Abstract
The first four genes of the capsule locus (cps) of Streptococcus pneumoniae (cpsA to cpsD) are common to most serotypes. We have previously determined that CpsD is an autophosphorylating protein-tyrosine kinase, demonstrated that CpsC is required for CpsD tyrosine-phosphorylation, and shown that CpsB is required for dephosphorylation of CpsD. In the present study we show that CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase that belongs to the PHP (polymerase and histidinol phosphatase) family of phosphoesterases. We also show that an S. pneumoniae strain with point mutations in cpsB, affecting one of the conserved motifs of CpsB, is unencapsulated and appears to be morphologically identical to a strain in which the cpsB gene had been deleted.
Streptococcus pneumoniae (the pneumococcus) is an important cause of invasive disease in human populations throughout the world, resulting in high morbidity and mortality. An important feature of S. pneumoniae is its capacity to produce a polysaccharide capsule which is structurally distinct for each of the 90 known serotypes of the organism (2, 8). The capsular polysaccharide (CPS) is essential for pneumococcal virulence (2); all fresh clinical isolates of S. pneumoniae are encapsulated (smooth), and spontaneous nonencapsulated (rough) derivatives of such strains are almost completely avirulent.
The capacity to regulate CPS production in S. pneumoniae may be very important for the survival of the pneumococcus in different host environments. Maximal expression of CPS is essential for systemic virulence because of its antiphagocytic properties (2). However, invasive disease is invariably preceded by asymptomatic colonization of the nasopharynx, and the thickness of the capsule influences the degree of exposure of other important pneumococcal surface structures, such as the adhesins, which are required during this initial colonization phase. Pneumococci have been shown to undergo a bidirectional phase variation between two distinct colonial morphologies, described as “opaque” and “transparent.” The transparent phenotype exhibits an enhanced capacity to colonize the nasopharynx of infant rats (22), and the opaque form is associated with massively increased virulence in animal models of systemic disease. This correlates with increased production of CPS relative to cell wall teichoic acid in the opaque phenotype (11).
The first four genes of the cps locus (cpsA to cpsD) are associated with two distinct CPS regulatory mechanisms. These four genes are common to all pneumococcal serotypes, except types 3 and 37 (13, 18). Moreover, homologues of all of these genes are found in the cps loci of most streptococci, and homologues of cpsB, cpsC, and cpsD are found in capsule loci from other gram-positive genera. CpsA has homology to transcriptional regulators, and Cieslewicz et al. (5) have demonstrated that CpsA is a transcriptional activator that regulates the level of cps gene expression in Streptococcus agalactiae. We have shown that CpsB, -C, and -D are essential for encapsulation and regulate CPS production (17). CpsC and CpsD are predicted to function together in chain-length regulation and export of CPS, in a fashion similar to ExoP in exopolysaccharide (EPS) production in Sinorhizobium meliloti (6, 7) and Wzc in Escherichia coli (21, 23). We determined that CpsD, like Wzc, is an autophosphorylating protein-tyrosine kinase and demonstrated that CpsC is required for CpsD tyrosine phosphorylation, and we have shown that CpsB is required for dephosphorylation of CpsD (17). We have also shown that autophosphorylation of CpsD at tyrosines located near the carboxy terminus attenuates its activity and reduces the level of encapsulation. Thus, tyrosine phosphorylation of CpsD negatively regulates CPS production. As CpsB has no sequence similarity to known phosphotyrosine-protein phosphatases, such as Wzb, which is associated with gram-negative cps loci (21, 23), we have further investigated its role in dephosphorylation of CpsD and in encapsulation.
Bacterial strains and plasmids.
The S. pneumoniae strain Rx1-19F is a derivative of Rx1 (a rough derivative of the type 2 strain D39) expressing type 19F capsule (16). Rx1-19F-B, in which the cps19fB gene has been interrupted with pVA891, has been described previously (7). Rx1-19F-BΔ and Rx1-19F-DΔ, containing defined in-frame deletion mutations in cps19fB and cps19fD, respectively, have also been described previously (17). Pneumococci were routinely grown either in Todd-Hewitt broth (Oxoid, Basingstoke, Hampshire, England) with 0.5% yeast extract (Difco Laboratories, Detroit, Mich.) or on blood agar. Where appropriate, erythromycin was added to medium at a concentration of 0.2 μg/ml. The S. pneumoniae strains were transformed as described previously for strain D39 (3). The production of type 19F capsule by pneumococci was assessed by quellung reaction with monospecific antisera obtained from Statens Seruminstitut, Copenhagen, Denmark.
CpsB is a member of the PHP family of phosphoesterases.
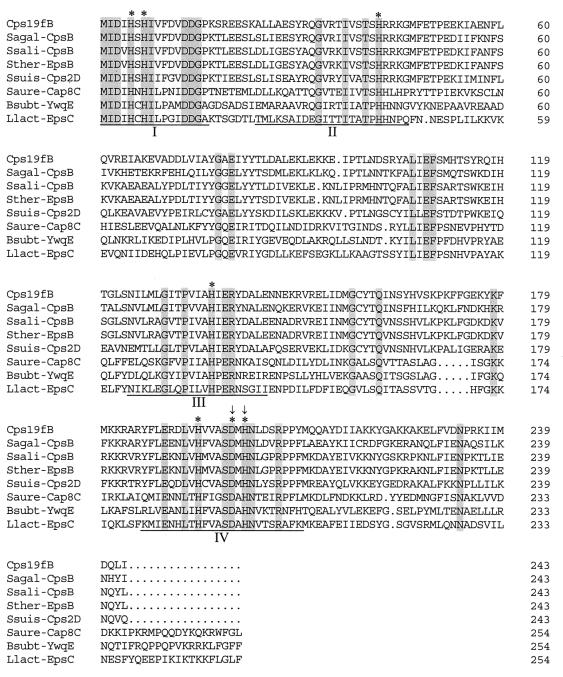
A database search using CD-Search (National Center for Biotechnology Information) identified CpsB as a member of the PHP (polymerase and histidinol phosphatase) family of phosphoesterases. Aravind and Koonin (1) divided the members of the PHP superfamily into five distinct protein families, which include phosphoesterases with distinct functions: the N-terminal domains of bacterial DNA polymerase III α subunits, stand-alone proteins from various bacteria, C-terminal domains of family X DNA polymerases from various bacteria, histidinol phosphatases, and CpsB homologues encoded by cps loci from gram-positive bacteria. The only member of this family of proteins for which phosphoesterase activity has actually been demonstrated is the yeast histidinol phosphatase. This enzyme is involved in the ninth step of histidine biosynthesis and cleaves the phosphomonoester bond in histidinol phosphate to release histidinol (10). Aravind and Koonin (1) identified four conserved motifs in all members of the PHP superfamily. Figure 1 shows an alignment of Cps19fB with several homologous proteins with the multiple sequence alignment settings of the program DNAMAN (Lynnon BioSoft, Quebec, Canada). The four conserved motifs and the amino acids predicted to be essential for function are highlighted. Each of the four motifs has conserved histidine residues which were predicted to be involved in metal binding (possibly zinc or nickel). These proteins were predicted to require two coordinated metal cations for enzymatic activity. Indeed, a mutation affecting the conserved histidine in motif 3 in the yeast histidinol phosphatase resulted in a histidine-requiring phenotype (14). The conserved aspartate residues are predicted to be essential for catalysis by participating in electron transfer (1).
FIG. 1.
Alignment of Cps19fB with Streptococcus agalactiae CpsB (Sagal-CpsB; GenBank accession number AF163833), Streptococcus salivarius CpsB (Ssali-CpsB; Y17900), Streptococcus thermophilus EpsB (Sther-EpsB; U40830), Streptococcus suis Cps2D (Ssuis-Cps2D; AF118389), Staphylococcus aureus Cap8C (Saure-Cap8C; U73374), Bacillus subtilis YwqE (Bsubt-YwqE; Z92952), and Lactococcus lactis EpsC (Llact-EpsC; AF036485). The four conserved motifs are underlined, and identical residues are shaded; the asterisks indicate amino acids predicted to be essential for function. The arrows indicate amino acids mutated in Rx1-19F-BNQ, and the absence of a residue is indicated by a period.
Construction of S. pneumoniae containing a defined mutation in cpsB.
An S. pneumoniae strain containing defined mutations in motif 4 of CpsB, in which the aspartate at position 199 was changed to asparagine (D199N) and the histidine at position 201 was changed to glutamine (H201Q) (Fig. 1), was constructed by using overlap-extension PCR as described by Horton (9). The required changes were incorporated into overlapping primers J194 (GTTGCAAGTAACATGCAGAATTTAGAC; nucleotides [nt] 2201 to 2227 of the cps19f sequence, GenBank accession no. U09239) and J195 (GTCTAAATTCTGCATGTTACTTGCAAC; complementary to nt 2201 to 2227). The nucleotide changes relative to the wild-type sequence are underlined. Separate PCRs were set up using primers J195 and CPS5′ (TGATGTTCAAGGTATAGGTGTTAATCA; nt 146 to 169) as well as primers J194 and J6 (TTGGATATGACCAGACAAGAC; complementary to nt 5704 to 5724). A second round of PCR amplification was then performed using primers CPS5′ and J6 and both purified PCR products as a template. The final 6-kb CPS5′-J6 PCR product incorporating the specific mutations was transformed directly into the insertion-duplication mutant Rx1-19F-B (in which the cps19fB gene has been interrupted with pVA891). The transformants were initially screened for loss of erythromycin resistance, and the presence of the specific point mutations was confirmed by the sequencing of PCR products. This mutant was designated Rx1-19F-BNQ.
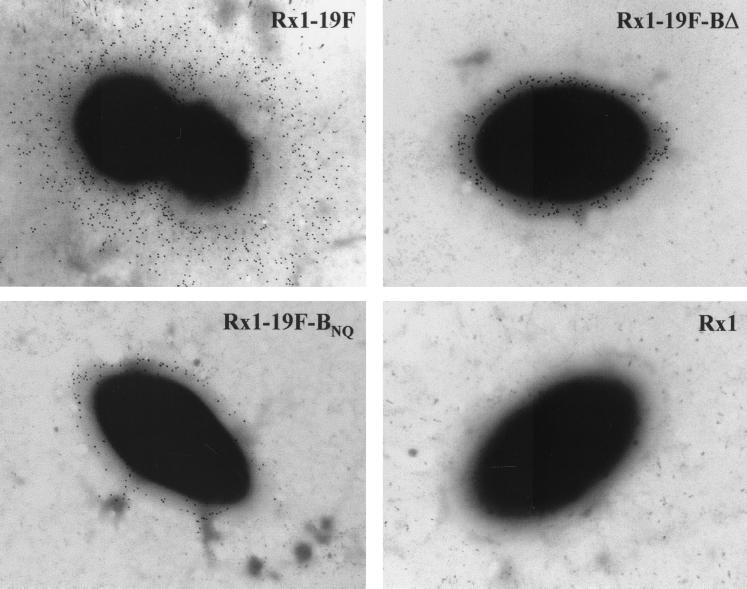
The phenotype of Rx1-19F-BNQ was then compared with that of Rx1-19F-BΔ, in which the entire cpsB gene has been deleted. When grown on blood agar plates, both strains exhibited a “rough” colony morphology. Further examination of capsule production by immunogold electron microscopy, performed as described elsewhere (19), could not distinguish between Rx1-19F-BNQ and Rx1-19F-BΔ. The bound polyclonal rabbit anti-type 19F antiserum was detected with protein A-gold conjugate (10 nm; Sigma), and the grids were examined with a Phillips CM100 transmission electron microscope operated at 80-kV accelerating voltage. As shown in Fig. 2, both strains have a small amount of anti-type 19F CPS-reactive material covering the cell surface as previously described for Rx1-19F-BΔ (17).
FIG. 2.
Detection of CPS on the cell surface of S. pneumoniae Rx1-19F and the two S. pneumoniae cpsB mutants by immunogold electron microscopy. Digital images were taken at ×20,000 magnification. A single representative bacterium is shown for Rx1-19F, Rx1-19F-BΔ, Rx1-19F-BNQ, and the nonencapsulated strain Rx1.
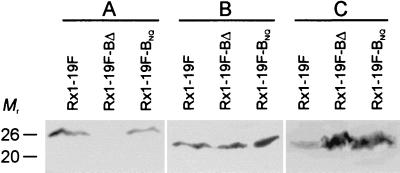
Western blot analysis of Rx1-19F, Rx1-19F-BΔ, and Rx1-19F-BNQ was also undertaken. Bacterial cell lysates from 1 ml of log-phase culture (A600 = 0.27; containing 5 × 108 CFU/ml) were separated on sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis gels (12) prior to transfer onto nitrocellulose or polyvinylidene difluoride (PVDF) membranes (20). Membranes were probed with either mouse antiphosphotyrosine monoclonal antibody clone 4G10 (Upstate Biotechnology, Lake Placid, N.Y.) used at a dilution of 1:2,000, mouse anti-CpsB antiserum used at 1:500, or mouse anti-CpsD antiserum used at 1:100, followed by the addition of a 1:5,000 dilution of goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Bio-Rad Laboratories, Hercules, Calif.). Enzyme-labeled bands were visualized by using Lumi-Light Western blotting substrate (Roche Molecular Biochemicals, Mannheim, Germany). These results confirm that Rx1-19F-BNQ produces a mutant CpsB protein, which is detectable by Western blotting, but this protein is absent in Rx1-19F-BΔ (Fig. 3A). Although all three strains contain similar amounts of CpsD (Fig. 3B), both Rx1-19F-BNQ and Rx1-19F-BΔ contain elevated levels of the phosphorylated form of CpsD (CpsD-P) compared to Rx1-19F (Fig. 3C). These results indicate that mutations D199N and H201Q collectively inactivate CpsB, demonstrating that the conserved motifs are indeed important for function, as predicted above.
FIG. 3.
Western blot analysis. S. pneumoniae Rx1-19F, Rx1-19F-BΔ, and Rx1-19F-BNQ lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto nitrocellulose (A and B) or PVDF (C) membranes. Filters were probed with mouse polyclonal anti-CpsB antibody (A), mouse polyclonal anti-CpsD antibody (B), or mouse antiphosphotyrosine monoclonal antibody clone 4G10 (C) to detect CpsB, CpsD, and CpsD-P, respectively. The mobility of molecular size markers is also indicated in kilodaltons.
Detection of phosphatase activity associated with CpsB.
Phosphatase activity was initially investigated in deoxycholate cell lysates of Rx1-19F by assaying its ability to cleave p-nitrophenyl phosphate at different temperatures and at different pH values. Deoxycholate cell lysates were prepared by resuspending pneumococcal cells, grown on blood agar plates overnight at 37°C, in 150 mM Tris-HCl (pH 7.0), such that the absorbance at 600 nm (A600) was 0.6. The cells were then pelleted in a bench centrifuge and resuspended in a one-fifth volume of 100 mM sodium citrate (pH 6.5), 1 M Tris-HCl (pH 7.0), or 1 M Tris-HCl (pH 8.0), and 100-μl aliquots were then used in the assay. The cells were lysed by the addition of 0.1% (wt/vol) deoxycholate (final concentration), followed by incubation at 37°C for 10 min. The reaction was started by addition of 10 μl of 5 mg of p-nitrophenyl phosphate (Sigma)/ml in 100 mM Tris-HCl (pH 7.0). After incubation for 1 h at 37°C, the cell debris was pelleted, and the A410s of the supernatants were measured. The phosphatase activity was expressed as units, where one unit is the amount of enzyme that produces 1 nmol of p-nitrophenol per h per ml of cell lysate at 37°C (15). No phosphatase activity was detected when the assay was carried out in 100 mM sodium citrate (pH 6.5). In 1 M Tris-HCl (pH 7.0) buffer, phosphatase activity was detectable, but activity was substantially higher when assayed in 1 M Tris-HCl (pH 8.0) (data not shown). Phosphatase activity at 37°C was approximately double that detectable at room temperature (data not shown).
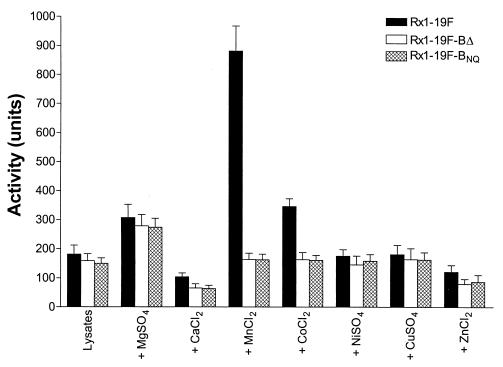
Phosphatase activity in lysates of Rx1-19F, Rx1-19F-BΔ, and Rx1-19F-BNQ was then compared by using 1 M Tris-HCl (pH 8.0) buffer, with incubation at 37°C for 1 h, in the presence or absence of various metal ions (Fig. 4). MgSO4, CaCl2, MnCl2, CoCl2, NiSO4, CuSO4, or ZnCl2 was added to a final concentration of 1 mM where appropriate. The results of three independent experiments were analyzed by using GraphPad Prism version 3.00 for Windows (GraphPad Software, San Diego, Calif.). A baseline level of phosphatase activity of up to 200 U was observed for all three strains with or without 1 mM CaCl2, NiSO4, CuSO4, or ZnCl2. The addition of 1 mM MgSO4 stimulated phosphatase activity in lysates from all three strains ca. 1.5-fold. However, in the presence of 1 mM MnCl2 and, to a lesser extent, 1 mM CoCl2, there was a marked and statistically significant elevation of phosphatase activity in the Rx1-19F lysate (ca. 900 and 350 U, respectively), relative to the levels in either of the cpsB mutant lysates (<200 U in all cases; P = 0.0013 and P = 0.0076 for MnCl2 and CoCl2, respectively, as determined by unpaired t test in the GraphPad Prism software package). These results suggest that CpsB is a metal-binding phosphatase requiring manganese for optimal activity.
FIG. 4.
Phosphatase assay. The ability of deoxycholate lysates from Rx1-19F, Rx1-19F-BΔ, and Rx1-19F-BNQ to cleave p-nitrophenyl phosphate in 1 M Tris-HCl (pH 8.0) in the presence or absence of various metal salts was assayed by measuring the A410 and expressing the activity in units (per milliliter of lysate) after incubation at 37°C for 1 h. Data are the means of three independent assays ± the standard errors of the means (SEM).
Detection of tyrosine phosphatase activity associated with CpsB.
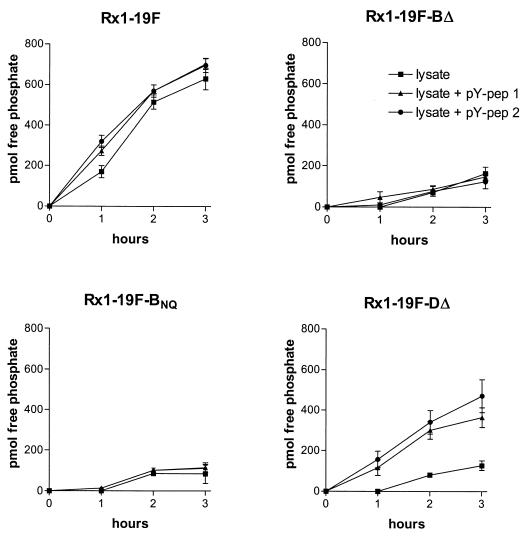
Protein phosphotyrosine phosphatase activity in cell lysates from Rx1-19F, Rx1-19F-BΔ, Rx1-19F-BNQ, and Rx1-19F-DΔ (a cpsD deletion mutant that does not express CpsD) was investigated by using the Tyrosine Phosphatase Assay System (Promega Corporation, Madison, Wis.). Rx1-19F-DΔ was included in this assay because CpsD-P, the natural substrate for CpsB, cannot be produced in this strain, and phosphate should only be released when a synthetic peptide substrate is added to the cell lysate (see below). Cell lysates were prepared by resuspending pneumococcal cells, grown on blood agar plates overnight at 37°C, in 150 mM Tris-HCl (pH 8.0) such that the A600 was 0.6. The cells were pelleted in a bench centrifuge, resuspended in one-fifth volume of 150 mM Tris-HCl (pH 8.0), and then lysed by using a French pressure cell (deoxycholate interferes with this assay system). The lysates were centrifuged to remove the insoluble cell wall material, and the protein concentration of the supernatant was determined according to the method of Bradford (4). The protein concentrations of the lysates were consistently ca. 900 μg/ml. The lysates were treated to deplete endogenous free phosphate and ATP according to the manufacturer’s instructions. The lysates were then assayed for tyrosine phosphatase activity by incubation at 37°C, in the presence of 1 mM MnCl2, for up to 3 h with or without the addition of the synthetic phosphotyrosine peptides pY-pep 1 (END[pY]INASL) and pY-pep 2 (DADE[pY]LIPQQG). The amount of phosphate released in 50 μl of cell lysate was determined by measuring the A600 after addition of molybdate dye solution (Promega). Free phosphate was quantitated by using a standard curve generated from samples containing known free phosphate concentrations. No phosphate was released in the absence of MnCl2 (data not shown).
After 3 h, the wild-type Rx1-19F lysate liberated 600 to 750 pmol phosphate per 45 μg of soluble cellular protein compared to less than 200 pmol for the lysates of CpsB mutants Rx1-19F-BΔ and Rx1-19F-BNQ (P < 0.0001) (Fig. 5). The addition of phosphotyrosine-containing peptides did not affect the amount of phosphate released by lysates from these strains. All three of these strains contain CpsD-P, which is the natural substate for CpsB. Thus, the phosphate release observed in the Rx1-19F lysate is likely to be due to CpsB acting on CpsD-P. This activity is greatly reduced in the Rx1-19F-BΔ and Rx1-19F-BNQ lysates due to the lack of functional CpsB proteins in these strains. The background activity observed in the Rx1-19F-BΔ and Rx1-19F-BNQ lysates is probably due to different phosphatases acting on endogenous cellular substrates. In contrast, when Rx1-19F-DΔ was assayed, the addition of the phosphotyrosine-containing peptides had a significant impact on the amount of phosphate liberated. The CpsB present in the Rx1-19F-DΔ lysate was able to liberate phosphate from pY-pep 2 (470 pmol of phosphate) more efficiently than from pY-pep1 (360 pmol of phosphate). In both cases phosphate release was significantly greater than in the absence of substrate (P = 0.0154 and P = 0.0121, respectively). This demonstrates that CpsB is able to dephosphorylate the synthetic phosphotyrosine-containing peptide substrates in the absence of CpsD-P, its natural substrate.
FIG. 5.
Phosphotyrosine peptide-based phosphatase assay. The amount (in picomoles) of free phosphate generated at 37°C by 45 μg of cellular protein (50 μl cell lysate) from Rx1-19F, Rx1-19F-BΔ, Rx1-19F-BNQ, and Rx1-19F-DΔ was measured at the indicated time points. The phosphopeptides pY-pep 1 (END[pY]INASL) and pY-pep 2 (DADE[pY]LIPQQG) were added at a final concentration of 100 μM to the lysates where indicated. Data are the means of three independent assays ± the SEM.
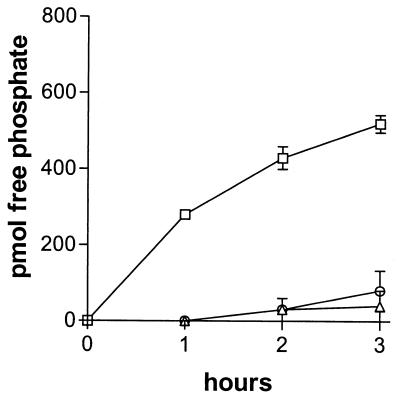
To directly demonstrate the dephosphorylation of CpsD-P by CpsB, the Rx1-19F-BΔ and Rx1-19F-DΔ lysates were mixed and incubated at 37°C. The release of phosphate was significantly greater than that seen with either lysate on its own (P = 0.0009) (Fig. 6). Thus, CpsB in the Rx1-19F-DΔ lysate is able to dephosphorylate the CpsD-P in the Rx1-19F-BΔ lysate. Together, these results indicate that CpsB is able to dephosphorylate both CpsD-P and synthetic phosphotyrosine-containing peptides and is indeed a phosphotyrosine-protein phosphatase.
FIG. 6.
Dephosphorylation of CpsD-P by CpsB. Cell lysates (50 μl containing 45 μg of protein) from Rx1-19F-BΔ, Rx1-19F-DΔ, or both lysates combined were incubated at 37°C, and the amount of free phosphate (pmol) was determined at the indicated times. Symbols: □(, Rx1-19F-BΔ-Rx1-19F-DΔ lysate mix; ○, Rx1-19F-DΔ lysate; ▵, Rx1-19F-BΔ lysate. Data are the means of three independent assays ± the SEM.
Conclusions.
In this study we have demonstrated that CpsB is a novel phosphotyrosine-protein phosphatase. CpsB is only the second member of the PHP family of putative phosphoesterases for which such enzymatic activity has actually been demonstrated, strengthening the probability that other members of this family also have phosphoesterase activity. The four conserved motifs containing histidine residues present in all members of the PHP superfamily were predicted to be involved in metal binding (1). We have demonstrated here that CpsB is dependent on the presence of manganese for optimal activity. Moreover, inactivation of CpsB by mutagenesis of motif 4 confirms the important role of these conserved histidine residues.
We have previously hypothesized that CpsC, CpsD, and ATP interact and that tyrosine phosphorylation of CpsD negatively regulates CPS production (17). In the present study we have shown that CpsB is the phosphotyrosine-protein phosphatase required to dephosphorylate CpsD and thus plays an important role in the regulation of CPS production in S. pneumoniae. We are currently investigating how the degree of CpsD-P phosphorylation affects CPS synthesis and the role of phosphorylation in CpsC and CpsD function.
Acknowledgments
This work was supported by grants from the Channel 7 Children’s Research Foundation and the National Health and Medical Research Council of Australia (grant 157923).
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1998. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 26:3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austrian, R. 1981. Some observations on the pneumococcus and on the current status of pneumococcal disease and its prevention. Rev. Infect. Dis. 3(Suppl.):S1–S17. [DOI] [PubMed] [Google Scholar]
- 3.Berry, A. M., J. Yother, D. E. Briles, D. Hansman, and J. C. Paton. 1989. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect. Immun. 57:2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 5.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139–146. [DOI] [PubMed] [Google Scholar]
- 6.Glucksmann, M. A., T. L. Reuber, and G. C. Walker. 1993. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J. Bacteriol. 175:7045–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guidolin, A., J. K. Morona, R. Morona, D. Hansman, and J. C. Paton. 1994. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect. Immun. 62:5384–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton, R. M. 1993. In vitro recombination and mutagenesis of DNA. Methods Mol. Biol. 15:251–261. [DOI] [PubMed] [Google Scholar]
- 10.Jones, E. W., and G. R. Fink. 1982. Regulation of amino acid and nucleotide biosynthesis in yeast, p.181–299. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Kim, J. O., and J. N. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368–377. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 13.Llull, D., R. López, E. García, and R. Muñoz. 1998. Molecular structure of the gene cluster responsible for the synthesis of the polysaccharide capsule of Streptococcus pneumoniae type 33F. Biochim. Biophys. Acta 1443:217–224. [DOI] [PubMed] [Google Scholar]
- 14.Malone, R. E., S. Kim, S. A. Bullard, S. Lundquist, L. Hutchings-Crow, S. Cramton, L. Lutfiyya, and J. Lee. 1994. Analysis of a recombination hotspot for gene conversion occurring at the his2 gene of Saccharomyces cerevisiae. Genetics 137:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Morona, J. K., A. Guidolin, R. Morona, D. Hansman, and J. C. Paton. 1994. Isolation, characterization, and nucleotide sequence of IS1202, an insertion sequence of Streptococcus pneumoniae. J. Bacteriol. 176:4437–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine-phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431–1442. [DOI] [PubMed] [Google Scholar]
- 18.Paton, J. C., and J. K. Morona. 1999. Streptococcus pneumoniae capsular polysaccharide, p.201–213. In V. A. Fischetti, R. Novick, J. Ferretti, D. Portnoy, and J. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 19.Sharma, D. P., C. Thomas, R. H. Hall, M. M. Levine, and S. R. Attridge. 1989. Significance of toxin-coregulated pili as protective antigens of Vibrio cholerae in the infant mouse model. Vaccine 7:451–456. [DOI] [PubMed] [Google Scholar]
- 20.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vincent, C., B. Duclos, C. Grangeasse, E. Vaganay, M. Riberty, A. J. Cozzone, and P. Doublet. 2000. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J. Mol. Biol. 304:311–321. [DOI] [PubMed] [Google Scholar]
- 22.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wugeditsch, T., A. Paiment, J. Hocking, J. Drummelsmith, C. Forrester, and C. Whitfield. 2001. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J. Biol. Chem. 276:2361–2371. [DOI] [PubMed] [Google Scholar]