Abstract
Mutants of Salmonella enterica lacking the RecBC function are avirulent in mice and unable to grow inside macrophages (N. A. Buchmeier, C. J. Lipps, M. Y. H. So, and F. Heffron, Mol. Microbiol. 7:933–936, 1993). The virulence-related defects of RecBC− mutants are not suppressed by sbcB and sbcCD mutations, indicating that activation of the RecF recombination pathway cannot replace the virulence-related function(s) of RecBCD. Functions of the RecF pathway such as RecJ and RecF are not required for virulence. Since the RecBCD pathway, but not the RecF pathway, is known to participate in the repair of double-strand breaks produced during DNA replication, we propose that systemic infection by S. enterica may require RecBCD-mediated recombinational repair to prime DNA replication inside phagocytes. Mutants lacking both RecD and RecJ are also attenuated in mice and are unable to proliferate in macrophages, suggesting that exonucleases V and IX provide alternative functions for RecBCD-mediated recombinational repair during Salmonella infection.
Salmonella enterica is a gram-negative bacterial pathogen that causes gastrointestinal disorders and systemic infections in humans and livestock animals (29). A hallmark of Salmonella pathogenesis is the capacity of the bacterium to survive and proliferate within phagocytic cells (29). In vitro and in vivo studies have provided evidence that bacterial growth inside macrophages and neutrophils is required for systemic infection (3, 8, 18, 25, 31). Phagocytic cells synthesize DNA-damaging agents such as nitric oxide and oxygen radicals (37) as mammalian defense mechanisms against pathogens; hence, Salmonella must face the attack of compounds that challenge genome integrity (21, 33, 37). Active functions and mechanisms that protect Salmonella from oxidative stress inside phagocytes have been described recently (2, 7, 38). In addition, almost a decade ago Buchmeier et al. showed that mutants of S. enterica lacking RecA or RecBC functions are avirulent in the murine typhoid model and highly sensitive to oxidative compounds synthesized by macrophages, suggesting that recombination is required for the repair of oxidative DNA damage (4). This view is supported by the recent report that Escherichia coli uses recombinational repair for survival of exposure to nitric oxide (35).
Like E. coli and other bacterial species, S. enterica can perform recombination between homologous DNA molecules by means of two main processes or pathways, RecBCD and RecF (20). Each pathway has distinct DNA substrate preferences and requires a specific set of recombination functions (20). In this study, we have examined the ability of S. enterica recombination mutants to cause systemic infection in mice and to proliferate inside macrophages. The strains used lack recombination functions of the RecBCD pathway and/or the RecF pathway. Our data indicate that the RecBCD enzyme is the key function required for both systemic infection and growth inside macrophages. Suppressor mutations that restore recombination proficiency in RecBC− mutants by activating the RecF pathway do not restore virulence, indicating that a process different from standard recombination is affected. Because the RecF pathway cannot functionally replace RecBCD-mediated recombination for the repair of double-strand DNA breaks (15, 36), the inability of RecBC− SbcB− and RecBC− SbcB− SbcCD− mutants of S. enterica to grow inside macrophages may result from arrested chromosome replication upon DNA damage. We also show that the functions of the RecF pathway are not required for virulence; however, mutants lacking both RecD and RecJ are avirulent, suggesting that exonuclease IX (RecJ) can substitute for the exonuclease activity of RecBCD.
Tests for virulence of S. enterica serovar Typhimurium recombination mutants in BALB/c mice.
To identify recombination functions required for virulence in the murine typhoid model, BALB/c mice were challenged with recombination-deficient strains of S. enterica serovar Typhimurium (Table 1). In parallel, infections were performed with the virulent strain SV1445, a His+ derivative of the mouse virulent strain SL1344 (12). The insertion zeb-6312::Tn10dTc carried by strains SV4189 and SV4190 is a Tn10dTc insertion linked to the sbcB locus and has no effect on virulence (data not shown). The 50% lethal doses (LD50) shown in Table 2 support the following conclusions. (i) Among the single mutants, the most attenuated is the RecBC− mutant, which has an oral LD50 more than 104-fold higher than that of the parental strain. Actually, the RecBC− mutant was unable to cause the death of any animal, even at the higher doses used (7.2 × 109 CFU). We also observed that strain SV4191 (recA1) was able to kill mice at doses 10 to 100 times lower than those of SV4188 (recB497::MudJ) (data not shown). The difference observed between the RecA− and RecBC− mutants may reflect the pleiotropy of recBC mutations; i.e., they impair growth in standard culture media and cause various kinds of DNA repair defects (1, 16, 19, 22). RecF−, RecJ−, and RecF− RecJ− strains were all virulent, suggesting that, aside from the RecA function, functions of the RecF pathway of recombination are not required for systemic infection. LD50 trials by intraperitoneal inoculation confirmed that RecA− and RecBC− strains are the only avirulent strains among the single mutants tested (data not shown).
TABLE 1.
Strain lista
| Strain | Genotype |
|---|---|
| SV1445 | Wild type |
| SV4188 | recB497::MudJ |
| SV4189 | recB497::MudJ sbcB21 zeb-6312::Tn10dTc |
| SV4190 | recB497::MudJ sbcB21 sbcCD51 zeb-6312::Tn10dTc |
| SV4191 | recA1 |
| SV4212 | recD543::Tn10dCm |
| SV4213 | recF522::Tn5 |
| SV4192 | recJ504::MudJ |
| SV4363 | recD543::Tn10dCm recF522::Tn5 |
| SV4364 | recD543::Tn10dCm recJ504::MudJ |
| SV4496 | recA1 recF522::Tn5 |
| SV4497 | recA1 recJ504::MudJ |
| SV4498 | recB503::Tn10 recF522::Tn5 |
| SV4499 | recF522::Tn5 recJ504::MudCm |
All the recombination mutations used were null and had been isolated in strain LT2. The origin of each mutant allele is as follows: recA1 and recF522::Tn5, J. R. Roth, Department of Biology, University of Utah, Salt Lake City, Utah; sbcB21, L. Bossi, Centre de Génétique Moléculaire, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France; recB497::MudJ, reference 22; recD543::Tn10dCm, reference 27; recJ504::MudJ and recJ504::MudCm, reference 23; sbcCD51, laboratory collection. Transfer of the mutations to the mouse-virulent strain SV1445 was performed by transductional crosses with P22 HT (34), followed by lysogen disposal on green plates (5). The recombination-deficient phenotypes of the newly constructed strains were checked using tests described elsewhere (11, 22, 23, 26, 27).
TABLE 2.
Virulence of S. enterica recombination mutants in BALB/c mice
| Strain | Relevant genotype | LD50 (oral)a |
|---|---|---|
| SV1445 | Wild type | 1.0 × 105 |
| SV4191 | recA1 | <3.0 × 108 |
| SV4188 | recB497::MudJ | >7.2 × 109 |
| SV4212 | recD543::Tn10dCm | <7.6 × 106 |
| SV4213 | recF522::Tn5 | <7.4 × 106 |
| SV4192 | recJ504::MudJ | <2.0 × 106 |
| SV4496 | recA1 recF522::Tn5 | <6.5 × 108 |
| SV4497 | recA1 recJ504::MudJ | <3.5 × 108 |
| SV4498 | recB503::Tn10 recF522::Tn5 | >5.0 × 109 |
| SV4499 | recF522::Tn5 recJ504::MudCm | <8.7 × 105 |
| SV4363 | recD543::Tn10dCm recF522::Tn5 | <6.9 × 106 |
| SV4364 | recD543::Tn10dCm recJ504::MudJ | >2.6 × 108 |
Bacterial suspensions were prepared as described elsewhere (9). The acidic pH of the stomach was buffered by suspending bacteria in a 2.5% bicarbonate-0.2% lactose solution prior to administration. Survival of bacterium-infected mice was recorded for a minimum of 4 weeks. The LD50 was calculated by the method of Reed and Muench (30).
(ii) RecA− RecF− and RecA− RecJ− mutants were attenuated at levels similar to that of a RecA− strain, confirming that RecF and RecJ functions are not required for virulence, regardless of the presence of RecA.
(iii) A RecBC− RecF− mutant showed attenuation at a level similar to that of a RecBC− strain, thereby suggesting that the only recombination pathway required for systemic infection is that of RecBCD. A corollary is that the RecF pathway cannot provide the recombination functions required for virulence, not even when the RecBCD pathway is absent. LD50 trials with RecBC− RecJ− mutants were not feasible because of their low viability (11).
(iv) Unexpectedly, a RecD− RecJ− mutant turned out to be avirulent. This result suggests that either exonuclease V or IX is required for systemic infection and hence that RecJ can participate in RecBCD-mediated recombinational repair in the absence of the RecD subunit. Previous observations have suggested that RecD and RecJ provide alternative functions for recombinational repair of DNA damage in both Salmonella (11, 23, 27) and E. coli (39). Function redundancy may thus explain why RecD− and RecJ− single mutants are virulent (Table 2; see also reference 40).
Effect of suppressors of recB and recC mutations on the virulence of S. enterica.
Mutations that suppress the recombination defect of RecBC− mutants without restoring the activities of the RecB and RecBC enzymes have been characterized for both E. coli and Salmonella (1, 16, 19). In E. coli, sbc mutations are known to activate the RecF and RecE pathways of recombination (19, 20). The prophage-associated recE gene (13) does not exist in Salmonella. Mutations in the sbcB locus of S. enterica suppress the recombination deficiency and the UV sensitivity of RecBC− mutants by activating the RecF pathway (1). If combined with mutations in another locus, sbcCD, suppression of mitomycin C sensitivity is also observed (1). RecBC− SbcB− SbcCD− mutants also regain the ability to grow normally in standard media (1). With these facts in mind, we examined the ability of sbc mutations to suppress the virulence defect of RecBC− mutants of S. enterica. Both RecBC− SbcB− and RecBC− SbcB− SbcCD− strains remained avirulent by the oral and intraperitonal routes (Table 3). The avirulence of the RecBC− SbcB− SbcCD− mutant is specially significant, because these strains are recombination proficient in transductional tests, are resistant to both UV and mitomycin C, and do not exhibit growth defects (data not shown; see also reference 1). The inability of sbc suppressors to restore virulence in RecBC− mutants confirms the idea that the role of RecBC in Salmonella virulence cannot be performed by functions of the RecF pathway. The hypothetical nature of this role is discussed below.
TABLE 3.
Effect of sbc suppressor mutations on Salmonella virulence in BALB/c mice
| Strain | Relevant genotype | LD50 (oral)a | LD50 (intraperitoneal)a |
|---|---|---|---|
| SV1445 | Wild type | 1.0 × 105 | <50 |
| SV4188 | recB497::MudJ | >7.2 × 109 | >8.0 × 103 |
| SV4189 | recB497::MudJ sbcB21 | >1.5 × 108 | NTb |
| SV4190 | recB497::MudJ sbcB21 sbcCD51 | >1.4 × 109 | >2.9 × 103 |
See footnote a of Table 2. Buffering of bacterial suspensions was omitted for intraperitoneal inoculation.
NT, not tested.
Ability of recombination mutants of S. enterica to proliferate in macrophages.
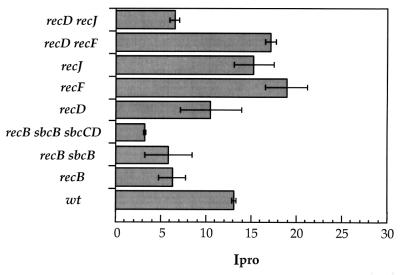
Growth of Salmonella inside phagocytes is a strict requirement for systemic infection (8). Furthermore, a correlation between the avirulence of S. enterica RecA− and RecBC− mutants and their impaired ability to repair DNA was established in the seminal study of Buchmeier et al.: RecA− and RecBC− mutants can grow within macrophages that do not synthesize reactive oxygen compounds (4). As expected, we also found a straight correlation between the virulence of S. enterica recombination mutants and their ability to proliferate in macrophages. Unlike the virulent recombination mutants, those with virulence defects when tested in mice (RecA−, RecBC−, RecD− RecJ−, RecBC− SbcB−, and RecBC− SbcB− SbcCD−) also showed defects for intracellular proliferation within J774.A1 mouse macrophages (Fig. 1). A noteworthy observation is, however, that sbc suppressor mutations were unable to restore intracellular proliferation within macrophages. Since sbc mutations restore recombination proficiency by activating pathways other than RecBCD, our observations indicate that S. enterica requires recombinational repair via the RecBCD pathway to grow inside phagocytes. Because of the unique ability of the RecBCD pathway to repair double-strand DNA breaks (15, 17, 36), it seems logical to conclude that S. enterica relies on the RecBCD recombination pathway to repair this kind of DNA lesion inside macrophages. In fact, a recent study has shown the occurrence of double-strand breaks in bacteria growing inside J774.A1 macrophages (33).
FIG. 1.
Ability of recombination mutants of S. enterica to proliferate in J774.A1 macrophages. Growth, infection, and lysis of J774.A1 mouse macrophages were carried out as described elsewhere (9, 10). The bacterial strains used for infection were SV1445, SV1488, SV4189, SV4190, SV4212, SV4213, SV4192, SV4363, and SV4364. The intracellular proliferation rate (Ipro) was calculated as the ratio of the number of viable intracellular bacteria present at 24 h to the number present at 2 h postinfection. Data are means ± standard deviations of results from two representative experiments. The assay was repeated three times for each strain. wt, wild type.
What is the role of the RecBCD pathway in Salmonella virulence?
A tight association between DNA replication and recombination in phage T4 has long been known (28). In the last 5 years, the idea that DNA replication requires recombinational repair has been extended to both bacteria (14, 32) and Saccharomyces cerevisiae (24). Whenever a double-strand DNA break is produced, recombinational repair via the RecBCD pathway is required to prime DNA synthesis and hence for the resumption of chromosome replication (17, 36). Our observation that the RecBC enzyme is required for growth inside macrophages correlates with the extreme sensitivity of RecBCD− mutants to NO (35) and with the occurrence of double-strand breaks among the DNA lesions found in E. coli cells grown inside macrophages (33). Since the RecF pathway cannot functionally replace RecBCD-mediated recombination for the repair of double-strand DNA breaks, sbcB and sbcCD suppressor mutations do not restore recombinational repair (17). Hence, the inability of RecBC− SbcB− and RecBC− SbcB− SbcCD− mutants of S. enterica to grow inside macrophages may result from arrested chromosome replication upon DNA damage.
Our finding that RecD− RecJ− mutants are avirulent and unable to grow inside macrophages seems to indicate that, in the absence of the RecD exonuclease, recombinational repair via the RecBCD pathway can be carried out with the participation of RecJ. This situation is similar to that of Rep− mutants of E. coli, which require either RecD or RecJ for double-strand break repair (36). Furthermore, there is evidence that RecJ is involved in the recovery of replication forks upon DNA damage (6) and in other RecBC-dependent recombination processes (27, 39).
In summary, we propose that S. enterica uses the RecBCD recombination pathway to repair DNA double-strand breaks produced during growth inside macrophages and that RecD and RecJ provide alternative exonuclease activities for the repair process. The absence of RecBC or both RecD and RecJ renders Salmonella avirulent. A tentative explanation, supported by abundant literature (reviewed in reference 15), may be that Salmonella needs RecBC-mediated DNA repair to prime DNA replication inside phagocytes.
Acknowledgments
This work was supported by grants from the Dirección General de Enseñanza Superior of the Government of Spain (PM97-0148-CO2), the European Union (QLK2-1999-00310), and the Comunidad de Madrid (08.2/0045.1/2000). M.G.P. is supported by a postdoctoral fellowship from the Comunidad de Madrid.
We thank Nello Bossi and John Roth for providing strains, Mike Mahan for discussions on recombination, and Andrés Aguilera for critical reading of the manuscript.
REFERENCES
- 1.Benson, N. R., and J. Roth. 1994. Suppressors of recB mutations in Salmonella typhimurium. Genetics 138:11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchmeier, N., S. Bossie, C. Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmeier, N. A., and F. Heffron. 1991. Inhibition of phagosome-lysosome fusion by Salmonella typhimurium. Infect. Immun. 59:2232–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., C. J. Lipps, M. Y. H. So, and F. Heffron. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7:933–936. [DOI] [PubMed] [Google Scholar]
- 5.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline by phage P22 in Salmonella typhimurium. II. Properties of a high frequency transducing lysate. Virology 50:883–898. [DOI] [PubMed] [Google Scholar]
- 6.Courcelle, J., and P. C. Hanawalt. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated E. coli. Mol. Gen. Genet. 262:543–551. [DOI] [PubMed] [Google Scholar]
- 7.DeGroote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields, P. I., R. W. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-del Portillo, F., M. Puciarelli, and J. Casadesús. 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion and M cell cytotoxicity. Proc. Natl. Acad. Sci. USA 96:11578–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-del Portillo, F., M. B. Zwick, K. Y. Leung, and B. B. Finlay. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. USA 90:10544–10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garzón, A., C. R. Beuzón, M. J. Mahan, and J. Casadesús. 1996. recB recJ mutants of Salmonella typhimurium are deficient in transductional recombination, DNA repair and plasmid maintenance. Mol. Gen. Genet. 270:570–580. [DOI] [PubMed] [Google Scholar]
- 12.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are nonvirulent and effective as live vaccines. Nature 291:238–239. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser, K., and N. E. Murray. 1979. Physical characterization of the “Rac prophage” in E. coli K-12. Mol. Gen. Genet. 175:159–174. [DOI] [PubMed] [Google Scholar]
- 14.Kogoma, T., G. W. Caldwell, K. G. Barnard, and T. Asai. 1996. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J. Bacteriol. 178:1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156–165. [DOI] [PubMed] [Google Scholar]
- 16.Kushner, S. R., H. Nagaishi, and A. J. Clark. 1972. Indirect suppression of recB recC mutations by exonuclease I deficiency. Proc. Natl. Acad. Sci. USA 69:1366–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzminov, A., and F. W. Stahl. 1999. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 13:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung, K. Y., and B. B. Finlay. 1991. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 88:11470–11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd, R. G., and K. B. Low. 1996. Homologous recombination, p.2236–2255. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 21.MacMicking, J., Q. W. Xie, and C. Nathan. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323–350. [DOI] [PubMed] [Google Scholar]
- 22.Mahan, M. J., and J. R. Roth. 1989. The recB and recC genes of Salmonella typhimurium. J. Bacteriol. 171:612–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahan, M. J., J. Casadesús, and J. R. Roth. 1992. The Salmonella typhimurium RecJ function permits growth of P22 abc phage on recBCD+ hosts. Mol. Gen. Genet. 232:470–478. [DOI] [PubMed] [Google Scholar]
- 24.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui, H., M. Eguchi, and Y. Kikuchi. 2000. Use of confocal microscopy to detect Salmonella typhimurium within host cells associated with Spv-mediated intracellular proliferation. Microb. Pathog. 29:53–59. [DOI] [PubMed] [Google Scholar]
- 26.Miesel, L., and J. R. Roth. 1994. Salmonella typhimurium recD mutations increase recombination in a short-sequence transduction assay. J. Bacteriol. 176:4092–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miesel, L., and J. R. Roth. 1996. Evidence that SbcB and RecF pathway functions contribute to RecBCD-dependent transductional recombination. J. Bacteriol. 178:3146–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosig, G. 1998. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 32:379–413. [DOI] [PubMed] [Google Scholar]
- 29.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259–274. [DOI] [PubMed] [Google Scholar]
- 30.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497. [Google Scholar]
- 31.Richter-Dahlfors, A., A. M. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandler, S. J., H. S. Samra, and A. J. Clark. 1996. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA and dnaC. Genetics 143:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlosser-Silverman, E., M. Elgrably-Weiss, I. Rosenshine, R. Kohen, and S. Altuvia. 2000. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 182:5225–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transducing abilities. Mol. Gen. Genet. 119:75–88. [DOI] [PubMed] [Google Scholar]
- 35.Spek, E. J., T. L. Wright, M. S. Stitt, N. R. Taghizadeh, S. R. Tannenbaum, M. G. Marinus, and B. P. Engelward. 2001. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J. Bacteriol. 183:131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uzest, M., S. D. Ehrlich, and B. Michel. 1995. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol. Microbiol. 17:1177–1188. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez-Torres, A., and F. C. Fang. 2001. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 9:29–33. [DOI] [PubMed] [Google Scholar]
- 38.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655–1658. [DOI] [PubMed] [Google Scholar]
- 39.Viswanathan, M., and S. T. Lovett. 1998. Single-strand DNA-specific exonucleases in E. coli: roles in repair and mutation avoidance. Genetics 149:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zahrt, T. C., N. Buchmeier, and S. Maloy. 1999. Effect of mutS and recD mutations on Salmonella virulence. Infect. Immun. 67:6168–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]