI] [PubMed] [Google Scholar]
- 31.Settlage, S. B., R. F. Wilson, and P. Kwanyuen. 1995. Localization of diacylglycerol acyltransferase to oil body associated endoplasmic reticulum. Plant Physiol. Biochem. 33:399–407. [Google Scholar]
- 32.Smith, S. J., S. Cases, D. R. Jensen, H. C. Chen, E. Sande, B. Tow, D. A. Sanan, J. Raber, R. H. Eckel, and R. V. Farese, Jr. 1999. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 25:87–90. [DOI] [PubMed] [Google Scholar]
- 33.Triki, S., C. Demandre, and P. Mazliak. 1999. Biosynthesis of triacylglycerols by developing sunflower seed microsomes. Phytochemistry 52:55–62. [Google Scholar]
- 34.Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- 35.Wach, A., A. Brachat, R. Pohlmann, and P. Phillipsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808. [DOI] [PubMed] [Google Scholar]
- 36.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruption in S. cerevisiae. Yeast 12:259–265. [DOI] [PubMed] [Google Scholar]
- 37.Wagner, S., and F. Paltauf. 1994. Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast 10:1429–1437. [DOI] [PubMed] [Google Scholar]
- 38.Wiggins, D., and G. F. Gibbons. 1992. The lipolysis/esterification cycle of hepatic triacylglycerol. Its role in the secretion of very-low-density lipoprotein and its response to hormones and sulphonylureas. Biochem. J. 284:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, L.-Y., A. Kuksis, J. J. Myher, and G. Steiner. 1995. Origin of triacylglycerol moiety of plasma very low density lipoproteins in the rat: structural studies. J. Lipid Res. 36:125–136. [PubMed] [Google Scholar]
- 40.Zinser, E., and G. Daum. 1995. Isolation and biochemical characterization of organelles from the yeast Saccharomyces cerevisiae. Yeast 11:493–536. [DOI] [PubMed] [Google Scholar]
- 41.Zinser, E., C. D. M. Sperka-Gottlieb, E.-V. Fasch, S. D. Kohlwein, F. Paltauf, and G. Daum. 1991. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173:2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou, J., Y. Wei, C. Jako, A. Kumar, G. Selvaraj, and D. C. Taylor. 1999. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 19:645–653. [DOI] [PubMed] [Google Scholar]
- 43.Zweytick, D., K. Athenstaedt, and G. Daum. 2000. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta 1469:101–120. [DOI] [PubMed] [Google Scholar]