Abstract
The steps involved in the biosynthesis of the ADP-l-glycero-β-d-manno-heptose (ADP-l-β-d-heptose) precursor of the inner core lipopolysaccharide (LPS) have not been completely elucidated. In this work, we have purified the enzymes involved in catalyzing the intermediate steps leading to the synthesis of ADP-d-β-d-heptose and have biochemically characterized the reaction products by high-performance anion-exchange chromatography. We have also constructed a deletion in a novel gene, gmhB (formerly yaeD), which results in the formation of an altered LPS core. This mutation confirms that the GmhB protein is required for the formation of ADP-d-β-d-heptose. Our results demonstrate that the synthesis of ADP-d-β-d-heptose in Escherichia coli requires three proteins, GmhA (sedoheptulose 7-phosphate isomerase), HldE (bifunctional d-β-d-heptose 7-phosphate kinase/d-β-d-heptose 1-phosphate adenylyltransferase), and GmhB (d,d-heptose 1,7-bisphosphate phosphatase), as well as ATP and the ketose phosphate precursor sedoheptulose 7-phosphate. A previously characterized epimerase, formerly named WaaD (RfaD) and now renamed HldD, completes the pathway to form the ADP-l-β-d-heptose precursor utilized in the assembly of inner core LPS.
Lipopolysaccharide (LPS) is a major component of the outer membrane of gram-negative bacteria (28). It has a tripartite structural organization consisting of lipid A, a conserved core oligosaccharide region, and an O-specific polysaccharide chain or O antigen. In the majority of gram-negative bacteria, the core oligosaccharide can be subdivided into an outer core, generally composed of hexoses and hexosamines, and an inner core made of 3-deoxy-d-manno-oct-2-ulosonic acid and l,d-heptose units. LPS plays an important role in maintaining the structural integrity of the bacterial outer membrane by interacting with outer membrane proteins and divalent cations (15), thereby providing a barrier against the entry of toxic hydrophobic compounds into the bacterial cell (27). Escherichia coli mutants defective in the biosynthesis of 3-deoxy-d-manno-oct-2-ulosonic acid are nonviable, whereas those impaired in l,d-heptose synthesis survive in vitro, although they display a pleiotropic phenotype referred to as “deep rough” (17). This phenotype is characterized by an extreme sensitivity to very low concentrations of novobiocin, detergents, and bile salts (32). Deep rough mutants also have defects in F plasmid conjugation and generalized transduction by the bacteriophage P1 (6, 16). Haemophilus influenzae heptose-deficient mutants were found to be serum sensitive and displayed a reduced virulence in vivo (18, 36).
The complete biosynthesis pathway of the l,d-heptose precursor has not been elucidated. Eidels and Osborn (11) proposed a four-step pathway for the synthesis of NDP-l,d-heptose, which is still widely accepted in the literature (see reference 13 for a review). It includes (i) conversion of d-sedoheptulose 7-phosphate to d,d-heptose 7-phosphate by a phosphoheptose isomerase; (ii) formation of d,d-heptose 1-phosphate by a phosphoheptose mutase; (iii) activation of the d,d-heptose 1-phosphate intermediate to NDP-d,d-heptose by an NDP-heptose synthetase; and (iv) epimerization of the NDP-heptose to form the final product, NDP-l,d-heptose. Subsequent studies involving the isolation of ADP-d,d-heptose and ADP-l,d-heptose from Shigella sonnei and Salmonella enterica serovar Typhimurium indicated that ADP is the activating nucleotide (20–22). In the absence of purified ADP-heptose, Kadrmas and Raetz (19) used ADP-mannose as a substrate for the E. coli heptosyltransferase I (WaaC). More recently, it has been clearly demonstrated that heptosyltransferases I and II (WaaF) from E. coli accept ADP-l-β-d-heptose and ADP-d-β-d-heptose as substrates, although the efficiency of the transfer reactions with the d-β-d isomer is markedly reduced (14, 35).
In gram-negative bacteria, functional studies have only been performed for the isomerization reaction and the epimerization step (3, 9, 26), while the conversion of d,d-heptose 7-phosphate to d,d-heptose 1-phosphate and a functional proof of the activating step have not been demonstrated. The d-sedoheptulose 7-phosphate isomerase activity was described in S. enterica serovar Typhimurium (12), and the corresponding gene, gmhA, has been cloned both from E. coli and from H. influenzae (3, 4). The amino acid sequence of the GmhA polypeptide is highly conserved in different gram-negative bacteria (33). The epimerization step is catalyzed by the WaaD (formerly RfaD) protein (5), which has also been crystallized (8). We have recently shown that the E. coli rfaE gene product consists of two distinct domains that may be involved in the biosynthesis of d,d-heptose 1-phosphate, as well as the activating step (34). It was demonstrated that one of the RfaE domains shares structural features with members of the ribokinase family, while the other domain has conserved features present in nucleotidyltransferases (34). The demonstration of a protein domain corresponding to a putative sugar kinase suggested that the original pathway for NDP-heptose biosynthesis as proposed by Eidels and Osborn may not be accurate and, at the same time, predicted the existence of an additional phosphatase step (33).
The complete biosynthesis pathway of GDP-d-α-d-heptose from d-sedoheptulose 7-phosphate in the gram-positive bacterium Aneurinibacillus thermoaerophilus DSM 10155 was recently characterized (20). We demonstrated that two independent enzymes catalyze the originally proposed mutase step. A d,d-heptose 7-phosphate kinase adds a phosphate group at the C-1 position, and subsequently a d,d-heptose 1,7-bisphosphate phosphatase removes the phosphate group at the C-7 position. The GDP-activated d,d-isomer serves as a precursor for the incorporation of the heptose into the glycan moiety of a surface layer (S-layer) glycoprotein produced by A. thermoaerophilus (20). Amino acid sequence analysis of completely sequenced genomes revealed that the A. thermoaerophilus phosphatase is highly conserved among different gram-negative bacteria (20), in agreement with a previous suggestion that a phosphatase reaction is also required for the synthesis of ADP-d,d-heptose and ADP-l,d-heptose in these microorganisms (34). In the present study, we report the reconstruction in vitro with purified enzyme components of the complete biosynthesis pathway for ADP-d-β-d-heptose in E. coli. We also provide genetic evidence demonstrating that the function of a novel phosphatase gene in E. coli K-12, now designated gmhB (formerly yaeD), is required for the synthesis of ADP-d-β-d-heptose. Furthermore, we propose a new gene nomenclature to account for the differences and similarities between the components of the pathways leading to the formation of ADP-l-β-d-heptose and GDP-d-α-d-heptose in gram-negative and gram-positive bacteria, respectively.
MATERIALS AND METHODS
Chemicals.
ATP, GTP, d-sedoheptulose 7-phosphate and dithiothreitol were obtained from Sigma (Vienna, Austria). HiTrap Chelating, HiPrep Desalting and MonoQ HR5/5 columns were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). The Gateway system was obtained from Life Technologies (Vienna, Austria). d-α-d-Heptose 1-phosphate, d-β-d-heptose 1-phosphate, ADP-d-α-d-heptose, and ADP-d-β-d-heptose were synthesized as described elsewhere (35).
Bacterial strains and growth conditions.
E. coli MG1655 (K-12 F− λ−) was purchased from the American Type Culture Collection (Manassas, Va.). E. coli DH5α [K-12 F− φ80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK−) supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF) U169] and E. coli TG1 [supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5 (rK− mK−) (F′ traD36 proAB lacI qZΔM15)] (Stratagene, La Jolla, Calif.) were used for plasmid propagation. For enzyme overexpression E. coli BL21-SI [F− ompB hsdSB(rB− mB−) gal dcm endA1 proUp::T7 RNAP::malQ-lacZ Tets] (Life Technologies) was used. E. coli KCS2926 (rfaE::Tn10) has been described elsewhere (34). Bacteria were cultured in Luria broth, supplemented when necessary with ampicillin, kanamycin, or tetracycline at final concentrations of 100, 50, and 20 μg/ml, respectively.
Analytical techniques.
Nucleotide-activated sugars and monosaccharide phosphates were analyzed by high-performance anion-exchange chromatography (HPAEC) on a CarboPac PA-1 column (Dionex, Sunnyvale, Calif.) as previously described (20). Sugar phosphates were investigated by using pulsed electrochemical detection, and UV detection at 254 nm was used for nucleotide-activated sugars. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed for proteins according to the original method of Laemmli (23) with minor modifications. Gels were stained with Coomassie brilliant blue R250. Protein concentrations were determined by the method of Bradford (2). LPS was isolated as described by Marolda et al. (24), and analyzed by Tricine-SDS-PAGE (30). Commercially cast 16% polyacrylamide gels were purchased from Novex (San Diego, Calif.), and LPS bands were visualized by silver staining (24).
Plasmid constructions.
DNA recombinant procedures were performed according to standard methods described by Sambrook et al. (29). PCR was carried out by using a PCR Sprint thermocycler (Hybaid, Ashford, United Kingdom). Oligonucleotide primers for the amplification of DNA fragments containing the E. coli heptose biosynthesis genes were designed with attB1 or attB2 sites for the insertion into the Gateway donor vector pDONR201 (Life Technologies) by homologous recombination. Primers CGBD1 (5′-attB1-GCGAGAATCTCTATTTCCAAGGAATGAAAGTAACGCTGCCAGAG-3′) and CGBD2 (5′-attB2-ATCTGTGAACCGCTTTCC-3′) were used for the amplification of the putative bifunctional kinase/adenylyltransferase gene hldE (formerly rfaE). Primers CGC1 (5′-attB1-GCGAGAATCTCTACTTCCAAGGAAAGGTGGCGAAGAGCGTACC-3′) and CGC2 (5′-attB2-GCGCATTATAGGGAGTCG-3′) were used for the amplification of the putative phosphatase gene gmhB (formerly yaeD). attB1 and attB2 are the minimal 25-bp sequences required for efficient homologous recombination. PCR products were cloned into pDONR201, and the resulting plasmids, pCHE1 and pCHB1, were used to transfer the gene sequences into pDEST17 (His fusion vector) via homologous recombination. These experiments gave rise to pCHLDE3 and pCGMHB3, encoding hldE and gmhB, respectively, which were used for overexpression of the corresponding His-tagged protein products in E. coli BL21-SI. The gmhB gene was also amplified with primer 575 (5′-TCCCCCGGGAGTGGCGAAGAGCGTACCCGCA-3′) that contains an SmaI site (underlined) and primer 558 (5′-GATCCGAATTCTGCCGGTTTTTGCTGCTT-3′). The amplified DNA fragment was digested with SmaI and cloned into the glutathione S-transferase (GST)-fusion vector pGEX-2T (Amersham Pharmacia Biotech), which was also cleaved with SmaI. The resulting plasmid, pFM4, expressed the GmhB protein N-terminally fused to the GST moiety. The A. thermoaerophilus DSM 10155 gmhB gene was amplified by using the primers ATC1F (5′-CGGAATTCATAGGAAGGCGCAAAAGG-3′) and ATC2R (5′-CGCGGATCCTTACTTTAGCTTTGCAACACC-3′), where the underlined sequences correspond to the EcoRI and BamHI restriction sites, respectively. The digested PCR fragment was ligated into EcoRI/BamHI-cut pEXT20 (10) to yield the plasmid pCAT1.
Construction of E. coli strains carrying a deletion of the gmhB gene.
The one-step replacement method described by Datsenko and Wanner (7) was used to construct a gmhB deletion in E. coli K-12 W3110. In this procedure, recombination requires the λ phage Red recombinase, which is synthesized by an inducible promoter on a curable thermosensitive plasmid, and PCR primers that provide the homology to the targeted gene. A PCR fragment was generated with pKD4 (7) as a template and the primers 342 (5′-GAGCGTACCCGCAATTTTTCTTGACCGTGATGGCACCATTAATGGTGTAGGCTGGAGCTGCTTCG-3′) and 343 (5′-CAGGCTATTTAACACCCAATCCGCCGCGTTTTCTGCTTCAGCATATGAATATCCTCCTTAG-3′). The underlined sequences correspond to 21 bases that are specific for pKD4, while the remaining sequence corresponds to the ends of gmhB. The amplified DNA fragment contained a kanamycin resistance (Kmr) gene flanked by FLP (protein encoded by the FLP gene of the 2μm plasmid of Saccharomyces cerevisiae) recognition target sites and 40 bases at each end that are specific for the ends of the gmhB gene. The PCR product was transformed into E. coli W3110 containing pKD46, which encodes the Red recombination system of the λ phage (7). One of the Kmr mutants, BD1, was isolated, and the replacement of gmhB for the Kmr gene was verified by PCR. A second derivative was obtained by inducing the excision of the antibiotic resistance gene by introducing a plasmid expressing the FLP recombinase (7). This experiment resulted in strain BD2, which carries a chromosomal unmarked deletion of gmhB.
Enzyme purification.
The growth conditions of E. coli BL21-SI cells carrying expression plasmids and the purification of proteins were essentially identical to those recently described for the purification of GmhA and GmhB (previously named GmhC) from A. thermoaerophilus DSM 10155 (20). After cell lysis and removal of the membranes by ultracentrifugation, the bifunctional kinase/pyrophosphorylase HldE (synonymous to l,d-heptose formation) was purified in two steps by using HiTrap Chelating and MonoQ columns, while the phosphatase GmhB was purified in one step by using the HiTrap chelating column. The proteins were concentrated by ultrafiltration and stored at 4°C or, after stabilization with 50% glycerol, at −20°C. The purity of the enzymes was verified by SDS-PAGE analysis.
Enzyme assays.
All assays were performed in 0.5-ml PCR tub (Axyg en Scientific, Union City, Calif.). Approximately 10 nmol of d-sedoheptulose 7-phosphate or d,d-heptose 1-phosphate was used for enzyme assays; 50 nmol of d-glucose 1,6-bisphosphate was used in a negative control to test the specificity of d,d-heptose 1,7-bisphosphate phosphatase GmhB. Equimolar amounts of ATP with respect to d-sedoheptulose 7-phosphate and d,d-heptose 1-phosphate were used in the kinase reaction, as well as in the pyrophosphorylase reaction. The assay buffer contained 20 mM Tris-HCl (pH 8.0) and 10 mM MgCl2. Enzymes (500 ng of each of the purified proteins) were added, and after incubation at 37°C for different reaction times (5 to 120 min), the samples were analyzed by HPAEC as described previously (20). The total sample volume was 10 μl for sugar phosphate analysis and 100 μl for investigation of nucleotide-activated sugars. Detailed descriptions of the reactions are given in the figure legends.
RESULTS AND DISCUSSION
Overexpression and purification of ADP-heptose biosynthesis enzymes from E. coli K-12.
In the gram-positive bacterium A. thermoaerophilus DSM 10155, the four genes encoding the enzymes involved in the biosynthesis of GDP-d-α-d-heptose are part of a single operon (20). In contrast, the proposed four biosynthesis genes for catalysis of the five reaction steps to produce nucleotide-activated l,d-heptose in E. coli K-12 are located at four different loci. The d-sedoheptulose 7-phosphate isomerase encoded by gmhA is a monocistronic gene near the proAB locus (3). The rfaE gene encoding the putative bifunctional d,d-heptose 7-phosphate kinase/d,d-heptose 1-phosphate adenylyltransferase (20), is part of an operon together with genes coding for enzymes involved in nitrogen assimilation (34). The putative d,d-heptose 1,7-bisphosphate phosphatase gene, previously referred to as gmhC in A. thermoaerophilus DSM 10155 (20) and yaeD in E. coli K-12 (1), has been identified by sequence homology comparisons and is located in E. coli next to the rrnH rRNA operon. The predicted gene products from A. thermoaerophilus and E. coli share 40% amino acid sequence identity and they are homologous to the histidinol-phosphate phosphatase family (data not shown). Finally, the ADP-l,d-heptose epimerase gene, waaD (formerly rfaD) is part of the waa gene cluster encoding enzymes for the biosynthesis of the LPS core oligosaccharide.
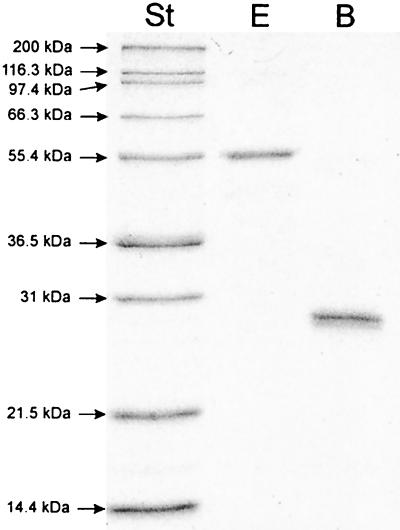
To elucidate the intermediate steps in the biosynthesis of ADP-l,d-heptose in E. coli, the enzymes putatively catalyzing steps 2 to 4 of the reaction cascade were overexpressed as histidine-tagged fusion proteins and purified as indicated in Materials and Methods. The molecular masses of the purified, denatured proteins, determined by SDS-PAGE analysis, were in agreement with the calculated molecular masses (54.6 and 24.9 kDa for RfaE and GmhB, respectively; Fig. 1). The purified proteins were utilized for functional characterization of the biosynthetic pathway of nucleotide-activated d,d-heptose in E. coli.
FIG. 1.
SDS-PAGE analysis of the purified enzymes HldE and GmhB. Lanes: lane St, molecular mass standards, myosin (200 kDa), β-galactosidase (116.3 kDa), phosphorylase b (97.4 kDa), bovine serum albumin (66.3 kDa), glutamic dehydrogenase (55.4 kDa), lactate dehydrogenase (36.5 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa); lane E, histidine-tagged d-β-d-heptose 7-phosphate kinase/d-β-d-heptose 1-phosphate adenylyltransferase HldE; lane B, histidine-tagged d,d-heptose 1,7-bisphosphate phosphatase GmhB.
In vitro synthesis of ADP-d-β-d-heptose.
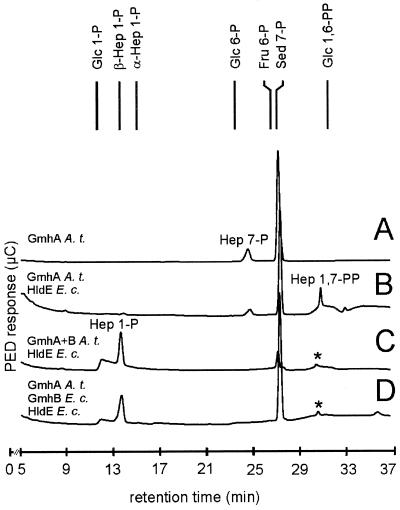
The biosynthetic steps leading to ADP-d-β-d-heptose were analyzed by HPAEC as described in an earlier study (20). Reaction mixtures were incubated at 37°C for different times and showed only marginal differences in the product yield. For practical reasons with regard to the HPAEC analysis, the duration of all reactions was standardized to 45 min. Due to the lack of commercially available d,d-heptose 7-phosphate, this sugar was prepared in situ from d-sedoheptulose 7-phosphate by using purified A. thermoaerophilus GmhA isomerase (20). Approximately 20% of d-sedoheptulose 7-phosphate was converted to d,d-heptose 7-phosphate in an equilibrium reaction (Fig. 2A). To analyze the second step, d-sedoheptulose 7-phosphate was incubated with A. thermoaerophilus GmhA, the putative bifunctional kinase-pyrophosphorylase RfaE from E. coli K-12, and ATP. Figure 2B shows that the peaks representing d-sedoheptulose 7-phosphate and d,d-heptose 7-phosphate decreased, while a new peak appeared at a retention time of 30.7 min. As a control for enzyme specificity, d-sedoheptulose 7-phosphate was incubated with only the RfaE protein and ATP, resulting in no change of the peak pattern (data not shown). Peaks corresponding to sugar bisphosphates are very small because the signal height corresponding to the free hydroxyl groups that are electrochemically detected by HPAEC decreases with a decreasing number of free OH groups. Synthetic d-β-d-heptose 1-phosphate eluted before synthetic d-α-d-heptose 1-phosphate, and the same was true for the corresponding d,d-heptose 1,7-bisphosphate anomers (20). Thus, the 30.7-min retention time observed for the heptose 1,7-bisphosphate product obtained with the A. thermoaerophilus GmhA and the E. coli RfaE was in agreement with d-β-d-heptose 1,7-bisphosphate. Since the retention time of the d-α-d-heptose 1,7-bisphosphate synthesized with GmhA and the kinase enzyme from A. thermoaerophilus was 31.2 min (20), we concluded that the anomeric configuration of the final product, d-β-d-heptose 1,7-bisphosphate or d-α-d-heptose 1,7-bisphosphate, is specified by the kinase reaction step. This conclusion is also in agreement with the lack of amino acid sequence homologies between the kinase domain of RfaE and the A. thermoaerophilus kinase enzyme (data not shown), suggesting that these two enzymes belong to different protein families.
FIG. 2.
HPAEC analysis of the enzymatic synthesis of d-β-d-heptose 1-phosphate by pulsed electrochemical detection. (A) d-Sedoheptulose 7-phosphate, converted with the isomerase GmhA from A. thermoaerophilus DSM 10155. (B) d-Sedoheptulose 7-phosphate, converted with GmhA from A. thermoaerophilus DSM 10155, the kinase/pyrophosphorylase HldE from E. coli K-12 and ATP. (C) d-Sedoheptulose 7-phosphate, converted with GmhA and the phosphatase GmhB from A. thermoaerophilus DSM 10155, HldE from E. coli K-12 and ATP. (D) d-Sedoheptulose 7-phosphate, converted with GmhA from A. thermoaerophilus DSM 10155, HldE and GmhB from E. coli K-12 and ATP. The peaks labeled with an asterisk are by-products not representing d-β-d-heptose 1,7-bisphosphate (for more details, see reference 20).
The third step of the reaction was proposed to be the removal of the phosphate group at the C-7 position by a phosphatase. This was first investigated by using the isomerase GmhA and the phosphatase GmhB from A. thermoaerophilus in combination with the bifunctional E. coli RfaE enzyme and ATP. The peaks corresponding to d-sedoheptulose 7-phosphate, d,d-heptose 7-phosphate and d,d-heptose 1,7-bisphosphate either decreased or disappeared completely, and a new peak was detected, displaying the same retention time as synthetic d-β-d-heptose 1-phosphate (13.6 min, Fig. 2C). Since ATP was present in equimolar amounts with respect to d-sedoheptulose 7-phosphate, minor conversion of d-β-d-heptose 1-phosphate to ADP-d-β-d-heptose can be expected. However, ADP-d-β-d-heptose shows no signal with pulsed amperometric detection used for detection of sugar phosphate. Another reaction, where d-sedoheptulose 7-phosphate was incubated with the isomerase GmhA from A. thermoaerophilus and the E. coli RfaE and GmhB proteins in the presence of ATP, resulted in the same change in the peak pattern as in the reaction with the phosphatase from A. thermoaerophilus (Fig. 2D). Again, the d,d-heptose 1-phosphate was in the β-anomeric configuration. A negative control was performed by using α-d-glucose 1,6-bisphosphate for the phosphatase reaction, showing that this sugar bisphosphate was not a substrate for the phosphatase from E. coli K-12 (data not shown). We concluded from these experiments that the phosphatases from A. thermoaerophilus and E. coli K-12 converted the d-β-d-heptose 1,7-bisphosphate intermediate into d-β-d-heptose 1-phosphate, indicating that these enzymes are insensitive to the anomeric configuration of the d,d-heptose 1,7-bisphosphate.
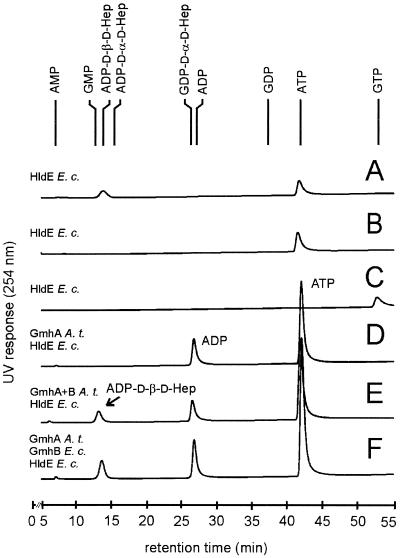
The next reaction step in the biosynthetic pathway leading to ADP-l-β-d-heptose is nucleotide activation. Synthetic d-β-d-heptose 1-phosphate was incubated with the RfaE protein and ATP, resulting in a peak comigrating with synthetic ADP-d-β-d-heptose (14.2 min, Fig. 3A). In contrast, no activation was detected when the same reaction was performed with either d-α-d-heptose 1-phosphate (Fig. 3B) or d-β-d-heptose 1-phosphate and GTP (Fig. 3C). Furthermore, other combinations of d,d-heptose 1-phosphates and standard nucleoside triphosphates different from ATP did not result in activation (data not shown). To determine whether the phosphatase activity is essential for nucleotide activation, d-sedoheptulose 7-phosphate was incubated with GmhA from A. thermoaerophilus and the E. coli RfaE in the presence of ATP. The nucleotide-HPAEC results did not show a new peak in the range of ADP-activated sugars (Fig. 3D). The ADP peak in the chromatogram results from the phosphate transfer, catalyzed by the kinase. ADP-d-β-d-heptose was also synthesized directly from d-sedoheptulose 7-phosphate involving the three enzymes analogous to the reactions described in Fig. 2C and 2D. ADP-d-β-d-heptose could be detected by using the GmhB phosphatase from either A. thermoaerophilus DSM 10155 (Fig. 3E) or E. coli K-12 (Fig. 3F).
FIG. 3.
HPAEC analysis of the activation of d,d-heptose using UV detection. (A) d-β-d-Heptose 1-phosphate, converted with the kinase/pyrophosphorylase HldE from E. coli K-12 and ATP. (B) d-α-d-Heptose 1-phosphate, converted with HldE and ATP. (C) d-β-d-Heptose 1-phosphate, converted with HldE from E. coli K-12 and GTP. (D) d-Sedoheptulose 7-phosphate, converted with the isomerase GmhA from A. thermoaerophilus DSM 10155, HldE from E. coli K-12, and ATP. (E) d-Sedoheptulose 7-phosphate, converted with GmhA and the phosphatase GmhB from A. thermoaerophilus DSM 10155, HldE from E. coli K-12, and ATP. (F) d-Sedoheptulose 7-phosphate, converted with GmhA from A. thermoaerophilus DSM 10155, HldE and GmhB from E. coli K-12, and ATP.
A deletion of the E. coli K-12 gmhB gene is associated with an altered LPS core phenotype.
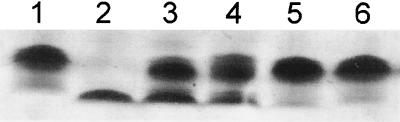
Based on the biochemical data obtained from the in vitro synthesis of ADP-d-β-d-heptose, it was predicted that a mutation in the E. coli gmhB gene would express a heptoseless LPS core. To verify this hypothesis, we constructed derivatives from the wild-type E. coli K-12 strain W3110 carrying a deleted gmhB gene. In one of these strains, BD1, the deletion was marked with a Kmr gene cassette, while in the other strain, BD2, the Kmr gene was excised from the chromosome resulting in an unmarked gmhB deletion. The deletion of gmhB in both mutants was confirmed by PCR analysis and Southern blot hybridization (data not shown). LPS extracted from strains W3110, BD1, and BD2 was examined by Tricine-SDS-PAGE and silver staining. LPS from strains BD1 and BD2 (Fig. 4, lanes 3 and 4) displayed a fast-migrating band that is absent from the W3110 LPS sample (Fig. 4, lane 1). The fast-migrating band coincided in molecular mass with that of the heptoseless LPS formed by the hldE::Tn10 mutant KCS2926 (Fig. 4, lane 2) (34). The results suggest that the deletion of gmhB caused a partial defect in the synthesis of the LPS core, resulting in the formation of heptoseless and heptose-rich forms. This phenotype could be corrected by the introduction of plasmids pCAT1 and pFM4 (Fig. 4, lanes 5 and 6, respectively), which encode cloned gmhB genes from A. thermoaerophilus and E. coli K-12, respectively. The genetic complementation experiment is in agreement with the biochemical function proposed for GmhB. However, the lack of a complete heptoseless phenotype suggests the existence of another function in E. coli K-12 that can at least partially compensate for the synthesis of a complete LPS core oligosaccharide. One of the homologs of the GmhB protein is the bifunctional protein HisB that carries a histidinol-phosphate phosphatase activity. Construction of a gmhB deletion in E. coli SØ874, which carries a deletion of the his operon (25), showed the same LPS phenotype as in strains BD1 and BD2. Therefore, we postulate that an additional phosphatase activity distinct from HisB, may exist in E. coli K-12, which could partially compensate for the gmhB deletion.
FIG. 4.
LPS analysis of E. coli K-12 W3110 and the gmhB deletion mutant. LPS was extracted and processed from the following E. coli K-12 strains: lane 1, W3110 (wild-type); lane 2, KCS2926 (rfaE:: Tn10); lane 3, BD1 (ΔgmhB:: Km); lane 4, BD2 (ΔgmhB); lane 5, BD2(pCAT1); and lane 6, BD2(pFM4). Plasmids pCAT1 and pFM4 encode gmhB genes from A. thermoaerophilus DSM 10155 and E. coli K-12, respectively.
Concluding remarks and proposed new nomenclature for genes involved in nucleotide-activated heptose synthesis.
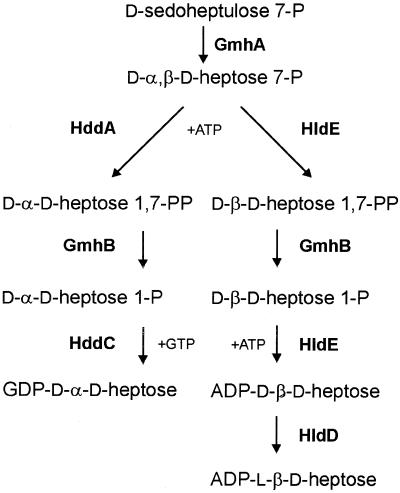
To our knowledge, this is the first report of the elucidation of the complete pathway for ADP-d-β-d-heptose biosynthesis in E. coli (Fig. 5), which involves kinase and phosphatase intermediary steps, as it was shown for the synthesis of GDP-d-α-d-heptose in A. thermoaerophilus (20). The phosphatase step, mediated by the gmhB gene product, is clearly essential for the nucleotide activation reaction, since treatment of d,d-heptose 7-phosphate with only the bifunctional enzyme and ATP did not yield ADP-activated sugar products. When the phosphatase was present in the system, the product resulting from the nucleotide transfer reaction comigrated with synthetic ADP-d-β-d-heptose. Despite efforts by several investigators in the past, the presence of the gmhB phosphatase gene in E. coli could not be detected by mutagenesis followed by the screening for novobiocin-sensitive mutants, which usually denotes the presence of a heptoseless LPS core. This may be explained in part by our finding that the deletion of gmhB did not confer a complete heptoseless LPS core phenotype, suggesting the presence of another yet-unidentified phosphatase activity that may partially compensate for the synthesis of a complete core. Current efforts are dedicated to the isolation and characterization of this gene and its product.
FIG. 5.
Comparison of the biosynthetic pathways for GDP-d-α-d-heptose in the gram-positive bacterium A. thermoaerophilus DSM 10155 (left) and ADP-l-β-d-heptose in E. coli K-12 and other gram-negative bacteria (right). GmhA, sedoheptulose 7-phosphate isomerase; GmhB, d-α,β-d-heptose 1,7-bisphosphate phosphatase; HddA, d-α-d-heptose 7-phosphate kinase; HddC, d-α-d-heptose 1-phosphate guanylyltransferase; HldA, d-β-d-heptose 7-phosphate kinase; HldE, bifunctional d-β-d-heptose 7-phosphate kinase/d-β-d-heptose 1-phosphate adenylyltransferase; HldD, ADP-d-β-d-heptose epimerase.
The pathways described in this work and also in a recent study (20) differ greatly from the majority of the classical pathways leading to the formation of nucleotide-activated sugars, which usually involve a mutase step catalyzing the intramolecular transfer of a phosphate group from the distal carbon to the C-1 position (11, 13). This phosphate subsequently reacts with NTP, resulting in the synthesis of an NDP-sugar precursor. In the majority of gram-negative bacteria examined to date, the genes encoding the enzymes for the synthesis of ADP-l-β-d-heptose are scattered throughout the genome. However, in Campylobacter jejuni NCTC 11168 and Helicobacter pylori strains 26695 and J99, these genes are found in discrete clusters. In the Neisseria meningitidis strains MC58 and Z2491, two separate genes encode each of the domains of the bifunctional RfaE enzyme (data not shown), and the gmhB phosphatase gene homolog has recently been identified, showing a distinct heptoseless phenotype, which suggests that there are no complementing phosphatase activities present in this organism (31). The presence of a cluster of genes in the gram-positive bacterium A. thermoaerophilus DSM 10155 encoding all of the enzymes for the biosynthesis of GDP-d-α-d-heptose greatly facilitated the assignment of the enzymes involved in this novel kinase-phosphatase cascade (20). The results of the present study in E. coli and our previous findings with A. thermoaerophilus (20) highlight not only similarities but also very important differences between the d,d-heptose and the l,d-heptose synthesis pathways (Fig. 5): (i) the kinase enzyme in each pathway has specificity for the anomeric form of d,d-heptose 7-phosphate, resulting in d-α-d-heptose 1,7-bisphosphate in the A. thermoaerophilus pathway and d-β-d-heptose 1,7-bisphosphate in E. coli; (ii) the phosphatase activity in both pathways is independent of the anomeric configuration of the bisphosphate sugar; (iii) the nucleotidylyltransferases in both pathways belong to two completely different protein families; and (iv) no epimerization step is required prior to the transfer of d-α-d-heptose onto the S-layer protein in A. thermoaerophilus. The β-anomeric configuration of the ADP-heptose is also in perfect agreement with the results of Zamyatina et al. (35) and Gronow et al. (14), indicating that heptosyltransferases from E. coli only process the β-anomers of ADP-d,d-heptose and ADP-l,d-heptose.
The elucidation of the pathway of nucleotide-activated d,d-heptose and l,d-heptose requires a reassessment of the gene nomenclature to develop a consistent nomenclature scheme, which takes into account the similarities and the differences among the two heptose pathways, and at the same time follows the general principles for bacterial polysaccharide gene nomenclature as outlined in the Bacterial Polysaccharides Genes Database (http://www.microbio.usyd.edu.au/BPGD/default.htm). We propose that the previous nomenclature of gmh (for glycero-mannose-heptose synthesis) should be maintained to design the genes gmhA (sedoheptulose 7-phosphate isomerase) and gmhB (d-α,β-d-heptose 1,7-bisphosphate phosphatase), which are common to both pathways (Fig. 5). The new nomenclature hdd (for d-α-d-heptose synthesis) is proposed for the genes hddA (d-α-d-heptose 7-phosphate kinase) and hddC (d-α-d-heptose 1-phosphate guanylyltransferase) of the gram-positive bacterium A. thermoaerophilus DSM 10155 and their homologs (Fig. 5, left). The nomenclature hld (for l-β-d-heptose synthesis) is proposed for the genes hldE (bifunctional d-β-d-heptose 7-phosphate kinase/d-β-d-heptose 1-phosphate adenylyltransferase, formerly rfaE) and hldD (ADP-d-β-d-heptose epimerase, formerly waaD or rfaD). In addition, in cases where the bifunctional d-β-d-heptose 7-phosphate kinase/d-β-d-heptose 1-phosphate adenylyltransferase is encoded by separate genes, such as in N. meningitidis and Ralstonia eutropha, they could be named hldA (d-β-d-heptose 7-phosphate kinase) and hldC (d-β-d-heptose 1-phosphate adenylyltransferase, formerly aut). This new nomenclature will permit a rational grouping of the homologs of all of these genes into gene and protein families and will facilitate comparative studies on the evolution of these genes, as well as future structure-function characterizations of the enzymes. This is especially important taking into account that the elucidation of the complete pathway for ADP-l,d-heptose biosynthesis and the characterization of each of these enzymes pave the way for the development of novel enzyme inhibitors with potential antimicrobial activity for the control of infections by gram-negative bacteria.
Acknowledgments
We thank Sonja Zayni and Andrea Scheberl for excellent technical assistance and Tracey Hunt for a critical reading of the manuscript. We also thank Peter Reeves for useful discussions regarding the heptose pathway gene nomenclature.
The construction of mutants BD1 and BD2 was part of an undergraduate research project conducted by Beth Dunn. This work was supported by the Austrian Science Fund (projects P12966-MOB and P14209-MOB to P.M.) and by the Natural Science and Engineering Research Council of Canada (to M.A.V).
REFERENCES
- 1.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 3.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608–3614. [DOI] [PubMed] [Google Scholar]
- 4.Brooke, J. S., and M. A. Valvano. 1996. Molecular cloning of the Haemophilus influenzae gmhA (lpcA) gene encoding a phosphoheptose isomerase required for lipooligosaccharide biosynthesis. J. Bacteriol. 178:3339–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, W. G., Jr. 1983. The rfaD gene codes for ADP-l-glycero-d-mannoheptose-6-epimerase. An enzyme required for lipopolysaccharide core biosynthesis. J. Biol. Chem. 258:1985–1990. [PubMed] [Google Scholar]
- 6.Curtiss, R., III, L. J. Charamella, D. R. Stallions, and J. A. Mays. 1968. Parental functions during conjugation in Escherichia coli K-12. Bacteriol. Rev. 32:320–348. [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deacon, A. M., Y. S. Ni, W. G. Coleman, Jr., and S. E. Ealick. 2000. The crystal structure of ADP-l-glycero-d-mannoheptose 6-epimerase: catalysis with a twist. Struct. Fold. Des. 8:453–462. [DOI] [PubMed] [Google Scholar]
- 9.Ding, L., B. L. Seto, S. A. Ahmed, and W. G. Coleman, Jr. 1994. Purification and properties of the Escherichia coli K-12 NAD-dependent nucleotide diphosphosugar epimerase, ADP-l-glycero-d-mannoheptose 6-epimerase. J. Biol. Chem. 269:24384–24390. [PubMed] [Google Scholar]
- 10.Dykxhoorn, D. M., R. St. Pierre, and T. Linn. 1996. A set of compatible tac promoter expression vectors. Gene 177:133–136. [DOI] [PubMed] [Google Scholar]
- 11.Eidels, L., and M. J. Osborn. 1971. Lipopolysaccharide and aldoheptose biosynthesis in transketolase mutants of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 68:1673–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eidels, L., and M. J. Osborn. 1974. Phosphoheptose isomerase, first enzyme in the biosynthesis of aldoheptose in Salmonella typhimurium. J. Biol. Chem. 249:5642–5648. [PubMed] [Google Scholar]
- 13.Gronow, S., and H. Brade. 2001. Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? J. Endotoxin Res. 7:3–23. [PubMed] [Google Scholar]
- 14.Gronow, S., C. Oertelt, E. Ervelä, A. Zamyatina, P. Kosma, M. Skurnik, and O. Holst. 2001. Characterization of the physiological substrate for lipopolysaccharide heptosyltransferases I and II. J. Endotoxin Res. 7:263–270. [PubMed] [Google Scholar]
- 15.Hancock, R. E. W., D. N. Karunaratne, and C. Bernegger-Egli. 1994. Molecular organization and structural role of outer membrane macromolecules, p.263–279. In J.-M. Ghuysen and R. Hackenbeck (ed.), Bacterial cell wall. Elsevier, New York, N.Y.
- 16.Havekes, L. M., B. J. J. Lugtenberg, and W. P. M. Hoekstra. 1976. Conjugation-deficient E. coli K-12 F− mutants with heptose-less lipopolysaccharide. Mol. Gen. Genet. 146:43–50. [DOI] [PubMed] [Google Scholar]
- 17.Heinrichs, D. E., M. A. Valvano, and C. Whitfield. 1999. Biosynthesis and genetics of lipopolysaccharide core, p.305–330. In H. Brade, D. C. Morrison, S. Vogel, and S. Opal (ed.), Endotoxin in health and disease. Marcel Dekker, New York, N.Y.
- 18.Helander, I. M., B. Lindner, H. Brade, K. Altmann, A. A. Lindberg, E. T. Rietschel, and U. Zähringer. 1988. Chemical structure of the lipopolysaccharide of Haemophilus influenzae I-69 Rd−/B+: description of a novel deep-rough chemotype. Eur. J. Biochem. 177:483–492. [DOI] [PubMed] [Google Scholar]
- 19.Kadrmas, J. L., and C. R. H. Raetz. 1998. Enzymatic synthesis of lipopolysaccharide in Escherichia coli: purification and properties of heptosyltransferase I. J. Biol. Chem. 273:2799–2807. [DOI] [PubMed] [Google Scholar]
- 20.Kneidinger, B., M. Graninger, M. Puchberger, P. Kosma, and P. Messner. 2001. Biosynthesis of nucleotide-activated d-glycero-d-manno-heptose. J. Biol. Chem. 276:20935–20944. [DOI] [PubMed] [Google Scholar]
- 21.Kocsis, B., and T. Kontrohr. 1984. Isolation of adenosine 5′-diphosphate-l-glycero-d-mannoheptose, the assumed substrate of heptose transferase(s), from Salmonella minnesota and Shigella sonnei Re mutants. J. Biol. Chem. 259:11858–11860. [PubMed] [Google Scholar]
- 22.Kontrohr, T., and B. Kocsis. 1981. Isolation of adenosine 5′-diphosphate-d-glycero-d-mannoheptose. An intermediate in lipopolysaccharide biosynthesis of Shigella sonnei mutants. J. Biol. Chem. 256:7715–7718. [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 24.Marolda, C. L., J. Welsh, L. Dafoe, and M. A. Valvano. 1990. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 172:3590–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuhard, J., and E. Thomassen. 1976. Altered deoxyribonucleotide pools in P2 eductants of Escherichia coli K-12 due to deletion of the dcd gene. J. Bacteriol. 126:999–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni, Y., P. McPhie, A. Deacon, S. Ealick, and W. G. Coleman, Jr. 2001. Evidence that NADP+ is the physiological cofactor of ADP-l-glycero-d-mannoheptose 6-epimerase. J. Biol. Chem. 276:27329–27334. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raetz, C. R. H. 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles, p.1035–1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1, 2nd ed. American Society for Microbiology Press, Washington, D.C.
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379. [DOI] [PubMed] [Google Scholar]
- 31.Shih, G. C., C. M. Kahler, R. I. W. Carlson, M. M. Rahman, and D. S. Stephens. 2001. gmhX, a novel gene required for the incorporation of l-glycero-d-manno-heptose into lipooligosaccharide in Neisseria meningitidis. Microbiology 147:2367–2377. [DOI] [PubMed] [Google Scholar]
- 32.Tamaki, S., T. Sato, and M. Matsuhashi. 1971. Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K-12. J. Bacteriol. 105:968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valvano, M. A. 1999. Biosynthesis and genetics of ADP-heptose. J. Endotoxin Res. 5:90–95. [Google Scholar]
- 34.Valvano, M. A., C. L. Marolda, M. Bittner, M. Glaskin-Clay, T. L. Simon, and J. D. Klena. 2000. The rfaE gene from Escherichia coli encodes a bifunctional protein involved in biosynthesis of the lipopolysaccharide core precursor ADP-l-glycero-d-manno-heptose. J. Bacteriol. 182:488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamyatina, A., S. Gronow, C. Oertelt, M. Puchberger, H. Brade, and P. Kosma. 2000. Efficient chemical synthesis of the two anomers of ADP-l-glycero- and d-glycero-d-manno-heptopyranose allows the determination of substrate specificities of bacterial heptosyltransferases. Angew. Chem. Int. Ed. 39:4150–4153. [DOI] [PubMed] [Google Scholar]
- 36.Zwahlen, A., L. G. Rubin, C. J. Connelly, T. J. Inzana, and E. R. Moxon. 1985. Alteration of the cell wall of Haemophilus influenzae type b by transformation with cloned DNA: association with attenuated virulence. J. Infect. Dis. 152:485–492. [DOI] [PubMed] [Google Scholar]