Abstract
In Shigella flexneri expression of the plasmid-encoded virulence genes is regulated via a complex mechanism involving both environmental signals and specific transactivators. The primary regulator protein, VirF, is a member of the AraC family of transcription factors and shares with other AraC-like proteins a conserved carboxy-terminal domain thought to be important for DNA binding. Random and site-directed mutagenesis of the virF gene encoding VirF yielded a number of mutations along the length of the protein which severely affected the ability of VirF to activate gene expression. The mutant proteins were shown to be affected in their ability to activate the virulence genes virB and icsA, both known to be regulated directly by VirF, as well as the virB-dependent virulence gene mxiC. Mutating key residues predicted to be important for DNA recognition had a significant negative effect, thereby suggesting that VirF interacts with its target sequence via two helix-turn-helix motifs. Two mutants that were dominant negative when coexpressed with the wild-type VirF protein were also isolated, indicating a role for protein-protein oligomerization in normal VirF function.
The gram-negative facultative intracellular pathogen Shigella flexneri is the causative agent of bacillary dysentery. It carries a 230-kb virulence plasmid, of which a 31-kb region termed the entry region encodes proteins required for invasion of mammalian epithelial cells, intra- and intercellular spread of the bacteria, and surface presentation and secretion of the invasion proteins via a type III secretion system (8, 22). This high-molecular-weight plasmid also carries two regulatory genes: virB, located within the entry region, and virF, outside this region. The primary regulator VirF is required for transcription of the virB gene; the virB gene product (VirB) then activates transcription of the functional genes of the entry region (reviewed in reference 4). The genes of the entry region are transcribed at 37°C but not at 30°C, and the VirF-VirB regulatory cascade is important for this temperature control (4). VirF also regulates positively expression of a gene outside the entry region (icsA, previously called virG) involved in intercellular spreading (20) and negatively autoregulates its own transcription (16).
The VirF protein is a member of the AraC family of transcriptional regulators, of which more than 100 different proteins have to date been identified by virtue of a conserved C terminus (6). These proteins regulate a wide range of cellular processes involved with carbon metabolism, stress response, and virulence. With the exception of CelD, which acts as a repressor, all family members are transcriptional activators and can be divided into three classes (6, 12). The first class consists of proteins which, like AraC itself, regulate transcription in response to a chemical signal, usually a carbohydrate molecule. The best studied is the Escherichia coli AraC protein, which controls expression of the genes necessary for uptake and catabolism of arabinose (reviewed in reference 25). AraC is composed of an amino-terminal domain that includes the dimerization and arabinose-binding regions (26, 27), a carboxy-terminal domain that contains two potential helix-turn-helix (HTH) motifs for sequence-specific DNA recognition and binding, and a linker region which connects the two functionally distinct domains (5). In contrast to family members involved in the utilization of sugars, the class of AraC proteins involved in stress response such as MarA, Rob, and SoxS are monomers (12). These proteins lack the N-terminal domain: MarA and SoxS consist of a DNA-binding domain alone, while Rob has an additional carboxy-terminal domain of unknown function with structural homology to the E. coli enzyme GalT (10).
The third group of proteins, to which VirF belongs, consists of proteins that regulate transcription in response to a physical signal, usually a change in temperature. Many of these are involved in virulence gene regulation, and unlike most AraC family members the virulence regulator subfamily shows homology throughout the length of the protein (4), raising the possibility that sequences found at the N termini are involved in a common mechanism of response to environmental stimuli. With one exception (UreR, which binds urea), the family members involved in virulence regulation do not bind specific effectors (6). Further evidence for common activation mechanisms among this subfamily is that some members, including VirF, can substitute for others in regulation of their cognate promoters (3, 14, 18, 23).
Among the AraC family, the degree of sequence conservation at the first HTH (HTH1) is low, perhaps reflecting the different DNA targets that each protein recognizes (6). This contrasts to the second HTH motif (HTH2), which is highly conserved, leading to the hypothesis that this motif serves a common role other than DNA binding, possibly contacting RNA polymerase (6). Recently the crystal structures of two AraC family proteins, MarA and Rob, have been solved (10, 19), and these have suggested conflicting roles for HTH2. Analysis of cocrystals of MarA and its mar promoter binding site (19) reveals that the protein is composed of seven α-helices comprising two HTH motifs (α2-T-α3 and α5-T-α6) connected by a linker α-helix (α4) and two flanking α-helices (α1 and α7). This structure confirms earlier predictions that AraC family proteins bind DNA via two DNA-binding determinants (15). Both HTH motifs form specific interactions with two adjacent segments of the major groove via nonidentical nucleotide contacts. The two HTH motifs are separated by the long α4 helix and because of the relative spacing of the recognition helices, binding of both motifs forms an ≈35° bend in the DNA. The conserved C termini of other AraC family proteins are likely to adopt the same tertiary structure as MarA, given the high sequence similarity (greater than 20% [6]). Indeed, a similar structure has been obtained for Rob (10), but here while the N-terminal HTH is inserted into the major groove of the binding site, the C-terminal HTH lies on the surface of the DNA helix, where it makes nonspecific contacts with the phosphodiester backbone. The DNA in the Rob cocrystal structure is unbent. Therefore, the Rob structure lends support to the idea that the second HTH has a function other than specific DNA recognition. However, crystal packing artifacts and/or its unique carboxy-terminal extension might make the Rob-DNA cocrystal structure different from that seen for MarA and expected for other AraC-like proteins (10, 12). Despite this, AraC itself is inferred to bind DNA via either one or two HTH motifs, depending on the binding site in question (9, 15), and the two structures may therefore represent genuine alternative binding modes (12). Thus, it is of interest to determine the role of the second HTH in the function of other AraC-like proteins.
How VirF responds to temperature is unknown, and it is not known if protein dimerization is required for activity. In addition to the general ignorance about the structure and mechanism of action of VirF, little is known about its interactions with other regulators of virulence gene expression in S. flexneri or with the transcriptional machinery, in particular RNA polymerase. Members of the AraC family are typically insoluble, making protein purification difficult and thereby hampering analysis of this protein family in vitro. Despite many attempts, we have failed to obtain purified, soluble VirF for use in in vitro studies (M. E. Porter and C. J. Dorman, unpublished data). As a first step in acquiring structural and functional information about the VirF polypeptide, we have therefore subjected its gene to both random and site-directed mutagenesis and assayed the functions of these mutant proteins in vivo. We find a significant negative effect of mutating the key residues in HTH1, and we also find that deletion of HTH2 and key mutations in its predicted DNA-interacting residues render VirF nonfunctional. Therefore, HTH2 seems to be important for DNA recognition by VirF. Mutations in some residues in the amino-terminal domain also have a major effect on VirF function. In addition, we have investigated oligomerization of VirF by analysis of the dominant negative effects of some of these mutant proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study were derivatives of S. flexneri strains BS184 (mxiC-lacZ) and CJD1006 (BS184 virF [17]) and of E. coli K-12 strain TG1. Plasmid pMEP31 (17) is a derivative of pACYC184, in which the virF gene has been cloned onto a 7.9-kb BamHI fragment. The virF gene on a 1.3-kb HpaII fragment cloned into pACYC184 was designated pMEP510 (18), while the same fragment in pBR322 resulted in pMEP532. pACYC184-based plasmids harboring the same virF fragment as pMEP510 but with point mutations within the virF coding region were designated by the single-letter amino acid code of the mutation.
Growth media and conditions.
Bacterial cultures were grown in L broth (13) or on L plates (L broth containing 1.5% [wt/vol] agar). The Lac phenotypes of strains were screened on either MacConkey-lactose (13) agar plates or L agar supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 20 μg/ml). Antibiotic selection was with ampicillin (50 μg/ml), chloramphenicol (15 μg/ml), or kanamycin (20 μg/ml).
Genetic techniques.
Transformation of plasmid DNA was by electroporation (Bio-Rad Gene Pulser, used according to the manufacturer’s instructions). Plasmid DNA was isolated and manipulated by standard methods (21). Restriction endonucleases and T4 DNA ligase were bought from Amersham (Little Chalfont, United Kingdom) and used according to the manufacturer’s recommendations.
Enzyme assays.
Expression of the mxiC-lacZ fusion was measured by β-galactosidase assays of overnight cultures grown at 30 or 37°C according to the method of Miller (13). Cultures were grown in duplicate, and each culture was assayed in duplicate; the data were expressed as the means of the two sets of measurements. Standard deviations were usually less than 10%. The experiments were repeated two to three times, and consistent results were obtained; the data from one experiment are shown.
RNA extraction.
Total cellular RNA was extracted by cell lysis in boiling REB buffer (20 mM sodium acetate [pH 5.2], 2% [wt/vol] SDS, 0.3 M sucrose) followed by phenol extraction and DNase I treatment, and the virB, icsA, and hupA mRNAs were detected on Northern blots with digoxigenin-dUTP-labeled RNA probes. Both methods have been described in detail previously (16, 17). The experiments were repeated three times, and typical data are shown.
Random mutagenesis.
Random mutagenesis with nitrosoguanidine was performed with the virF gene in plasmid pMEP31 essentially as described previously (13). Briefly, E. coli cells harboring pMEP31 were treated with 50 μg of nitrosoguanidine at 37°C for 30 min, and following washing the isolated plasmid DNA was electroporated into S. flexneri. These conditions yielded virF-containing plasmids with altered activation phenotypes at a frequency of approximately 1 in 200.
Site-directed mutagenesis.
Site-directed mutagenesis was performed with the virF gene cloned into pMEP510 using a Quikchange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s directions.
Western blotting.
Western blot analysis of VirF proteins was performed essentially by the method of Towbin et al. (29). The primary antibody had been raised in rabbits against a synthetic peptide equivalent to the last 12 amino acids of the VirF protein. The secondary antibody was a goat anti-rabbit immunoglobulin G alkaline phosphatase-conjugate, and its reaction with the primary antibody was detected by the Purple AP Precipitating Substrate (Roche) used according to the manufacturer’s directions. The experiment was repeated three times.
RESULTS
Random mutagenesis of the virF gene.
VirF positively regulates transcription of the structural gene icsA and the regulatory gene virB; VirB then activates expression of the genes of the entry region, including mxiC. As a first step towards a detailed structure-function analysis of S. flexneri VirF we randomly mutated a cloned copy of the virF gene and employed the VirB-regulated mxiC-lacZ fusion located on the S. flexneri virulence plasmid to monitor changes in β-galactosidase activity. The virF+ plasmid pMEP31 was subjected to nitrosoguanidine mutagenesis (see Materials and Methods) and transformed into the S. flexneri virF mutant strain CJD1006. This strain exhibits a Lac− phenotype on indicator plates at 37°C due to the absence of the VirF activator; a Lac+ phenotype can be restored by introducing the virF gene on a multicopy plasmid (18). Three independent rounds of mutagenesis (≈10,000 colonies screened) yielded 52 colonies with altered β-galactosidase activity on indicator plates. Of these, 21 may have acquired mutations in the virF gene of plasmid pMEP31, as assessed by retransformation into CJD1006. The β-galactosidase activity of the CJD1006 derivatives harboring the altered pMEP31 plasmid was determined (data not shown), allowing the mutated plasmids to be arranged into nine groups each with a different phenotype: a representative of each group was selected for further analysis. Each of the putative mutant virF genes was subcloned onto a 1.3-kb HpaII fragment into pACYC184 in order to eliminate any mutations occurring outside the virF open reading frame.
The mutant genes were sequenced, revealing a total of five different point mutations: L72F, A154T, V191A, E196K, and S236N (Table 1). L72F, located in the N-terminal region of the protein, results in a change of a residue which is a conserved hydrophobic in a number of AraC-like virulence gene regulators (Porter and Dorman, unpublished) but not in other members of the wider AraC family, which have an unrelated N-terminal domain. In contrast, A154T, located in the linker region between the N- and C-terminal domains, is poorly conserved among all members of the family (Porter and Dorman, unpublished). V191A is located in the recognition helix (helix 3) of the first HTH motif, and the conserved hydrophobic side chain of the equivalent residue (L44) contributes to the hydrophobic core of this subdomain in MarA (19). The residue at position 196 is generally polar in other members of the family (equivalent to K49 in MarA [6]) and is also located in helix 3 of the first HTH. In the MarA-DNA cocrystal structure, K49 makes an interaction with the phosphate backbone of DNA (19). Finally, the well-conserved serine S236 (6) is located at the end of the turn in the second HTH domain.
TABLE 1.
Characteristics and phenotypes of VirF point mutations studied
| Mutation | Sourcea | Phenotype of mutant proteinb | Location/characteristic of mutated residuec | Conservation of mutated residued |
|---|---|---|---|---|
| Y23S | SD | No activity | N terminus | Rns-FapR family |
| L72F | Random | Residual activity | N terminus | Conserved hydrophobic |
| S150A | SD | No activity (no protein) | Linker | Well- conserved |
| A154T | Random | Residual activity | Linker | Poorly conserved |
| I180N | SD | No activity; trans-dominant | Helix 2 (HTH1)/solvent exposed | Conserved hydrophobic |
| V191A | Random | No activity | Helix 3 (HTH1)/hydrophobic core | Conserved hydrophobic |
| K193A | SD | No activity | Helix 3 (HTH1)/base interacting | Conserved basic (E in Yersinia VirF) |
| E196K | Random | Residual activity | Helix 3 (HTH1)/backbone interacting | Mainly polar (K in MarA) |
| Y224Oche | SD | No activity; trans-dominant | Preceding helix 5/deletes HTH2 | NAf |
| V228T | SD | No activity | Helix 5 (HTH2)/hydrophobic core | Conserved hydrophobic |
| S236N | Random | Residual activity | End of HTH2 turn | Well conserved |
| Y239F | SD | No activity | Helix 6 (HTH2)/base interacting | Y (or T in MarA) |
| I241N | SD | No activity | Helix 6 (HTH2)/base interacting | Rns-FapR family |
From random or site-directed (SD) mutagenesis approaches.
Within the domain organization predicted for VirF or relative to the structural features of the C-terminal domain as defined for MarA and Rob.
Conservation of the mutated residue among related AraC family proteins (6; Porter and Dorman, unpublished).
Och, ochre stop codon.
NA, not applicable.
The effect of the VirF protein encoded by each plasmid on expression of the mxiC-lacZ fusion was assessed following transformation into the S. flexneri virF null mutant strain CJD1006 (Table 2). Although four out of the five mutant plasmids (pL72F, pA154T, pE196K, and pS236N) still possessed significant residual gene activation function, this was to levels lower than the unmutagenized plasmid pMEP510. The residual activity of the S236N mutant agrees with the moderate phenotype observed for MarA (S90A) by Gillette et al. (7), although the phenotype of the VirF E196K mutation was less severe than that produced by a mutation of MarA at the equivalent position (K49A) studied by these authors. As it is negatively charged, E196 in VirF may not contact the phosphate backbone as does K49 in MarA. It is interesting, however, that the charge reversal at this residue in the E196K mutant produces a protein which retains some function. One random mutation, pV191A, was completely unable to promote mxiC-lacZ expression in response to a thermal signal. This is as predicted for a mutation which disrupts the hydrophobic core of the first HTH subdomain (see above) and is in agreement with the results observed for the L44A mutation in MarA (7).
TABLE 2.
Effect of random and site-directed mutations of VirF on mxiC-lacZ expression
| Plasmida | Mean (SD) of β-galactosidase activity (Miller units)b
|
|
|---|---|---|
| 30°C | 37°C | |
| None | 7 (0.25) | 15 (0.24) |
| pMEP510 | 602 (2.40) | 1,055 (15.4) |
| pY23S | 11 (0.82) | 20 (0.68) |
| pL72F* | 25 (1.00) | 401 (28.0) |
| pS150A | 9 (0.64) | 27 (0.45) |
| pA154T* | 20 (0.65) | 279 (2.90) |
| pI180N | 7 (0.05) | 14 (0.46) |
| pV191A* | 9 (0.87) | 17 (0.71) |
| pK193A | 7 (0.42) | 15 (0.59) |
| pE196K* | 38 (0.39) | 350 (22.0) |
| pY224Och | 7 (0.86) | 15 (0.81) |
| pS236N* | 14 (0.20) | 124 (5.30) |
Transformed into the S. flexneri virF mutant strain CJD1006. pMEP510 is the wild-type virF+ plasmid; mutations encoded by the other plasmids are indicated by single-letter amino acid codes. Mutations isolated by random mutagenesis are indicated by asterisks; other mutations were engineered by site-directed mutagenesis. Och, ochre stop codon.
β-Galactosidase activity data are mean results of duplicate assays on duplicate cultures. High-level expression of mxiC-lacZ at 30°C in the presence of pMEP510 is an effect of overexpression of the VirF activator and is not seen when virF is present in single copy (16).
Site-directed mutagenesis of the virF gene.
While the random approach had yielded mutations throughout the virF gene, it was important to target specific residues in the N-terminal domain, the linker region, and the first HTH motif thought to be involved in DNA binding. Thus, a PCR-based technique was employed to mutagenize the virF gene in plasmid pMEP510. Two residues conserved in the related virulence regulators FapR, Rns, and AggR (6; Porter and Dorman, unpublished), Y23 (within the structurally uncharacterized N-terminal domain) and I180 (located in the positioning helix [helix 2] of the first HTH motif) were changed to serine and asparagine, respectively (Table 1). In the linker region between the N- and C-terminal domains, S150, highly conserved among both AraC-like virulence gene regulators and AraC itself (Porter and Dorman, unpublished), was changed to alanine, thus retaining the flexibility of the side chain but removing its functionality. Another residue highly conserved among AraC-like virulence gene regulators, K193, is located in the recognition helix (helix 3) of the first HTH motif, and in the MarA-DNA cocrystal structure the equivalent side chain, R46, makes multiple base-specific contacts in the major groove (19). Therefore K193 was mutated to alanine. Finally, in order to test the importance of the second HTH motif, a stop codon was introduced at Y224 immediately before the positioning helix (helix 5) of this motif, thus deleting it completely. Plasmids harboring the mutant copies of the virF gene were transformed into the virF null-mutant strain CJD1006, and their ability to promote expression of the mxiC-lacZ reporter gene fusion was assessed (Table 2). In all five cases the mutant proteins were completely unable to activate mxiC-lacZ expression. Therefore, it can be concluded that key residues in the positioning helix (I180) or recognition helix (K193) of the first HTH motif of VirF are critical for its function, as are conserved residues in the N-terminal domain (Y23). The second HTH motif also seems to be essential for function, although it is possible that its deletion might affect the folding of the rest of the C-terminal domain. A mutation in the linker region between the N- and C-terminal domains (S150A) also appeared to create a nonfunctional protein. However, production of the S150A protein was undetectable by Western blotting (see below).
Activation of virB and icsA transcription by the mutant virF gene products.
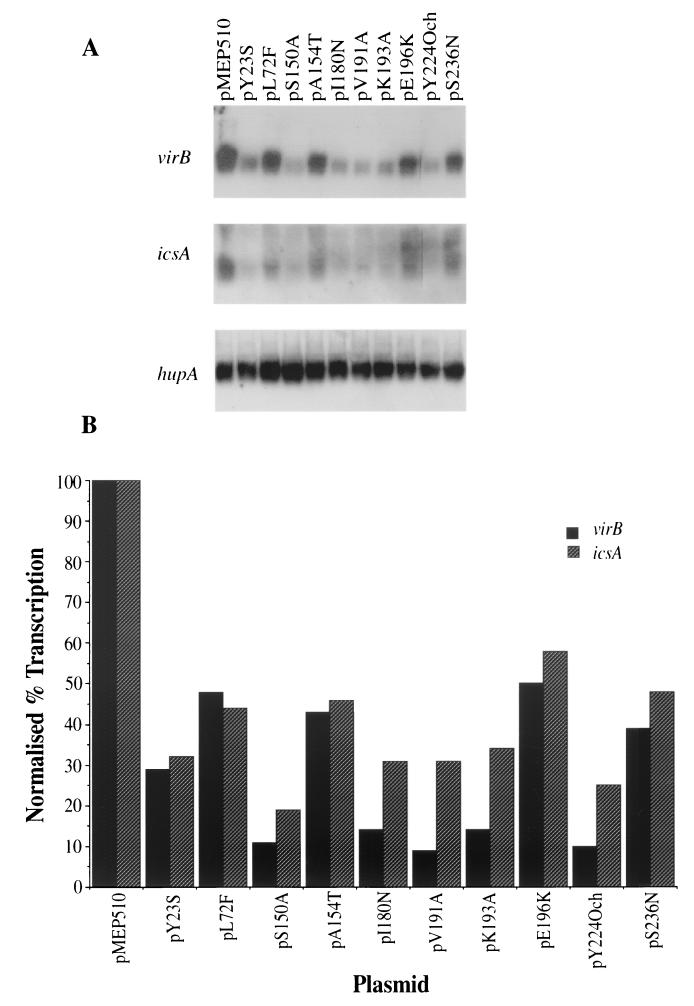
In the above work a lacZ fusion to the invasion gene mxiC was employed. However, this fusion is not activated directly by VirF; instead, VirF activates the virB gene and its product in turn activates the structural gene operon that includes mxiC. It was important, therefore, to determine the activation of virB and the unlinked VirF-dependent gene icsA directly by each of the mutant virF gene products. Transcript levels specified by the virB and icsA genes in CJD1006 derivatives harboring each of the virF mutant plasmids grown at 37°C were measured by Northern blotting (Fig. 1). As a control for RNA loading and integrity the hupA transcript, encoding the α subunit of the nucleoid protein HU, was also detected and used to normalize the levels of virB and icsA transcripts. The transcription patterns of both virB and icsA were qualitatively similar in the presence of each of the mutant virF plasmids used, although the effects of the mutations were generally greater at virB. Moreover, the expression pattern of virB was directly analogous to that obtained by assaying β-galactosidase activity of the mxiC-lacZ reporter gene fusion (Table 2). These data support the hypothesis that the reduction in mxiC-lacZ expression seen in the presence of mutant virF genes was due to a decrease in virB gene transcription and confirm that the mxiC-lacZ fusion is an accurate reporter of VirF activity.
FIG. 1.
(A) Northern blot analysis of the virB, icsA, and hupA transcripts in cultures of CJD1006 transformed with the virF+ plasmid pMEP510 or with mutant VirF-encoding plasmids as indicated. (B) Densitometric analysis of the data from panel A showing the relative levels of the virB (black bars) and icsA (hatched bars) transcripts. The data have been normalized to the level of the hupA transcript in each culture, and the level of transcription in the presence of the wild-type plasmid pMEP510 is taken as 100%.
Coexpression of mutant and wild-type virF genes.
Several members of the AraC family have been shown to form homodimers including Yersinia enterocolitica VirF and AraC itself (30, 31). It is known that residues important for AraC protein-protein interactions are contained within the N-terminal domain, being localized to a coiled-coil-forming α-helix located immediately before the linker which joins the N- and C-terminal domains (26, 27). It is frequently observed that non-DNA-binding mutant derivatives of a protein which retain dimerization function have a dominant negative effect on the wild-type protein. Given that most of the virF mutations studied here are within the latter half of the gene (the DNA-binding domain), it might be predicted that many of these mutant proteins would be dominant negative over the wild type. The possibility of VirF-VirF protein-protein interactions was therefore investigated in vivo by coexpressing mutant and wild-type copies of the virF gene. Wild-type strain BS184 was transformed with the mutant plasmids, and the effect on mxiC-lacZ expression was measured. As a control, the effect of the unmutagenized plasmid pMEP510 was also measured in BS184. Two of the mutant plasmids, pI180N and pY224Och, exerted a clear trans-acting dominant negative effect on VirF-dependent mxiC activation (Table 3), reducing expression by approximately 50%. The rest of the mutants were recessive to the presence of a wild-type copy of the virF gene. Therefore, although removal of the second helix-turn-helix motif in the Y224Och mutant does create a dominant negative protein, other mutations predicted to affect DNA binding (V191A, K193A) do not have such an effect. The I180N mutation may have only a minor effect on DNA binding (see Discussion). It was important to determine if the mutant virF genes in plasmids pI180N and pY224Och were still trans-dominant in cells possessing the wild-type virF gene on a vector with a similar copy number. Therefore, the virF gene was cloned on a 1.3-kb HpaII fragment into pBR322. The resulting construct, pMEP532, was transformed into BS184 and into its derivatives that contained plasmids pI180N and pY224Och. The results given in Table 3 show that, despite the presence of multiple copies of the wild-type virF allele, plasmids pI180N and pY224Och still exerted a strong dominant negative effect on mxiC expression.
TABLE 3.
trans-dominant effects of mutant VirF proteins on the wild-type virF gene
| Plasmida | Mean (SD) of β-galactosidase activity (Miller units)b
|
|
|---|---|---|
| BS184 | BS184 + pMEP532c | |
| None | 1,030 (34) | 1,209 (52) |
| pMEP510 | 1,135 (47) | ND |
| pY23S | 883 (46) | ND |
| pL72F | 1,261 (53) | ND |
| pS150A | 1,017 (38) | ND |
| pA154T | 1,225 (6.0) | ND |
| pI180N | 596 (50) | 652 (37) |
| pV191A | 987 (53) | ND |
| pK193A | 923 (68) | ND |
| pE196K | 1,189 (29) | ND |
| pY224Och | 504 (36) | 735 (30) |
| pS236N | 1,196 (36) | ND |
Transformed into the S. flexneri wild-type strain BS184. pMEP510 is the wild-type virF+ plasmid; mutations encoded by the other plasmids are indicated by single-letter amino acid codes. Och, ochre stop codon.
β-Galactosidase activity data of the mxiC-lacZ fusion at 37°C are mean results of duplicate assays on duplicate cultures.
Plasmid pMEP532 carries the wild-type virF gene, and mutant plasmids which had dominant negative effects were also tested in the presence of this plasmid. ND, not determined.
Detection of mutant and wild-type proteins by Western blotting.
To confirm that the activation phenotypes of the mutant proteins were not due to lack of production of the mutated regulator, VirF protein was detected in extracts from the virF-deleted strain CJD1006 transformed with the respective plasmids by using an antiserum raised against the extreme C terminus of VirF (see Materials and Methods). All mutant proteins were detectable except for S150A and Y224Och (data not shown). Y224Och is not detectable by this assay, as the antibody was raised against a portion of the protein deleted in this mutant, but the protein must be produced in vivo because the plasmid pY224Och has a trans-dominant phenotype (Table 3). The S150A protein may be unstable due to the mutation in the linker region making VirF susceptible to proteolysis, and the lack of activation by this mutant probably therefore reflects the absence of protein rather than a defect in function. For all other mutants tested, the observed effects on mxiC expression are most likely due to disruption of the DNA-binding or activation functions of VirF, as predicted.
Site-directed mutagenesis of the second helix-turn-helix motif.
The phenotype of the Y224Och truncation mutant suggests that the second HTH motif plays an important role in VirF function, probably by contributing to DNA binding. However, the truncation at Y224 might affect the folding of the whole C-terminal domain. The one random mutation isolated within this subdomain, S236N, had only a relatively mild effect on VirF function despite altering a well-conserved residue. In order to test the role of HTH2 in specific DNA recognition, we therefore generated an additional three site-directed mutations within the motif, selecting residues which should be critical for its putative DNA binding function based on the structural information available for MarA. The first mutation, V228T, targets a residue within the positioning helix of this motif (helix 5) which contributes to the hydrophobic core of the subdomain (19). It is a conserved hydrophobic in all closely related AraC-like virulence regulators, and in 95% of the overall AraC family it is a nonpolar residue (6). Mutation of the equivalent residue (L82) to alanine in MarA had a severe effect on function (7), although in the XylS protein, which seems to require predominantly its first HTH motif for DNA binding, the effect of an equivalent mutation (I285E) was less severe (11). Secondly, we introduced a mutation of Y239 to F in the recognition helix. While conserving the bulky aromatic side chain at this position, this mutation removes the hydroxyl group functionality and therefore the potential for hydrogen bond formation. This is significant because the equivalent residue in MarA, T93, forms water-mediated hydrogen bonds to DNA bases in the cocrystal structure (19). In the virulence-regulating AraC subfamily and in AraC itself, this residue is also a tyrosine (6). The final mutation, I241N, also lies in the recognition helix of the second HTH motif. This residue is a conserved isoleucine in the virulence regulators (6), and in MarA its equivalent (T95) forms a van der Waals interaction with a DNA base (19). A T95A mutant of MarA has significantly compromised function (7). These three additional mutations are summarized in Table 1.
The ability of each of the three mutant VirF proteins to activate mxiC expression was assessed by transforming each mutated plasmid into CJD1006 and measuring β-galactosidase expression (Table 4). In each case, no significant transcriptional activation by the mutant protein was seen, indicating that the second HTH motif plays a critical role in DNA binding by VirF. We also examined the trans-dominance of these point mutations in HTH2 on wild-type VirF. When transformed into BS184, plasmids expressing the mutated proteins were unable to disrupt the function of wild-type VirF produced from the chromosome or from the multicopy plasmid pMEP532 (Table 4). Therefore, as for mutations in HTH1, these mutations, which disrupt activation function severely, probably via loss of DNA-binding activity, are nevertheless unable to affect DNA binding and transcriptional activation by the wild-type protein. Western blotting analysis of the three mutant proteins confirmed that all three were detectable at significant levels in CJD1006 cell extracts transformed with the respective plasmids (data not shown).
TABLE 4.
Activation function and trans-dominant effects of VirF proteins with mutations in the second HTH motif
| Plasmida | Mean (SD) of β-galactosidase activity (Miller units)b
|
|||
|---|---|---|---|---|
| CJD1006 at 30°C | CJD1006 at 37°C | BS184 at 37°C | BS184 + pMEP532 at 37°Cc | |
| pMEP510 | 700 (28.0) | 1,247 (50.0) | 1,574 (65.5) | 1,292 (51.5) |
| pV228T | 15 (0.50) | 30.8 (1.05) | 1,296 (67.0) | 1,234 (48.0) |
| pY239F | 10 (0.20) | 16.4 (0.10) | 1,318 (34.0) | 1,202 (48.0) |
| pI241N | 10 (0.20) | 16.9 (1.00) | 1,501 (42.0) | 1,467 (59.5) |
Transformed into the S. flexneri virF mutant strain CJD1006 or wild-type strain BS184. pMEP510 is the wild-type virF+ plasmid; mutations encoded by the other plasmids are indicated by single-letter amino acid codes.
β-Galactosidase activity data of the mxiC-lacZ fusion are mean results of duplicate assays on duplicate cultures.
Plasmid pMEP532 carries the wild-type virF gene.
DISCUSSION
VirF plays a central role in controlling virulence gene expression in S. flexneri, but little is known of its structure or the mechanism by which it influences transcription. Because of the difficulties associated with working with VirF in vitro (AraC-like proteins are typically insoluble during protein purification procedures, as we have found for VirF), we have used random and site-directed mutagenesis to isolate mutations along the length of the virF gene and analyzed the ability of the mutated proteins to activate virulence gene expression in vivo by assaying β-galactosidase expression from an mxiC-lacZ gene fusion. As mxiC is regulated by the VirF-dependent virB gene product rather than by VirF itself, we also studied transcription of two genes regulated directly by VirF (virB and icsA) in the presence of the mutant proteins. By these approaches we characterized two mutations within the N-terminal domain, two within the linker region, four within the first HTH motif, and four within the second HTH motif, which affect transcriptional activation by VirF at 37°C to various degrees. We also examined the effect of deleting the second HTH motif and sequences downstream of it. All of the mutations also have a negative effect on VirF-dependent transcription at 30°C, but significant activation by VirF at this temperature is seen only when the protein is overexpressed and is therefore highly sensitive to alterations in the expression level of VirF. For this reason, the relative effects of the different mutations at 30°C are not considered meaningful and are not discussed further here. In contrast, activation by VirF at 37°C is not sensitive to the expression level of the activator (compare the effects of no plasmid [single copy] and pMEP510 [≈15 copies] in BS184 [Table 3]) and therefore provides an accurate measure of the level of impairment of the activation function of the mutant proteins.
The two mutations within the N-terminal domain analyzed, Y23S and L72F, affected activation severely and moderately, respectively. Both residues are well conserved in closely related AraC-like virulence regulators (FapR, Rns, and AggR) and lie in some of the better-conserved regions of the N-terminal domain (Porter and Dorman, unpublished), which may reflect an important contribution to the structural core of this domain or to the environmental responsiveness of these regulators. However, the lack of structural or functional information on the N terminus of this class of AraC-like proteins makes more-definite conclusions difficult to draw. Such mutations provide a basis for a more detailed study of the N-terminal domain, which may be aided by studying it in isolation from the C terminus, which is thought to account for the insolubility problems encountered within the AraC family (6). Of the two mutations studied within the linker region that connects the N- and C-terminal domains, S150A abolished activation function but also detectable expression of VirF, while A154T had a milder effect on transcriptional activation. As no other VirF mutation studied here abolished expression of the protein (data not shown), it is likely that the S150A mutation makes the protein sensitive to proteolysis at the exposed linker region rather than having a severe effect on translation. The linker region of VirF is rich in amino acid side chains such as alanine and serine that permit protein flexibility, and although the A154T mutation may restrict this slightly, its relatively mild effect is consistent with the hypothesis that the amino acids in the linker do not contribute critically to protein function and with the observation that the corresponding linker region in AraC is generally amenable to mutagenesis (5).
Within the first HTH motif, two mutations affecting hydrophobic residues, I180N and V191A, and two affecting charged residues, K193A and E196K, were studied. I180 was reported as being part of the hydrophobic core of this motif in MarA (equivalent to V33 [19]), but subsequent examination of the structure of MarA shows that the side chain is in fact largely solvent exposed (7). This correlates with the minor effect of an alanine substitution at this position in MarA on function (7). In contrast, the I180N mutation abolishes transcriptional activation by VirF; this may reflect a defect in a function not important in the MarA context, such as oligomerization. This is supported by the observation that this mutant protein exerts a dominant negative effect on wild-type VirF (see below). V191A, on the other hand, affects a residue which is genuinely part of the hydrophobic core of HTH1, and the lack of activation function seen for this mutant is probably due to disruption of the tertiary structure of this subdomain. Results of alanine scanning mutagenesis of MarA (7) suggest that base-specific contacts made by R46 (K193 in VirF) to bases within the major groove and phosphate backbone contacts made by K49 (E196) are major contributors to DNA-binding affinity. In MarA R46 penetrates into the major groove and makes hydrogen bonds with three bases (19); thus, the severe phenotype of the K193A mutant is expected. A mutation in H213 of AraC (equivalent to K193) also reduced binding to the araBAD promoter and affected the ability of the protein to bend DNA (24). The E196K mutation has a less severe effect on activation than did K193A (Table 2), suggesting that this residue is not so critical for DNA binding in VirF. However, a phosphate backbone interaction equivalent to that of K49 in MarA by E196 in the VirF-DNA complex is unlikely because of the charge-charge repulsion expected between the side chain and the DNA backbone. Overall, this set of four mutations confirm the importance of HTH1 in DNA recognition by VirF.
Data for mutants on the second HTH motifs of AraC family members give mixed conclusions: mutations of some residues in HTH2 of MelR or XylS result in activities similar to those of the wild type, and for AraC itself, some mutants lose contact with multiple bases or bind to DNA in a pattern not consistent with a canonical HTH motif (reviewed in reference 6). Studies on the second HTH in XylS reveal that the most conserved residues are involved in maintaining the tertiary structure (11). However, the crystal structure of MarA bound to its target DNA shows that this protein binds DNA via both HTH motifs, in agreement with studies suggesting that in AraC the second motif as well as the first can contact DNA (15). Alanine-scanning mutagenesis of MarA showed that both HTH motifs contribute to DNA binding in vivo, and the disruption of either nonspecific contacts with the sugar-phosphate backbone or specific contacts made by R46 (HTH1) and R96 (HTH2) to bases in the major groove severely reduced MarA activity (7). Mutagenesis studies have also revealed base-specific contacts by amino acids in the second HTH of the RhaS protein (1).
In this work, a VirF derivative lacking the extreme C terminus including HTH2 (Y224Och) was unable to activate gene expression, suggesting that this region has an important function. Given that introduction of a stop codon results in a deletion of the last 39 amino acids of the protein, the phenotype of this mutation could reflect effects on functions other than just DNA binding. The mutation also generates a dominant negative phenotype, while missense mutations targeting predicted DNA-interacting residues do not. Our site-directed mutants within HTH2 provide a clearer indication of the function of the motif. The V228T mutation disrupts the hydrophobic core of HTH2 and should therefore eliminate its function; as predicted, this mutant is inactive. Instead of targeting R242 in the VirF HTH2 recognition helix, which makes contacts to DNA in both the MarA and Rob binding modes, our mutations within this helix (Y239F and I241N) target residues which make specific base contacts in the MarA binding mode but are not involved in DNA contact in the Rob binding mode. Y239F is a very conservative substitution but removes the hydroxyl group from the side chain of this residue. The hydroxyl of the equivalent T93 in MarA makes water-mediated sequence-specific hydrogen bonds with two DNA bases (19). The fact that the Y239F mutant is inactive therefore suggests that Y239 (and hence HTH2) makes base-specific contacts in VirF. In MarA, the equivalent residue to I241, T95, makes a van der Waals contact with a DNA base, so the inactivity of the I241N mutant again suggests that a specific interaction also exists in the VirF-DNA complex.
The isolation of dominant negative mutants in this study is indicative of protein-protein interactions between VirF monomers; indeed the thermally responsive AraC-like protein VirF from Y. enterocolitica (30) and AraC itself (31) form dimers in solution. Only two mutations, I180N and Y224Och, resulted in a trans-dominant phenotype; other substitutions affecting HTH1 or HTH2 had no effect on mxiC expression in the presence of wild-type VirF. It is possible that, rather than dimerizing in solution, VirF binds to its target DNA as a monomer, following which a combination of DNA-bending and protein-protein interaction leads to the formation of a multisubunit nucleoprotein complex which activates transcription. Oligomerization is suggested by in vitro footprinting studies using a VirF-MalE fusion protein which yield a large, continuous footprint (≈100 bp) at the virB promoter (28); at least a VirF tetramer would be required to give such a footprint. Oligomerization following DNA binding is supported by the data of Bourgerie et al. (2) on the melibiose-dependent regulator MelR: both a full-length MelR protein and a derivative consisting of only the C terminus could bind DNA, but only the full-length protein could oligomerize and activate transcription. A mutation such as I180N on the solvent-exposed surface of HTH1 could alter the tertiary structure of the protein-DNA complex preventing efficient protein-protein interactions when bound to the DNA, thereby accounting for the trans-dominant effect. The dominant negative phenotype of the Y224Och mutant, which presumably cannot bind DNA on its own, is more difficult to account for. One possible explanation is that while a VirF protein containing a point mutation or deletion that affects DNA binding is unable to interact with the target sequence on its own, it could form a heteromeric complex on the DNA with wild-type VirF which was already bound, thereby appearing recessive to the wild-type protein. In the case of the Y224Och mutant the gross alteration caused by deletion of the last 39 amino acids may affect the overall structure of an oligomeric VirF complex containing VirFY224Och and/or alter contacts with the transcriptional machinery, thus resulting in a trans-dominant effect. Further work is required to determine how the DNA binding and oligomerization activities of VirF interact in vivo to generate a protein-DNA complex which is competent for transcriptional activation. Our studies provide a useful starting point for further mutagenesis work to investigate the details of transcriptional regulation by VirF and its close relatives in other bacterial pathogens.
Acknowledgments
This work was supported by grant 042380/Z/94/Z from the Wellcome Trust (United Kingdom).
We thank Bernt Eric Uhlin for his gift of the anti-VirF antibodies and Andrew Free for useful discussions and comments on the manuscript.
REFERENCES
- 1.Bhende, P. M., and S. M. Egan. 1999. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J. Bacteriol. 181:5185–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourgerie, S. J., C. M. Michan, M. S. Thomas, S. J. W. Busby, and E. I. Hyde. 1997. DNA binding and DNA bending by the MelR transcription activator protein from Escherichia coli. Nucleic Acids Res. 25:1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Haan, L. A., G. A. Willshaw, B. A. van der Zeijst, and W. Gaastra. 1991. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol. Lett. 67:341–346. [DOI] [PubMed] [Google Scholar]
- 4.Dorman, C. J., and M. E. Porter. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677–684. [DOI] [PubMed] [Google Scholar]
- 5.Eustance, R. J., and R. F. Schleif. 1996. The linker region of AraC protein. J. Bacteriol. 178:7025–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillette, W. K., R. G. Martin, and J. L. Rosner. 2000. Probing the Escherichia coli transcriptional activator MarA using alanine-scanning mutagenesis: residues important for DNA binding and activation. J. Mol. Biol. 299:1245–1255. [DOI] [PubMed] [Google Scholar]
- 8.Hale, T. L. 1991. Genetic basis of virulence in Shigella species. Microbiol. Rev. 55:206–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrickson, W., and R. Schleif. 1985. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc. Natl. Acad. Sci. USA 82:3129–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424–430. [DOI] [PubMed] [Google Scholar]
- 11.Manzanera, M., S. Marques, and J. L. Ramos. 2000. Mutational analysis of the highly conserved C-terminal residues of the XylS protein, a member of the AraC family of transcriptional regulators. FEBS Lett. 476:312–317. [DOI] [PubMed] [Google Scholar]
- 12.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132–137. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Munson, G. P., L. G. Holcomb, and J. R. Scott. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niland, P., R. Hühne, and B. Müller-Hill. 1996. How AraC interacts specifically with its target DNAs. J. Mol. Biol. 264:667–674. [DOI] [PubMed] [Google Scholar]
- 16.Porter, M. E., and C. J. Dorman. 1997. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol. Gen. Genet. 256:93–103. [DOI] [PubMed] [Google Scholar]
- 17.Porter, M. E., and C. J. Dorman. 1997. Virulence gene deletion frequency is increased in Shigella flexneri following conjugation, transduction, and transformation. FEMS Microbiol. Lett. 147:163–172. [DOI] [PubMed] [Google Scholar]
- 18.Porter, M. E., S. G. J. Smith, and C. J. Dorman. 1998. Two highly related regulatory proteins, Shigella flexneri VirF and enterotoxigenic Escherichia coli Rns, have common and distinct regulatory properties. FEMS Microbiol. Lett. 162:303–309. [DOI] [PubMed] [Google Scholar]
- 19.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413–10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai, T., C. Sasakawa, and M. Yoshikawa. 1988. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton VirF protein. Mol. Microbiol. 2:589–597. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Sansonetti, P. J. 1992. Molecular and cellular biology of epithelial invasion by Shigella flexneri and other enteroinvasive pathogens, p.47–60. In C. E. Hormaeche, C. W. Penn, and C. J. Smyth (ed.), Molecular biology of bacterial infection: current status and future perspectives. Cambridge University Press, Cambridge, United Kingdom.
- 23.Savelkoul, P. H., G. A. Willshaw, M. M. McConnell, H. R. Smith, A. M. Hamers, B. A. van der Zeijst, and W. Gaastra. 1990. Expression of CFA/I fimbriae is positively regulated. Microb. Pathog. 8:91–99. [DOI] [PubMed] [Google Scholar]
- 24.Saviola, B., R. R. Seabold, and R. F. Schleif. 1998. DNA bending by AraC: a negative mutant. J. Bacteriol. 180:4227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schleif, R. 1996. Two positively regulated systems, ara and mal, p.1300–1309. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umberger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 26.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421–425. [DOI] [PubMed] [Google Scholar]
- 27.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. The 1.6 Å crystal structure of the AraC sugar-binding and dimerization domain complexed with D-fucose. J. Mol. Biol. 273:226–237. [DOI] [PubMed] [Google Scholar]
- 28.Tobe, T., M. Yoshikawa, T. Mizuno, and C. Sasakawa. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J. Bacteriol. 175:6142–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wattiau, P., and G. R. Cornelis. 1994. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 176:3878–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilcox, G., and P. Meuris. 1976. Stabilization and size of AraC protein. Mol. Gen. Genet. 145:97–100. [DOI] [PubMed] [Google Scholar]