Abstract
Trichloroethylene (TCE) is the most frequently detected groundwater contaminant, and 1-naphthol is an important chemical manufacturing intermediate. Directed evolution was used to increase the activity of toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4 for both chlorinated ethenes and naphthalene oxidation. When expressed in Escherichia coli, the variant TOM-Green degraded TCE (2.5 ± 0.3 versus 1.39 ± 0.05 nmol/min/mg of protein), 1,1-dichloroethylene, and trans-dichloroethylene more rapidly. Whole cells expressing TOM-Green synthesized 1-naphthol at a rate that was six times faster than that mediated by the wild-type enzyme at a concentration of 0.1 mM (0.19 ± 0.03 versus 0.029 ± 0.004 nmol/min/mg of protein), whereas at 5 mM, the mutant enzyme was active (0.07 ± 0.03 nmol/min/mg of protein) in contrast to the wild-type enzyme, which had no detectable activity. The regiospecificity of TOM-Green was unchanged, with greater than 97% 1-naphthol formed. The beneficial mutation of TOM-Green is the substitution of valine to alanine in position 106 of the α-subunit of the hydroxylase, which appears to act as a smaller “gate” to the diiron active center. This hypothesis was supported by the ability of E. coli expressing TOM-Green to oxidize the three-ring compounds, phenanthrene, fluorene, and anthracene faster than the wild-type enzyme. These results show clearly that random, in vitro protein engineering can be used to improve a large multisubunit protein for multiple functions, including environmental restoration and green chemistry.
Biological catalysts have become attractive for synthetic reactions because of their high efficiency and selectivity, their ability to produce relatively pure compounds compared with racemic mixtures, and their ability to produce regio-, chemo-, and stereospecific compounds (13, 17, 24). The greatest strength of biocatalysis is performing reactions under mild conditions with nontoxic reagents (3). An example of a transformation which may be beneficial to conduct biologically is the conversion of naphthalene to naphthols; naphthols are used widely in the manufacturing of many herbicides, insecticides, drugs, and dyes (43). Current methods of manufacturing naphthols require acids, bases, and metal catalysts (43), none of which is environmentally friendly, cheap, or disposable. With 1.5 × 104 tons per year of 1-naphthol made in the United States (43), the use of biocatalytic methods to manufacture naphthols could dramatically decrease waste.
Although enzymes usually have good turnover numbers, the productivity per unit mass is sometimes low (3). To generate novel enzymes, the DNA-shuffling method of Stemmer (42) has proven to be the most powerful. DNA shuffling randomly mutates gene sequences and recombines mutations within genes while also removing undesired mutations (42).
Improving biocatalysts for bioremediation is also advantageous, and directed evolution has been used previously to enhance polychlorinated biphenyl degradation by improving the large subunit of the biphenyl dioxygenase (20) and arsenate detoxification (three beneficial mutations found in the efflux transport protein ArsB [8]). We have sought to improve enzymes for compounds like trichloroethylene (TCE) because it is a suspected carcinogen and one of the most common groundwater pollutant at hazardous waste sites (23). Because of its toxicity, it is regulated under the Safe Drinking Water Act to a maximum contaminant level of 5 ppb.
In the environment, TCE is degraded anaerobically via reductive dehalogenation to the less chlorinated ethenes trans-1,2-dichloroethylene (trans-DCE), cis-1.2-dichloroethylene (cis-DCE), 1,1-dichloroethylene (1,1-DCE), vinyl chloride (VC), ethene, and ethane (36). However, the dehalogenation of TCE is often incomplete, yielding mostly cis-DCE and VC. Since VC is a known human carcinogen (23), and both VC and cis-DCE are U.S. Environmental Protection Agency priority pollutants (2), establishing an effective aerobic treatment strategy which does not produce toxic intermediates is critical.
The biocatalyst toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4 is encoded by the 4.6-kb operon tomA012345 (37) and is a three-component enzyme consisting of a 211-kDa hydroxylase (from tomA1A3A4) with two catalytic oxygen-bridged binuclear iron centers, a 40-kDa NADH-oxidoreductase (from tomA5), and a 10.4-kDa protein (from tomA2) involved in electron transfer between the hydroxylase and reductase (27). TOM has 65% overall DNA identity to the toluene/benzene-2-monooxygenase of Pseudomonas sp. strain JS150 (19) and 54% overall DNA identity to phenol hydroxylase of Pseudomonas CF600 (28).
TOM has evolved to convert toluene in a two-step process into methylcatechol (27). Fortuitously, TOM also oxidizes naphthalene, TCE, all three dichloroethylenes, and vinyl chloride (37, 39). The ability of TOM to mineralize TCE primarily to CO2 and Cl− in vivo has made this enzyme a useful biocatalyst for bioremediation (22, 26). Recent studies also indicate that strains expressing TOM can degrade mixtures of chlorinated aliphatics (41). Unfortunately, because the breakdown of chlorinated aliphatics yields no carbon or energy for the cell and because the breakdown intermediates, such as TCE epoxide, are toxic (44), TOM will never evolve naturally to breakdown these compounds faster.
The aim of this study was to evolve TOM using in vitro techniques and to isolate variants of TOM with improved degradation of chlorinated compounds and improved oxidation of naphthalene. This work is the first protein engineering of TOM and the first application of directed evolution to this class of nonheme O2-dependent diiron enzymes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain TG1 (15) was routinely cultivated at 37°C in Luria-Bertani (LB) (34) medium, and antibiotics were added to maintain plasmids as appropriate (ampicillin at 100 μg/ml and kanamycin at 100 μg/ml). Experiments were conducted using exponential-phase cultures obtained by diluting overnight cells to an optical density (OD) at 600 nm of 0.05 to 0.15 and growing to an OD of 0.5 to 1.0. The exponentially grown cells were washed either once for the naphthol experiments with 1 volume 50 mM Tris-HCl, pH 8.0 (to remove metabolic by-products), or three times for the TCE experiments with 1 volume of 50 mM Tris-HNO3, pH 7.0 (to remove all traces of chloride and metabolic by-products). The maximum specific growth rates of the mutants and the wild-type TOM host with the plasmids were determined using LB with 100 μg of kanamycin per ml.
Protein analysis and molecular techniques.
The Total Protein Kit (Sigma Chemical Co., St. Louis, Mo.) was used to determine the total cellular protein for calculation of whole-cell specific activities, and cellular protein samples were analyzed on standard 12% Laemmli (21) discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gels as previously described (18). Plasmid DNA was isolated using a Midi Kit (Qiagen, Inc., Chatsworth, Calif.), and DNA fragments were isolated from agarose gels using the GeneClean III Kit (Bio 101, Vista, Calif.). E. coli strains were electroporated using a Bio-Rad GenePulser/Pulse Controller (Hercules, Calif.) at 15 kV/cm, 25 μF, and 200 Ω.
Construction of pBS(Kan)TOM.
To stably and constitutively express the B. cepacia G4 gene tomA012345, the expression vector pBS(Kan)TOM was constructed by removing the kanamycin resistance cartridge from pKG1022 (14) by digestion with KpnI and BamHI, blunt-ending the fragments using T4 DNA polymerase, and then blunt-ligating into the ScaI site in the middle of the ampicillin resistance gene of pBluescript II KS− (Stratagene, La Jolla, Calif.) to create pBS(Kan). The tomA012345 genes were obtained from the plasmid pMS64 (38) using PCR with a mixture of Taq and Pfu polymerases (1:1) and the front primer (5′-TGGCACGGGAATTCCTTCGGAATAT-3′), which generates an EcoRI site, and rear primer 1 (5′-TGTCCATCACTTCTAGACTCGCAT G-3′), which generates an XbaI site. The PCR product was cloned into the multiple cloning site in pBS(Kan) after double digestion with EcoRI and XbaI to create pBS(Kan)TOM. In pBS(Kan)TOM, the lac promoter yields constitutive expression of TOM due to high copy number of the plasmid. Expression of wild-type TOM from pBS(Kan)TOM within E. coli strains produced brown-colored cells on agar plates and in broth cultures.
DNA shuffling of TOM.
DNA shuffling was performed using the procedure of Stemmer (42) modified by Zhao and Arnold (47), with DNA errors introduced primarily during the shuffling reaction. To isolate template DNA to be shuffled, PCR was performed on 0.5 μg of pBS(Kan)TOM in a 100-μl reaction containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 vol% dimethyl sulfoxide (DMSO), 2 mM MgCl2, 200 μM each of the deoxynucleoside triphosphates (dNTPs), 5 U of Taq polymerase (Promega), and 0.3 μM each of the front primer (see above) and rear primer 2 (5′-TGAAAACCATGGGCTGGTCGGCTG-3′). Amplification was carried out in the GeneAmp PCR system 2400 (Perkin Elmer, Norwalk, Conn.).
Fragments for shuffling were created by digesting the cleaned PCR product with DNase I in a 50-μl reaction containing 3 to 5 μg of DNA, 50 mM Tris-Cl, pH 7.4, 10 mM MnCl2, and 0.01 U of DNase I for 20 min at 25°C. The fragments of 20 to 50 bp were purified using Centri-Sep spin columns (Princeton Separations, Adelphia, N.J.). The fragments were reassembled by PCR without primers in a 50 μl reaction containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 10 vol % DMSO, 2 mM MgCl2, 200 μM each dNTP, 25 μl of Centri-Sep DNA fragments, and 2.5 U of Pfu polymerase (Stratagene, La Jolla, Calif.). The 4.3-kb TOM fragment was recovered by PCR with the front primer and rear primer 2 in a 100-μl reaction containing 1 to 3 μl of reassembled DNA along with a 1:1 ratio of Taq and Pfu polymerases (2.5 U each). The PCR product generated was then cloned into the plasmid pBS(Kan)TOM, replacing the 3.5-kb region between the natural AvrII and PpuMI sites of the wild-type TOM with shuffled DNA; this effectively replaced 57% of tomA1, all of tomA2A3A4, and 56% of tomA5. This shuffled TOM plasmid library was electroporated into E. coli TG1.
Screening for naphthol formation.
Synthesis of naphthol from naphthalene using whole cells was adapted for the 96-well plate format by modifying the assay of Phelps et al. (29). Colored colonies containing putative mutated TOM enzymes were grown in 300 μl of LB containing 100 μg/ml kanamycin at 37°C with shaking in Costar 96-well plates (Corning, Corning, N.Y.). The cells were harvested at mid-log phase by filtering 200 μl of the cell cultures using MultiScreen-GV 96-well filter plates (Millipore, Bedford, Mass.). The collected cells were washed with 200 μl 50 mM Tris-Cl, pH 7.4, then suspended in 200 μl of the same buffer.
Cell suspensions in the filter plates were contacted with naphthalene (99.8%; Fisher Chemical Co., Fair Lawn, N.J.) vapors in an airtight chamber (23 by 20 by 23 cm) overnight with shaking. The naphthol products were filtered into a new Costar 96-well plate (Corning). The naphthols in 200 μl of supernatant were reacted with 10 μl of 1% (wt) tetrazotized o-dianisidine (Sigma Chemical Co.) for 15 s with mixing, followed by 40 μl of glacial acetic acid (color stabilizer), forming purple diazo dyes that were spectrophotometrically detected at 540 nm using a Multiscan RC plate reader (Labsystems, Helsinki, Finland).
Extents and initial rates of naphthol synthesis and extents of polyaromatic hydrocarbon hydroxylation.
To determine the total naphthols synthesized by each strain, 10 ml of washed cells (OD of 1.0) were contacted with either 5 mM or 0.1 mM naphthalene (dissolved in dimethyl formamide [DMF]) in a 60-ml serum vial sealed with a Teflon-coated septum and aluminum crimp seal. The inverted vials were shaken at 37°C for 24 h at 300 rpm on an IKA-Vibrax-VXR shaker (IKA-Works, Inc., Cincinnati, Ohio). The naphthols synthesized were detected by reacting 800 μl of cell-free supernatant with 40 μl of 1 wt% tetrazotized o-dianisidine and 160 μl of glacial acetic acid.
The purple azo dye product was detected at an OD of 528 nm with a DU640 spectrophotometer (Beckman, Fullerton, Calif.). The molar amount of naphthols synthesized was calculated by comparison to a 1-naphthol standard curve (molar extinction coefficient of 8,500 M−1 cm−1). This was done at least three times for each strain tested. To determine the rates of naphthol synthesis, three 25-ml serum vials per strain, each containing 5 ml of washed whole cells adjusted to an OD of 5.0 and 0.1 mM naphthalene (dissolved in DMF), were used. A vial was sacrificed at 0, 15, and 30 min, and the amount of naphthols produced was quantified by reaction with tetrazotized o-dianisidine as done for the determination of naphthol total yields.
The extents of phenanthrene, fluorene, and anthracene (98%; Acros Organics) hydroxylation were determined at 0.1 mM and 0.2 mM (using 100 mM DMF stock solutions) with 10 ml of washed cells (OD of 5.0) contacted in 60-ml vials sealed with Teflon-coated septa and aluminum crimp seals. After 20 and 44 h of shaking at 37°C at 300 rpm on an IKA-Vibrax-VXR shaker, the amount of hydroxylated polycyclic aromatic hydrocarbons was determined using the same o-dianisidine-based assay used for naphthol detection.
HPLC product analysis.
To generate naphthols for analysis, 500 ml of log-phase TOM-expressing cells were harvested, washed, and suspended in 5 ml of 50 mM Tris-HCl, pH 7.4, and then contacted with naphthalene (25 mM overall) overnight with shaking at 37°C. The naphthols synthesized were extracted with 2 ml of chloroform, proteins were precipitated with ethanol, and products were redissolved in 1 ml of methanol. The naphthols were identified and quantitated using high-pressure liquid chromatography (HPLC) with photodiode array detection using Millenium32 Chromatography Manager software (Waters Corp., Milford, Mass.). A gradient system from 1:4 methanol-water to 100% methanol over 40 min on a Novapak C18 column (Waters Corp.) was used to separate the naphthol products. The products generated were compared to the standards 1-naphthol, 2-naphthol, 1,5-dihydroxynaphthol, 2,7-dihydroxynaphthol, 1,3-dihydroxynaphthol, and 2,3-dihydroxynaphthol (Sigma Chemical Co.).
Screening for TCE mineralization.
The inorganic chloride generated from the mineralization of TCE by whole cells was measured spectrophotometrically by adapting the procedure of Bergmann and Sanik (33) for 96-well plates. Colored colonies containing putative mutated TOM enzymes were grown in 270 μl of LB chloride-free medium (LB with 0.171 M NaNO3 and no NaCl) along with 100 μg/ml kanamycin with shaking at 37°C in Costar 96-well plates (Corning). The cells were harvested at mid-log phase by filtering all of the cell cultures using MultiScreen-GV 96-well filter plates (Millipore). The collected cells were washed three times with 200 μl of 50 mM Tris-HNO3, pH 7.0, and then suspended in 340 μl of the same buffer.
Cell suspensions (260 μl) were then transferred to a Costar 96-well plate and contacted with TCE vapors (800 μM) in an airtight chamber (23 by 20 by 23 cm) for 16 to 21 h with shaking. The entire cell suspension was transferred to another MultiScreen-GV 96-well plate (Millipore), and the supernatant was filtered into a new Costar 96-well plate. The chloride ions generated from the TCE mineralization were detected by adding 46 μl of 0.25 M Fe(NH4)(SO4)2 · 12H2O in 9 M HNO3 and 46 μl of saturated Hg(SCN)2 in 95% ethanol to the 260 μl of supernatant in each well of the 96-well plate. After 5 min, the absorbance of the Fe(SCN)2+ product was measured at 450 nm using a Multiscan RC plate reader.
Rates of degradation and extents of mineralization of chlorinated ethenes and toluene.
Ten milliliters of washed cell suspension adjusted to an OD of 5.0 were added to 60-ml glass serum vials which were sealed, and the cells were contacted with the chlorinated compounds (200 μM) or toluene (50 μM) at 37°C and 300 rpm. The headspace concentrations were determined every 5 min for the first 2 h as described previously using gas chromatography (41). At least two independent experiments for each chlorinated compound were analyzed.
The supernatant chloride ion concentrations generated from the mineralization of the chlorinated aliphatic compounds were measured after specific times of incubation (TCE, 1,1-DCE, and VC, 40 min; cis-DCE, 4.5 h; and trans-DCE, 3.5 h). After contact, the TOM activity was quenched by heating the vials in boiling water for 90 s. The chloride ion concentrations in 500 μl of supernatant were measured spectrophotometrically as indicated above (the minimum detectable chloride concentration with this method was 8 μM). The relatively stable cis-DCE-epoxide (44) was allowed to break down for 24 h before the chloride was measured.
DNA sequencing.
A dye terminator cycle sequencing protocol based on the dideoxy method of sequencing DNA developed by Sanger et al. (35) was used to sequence TOM DNA. Twenty-two primers of 18 to 22 bp in length were generated from the wild-type TOM sequence (U.S. patent 5,543,317) (37) for sequencing both strands of TOM. Sequence data generated were analyzed using the BioEdit sequencing alignment editor (16).
RESULTS AND DISCUSSION
Two goals of our laboratory have been to use the biocatalyst TOM for green chemistry as well as to harness it for rhizoremediation in a system using poplar trees and genetically engineered microorganisms. We have successfully obtained competitive bacteria which express TOM in the rhizosphere by engineering poplar-colonizing microorganisms and coating them onto poplar roots (40). To enhance this application, DNA shuffling was used to increase the oxidation activities of TOM toward chlorinated aliphatics (a related enzyme, toluene o-xylene monooxygenase of Pseudomonas stutzeri OX1, was found by us to attack tetrachloroethylene [33], so shuffling may be used to enhance these enzymes for all chlorinated aliphatics).
Shuffled TOM library construction.
Construction of a small stable vector constitutively expressing TOM, pBS(Kan)TOM, was vital for creating large libraries via electroporation and reliably screening them for improved TOM function; hence, the ampicillin resistance marker was replaced with one for kanamycin resistance to circumvent the initial plasmid segregational instability (β-lactamase is periplasmic and frequently leaks, whereas aminoglycoside 3′-phosphotransferase II is cytosolic [45]) and to avoid feeder colonies.
Starting with a low-fidelity (Taq) PCR fragment of TOM as the template, mutations in TOM were created using the random combinatorial method of DNA shuffling (42). By cloning the shuffled TOM DNA back into pBS(Kan)TOM (in place of the wild-type TOM sequence), a library of 26,705 mutants was generated. The mutant library contained a variety of different colored colonies in various shades of brown (wild-type color), green, blue, and white. The color of the colonies is presumably formed by various hydroxylation products of indole (formed as a breakdown product of tryptophan in E. coli) that spontaneously oxidize into colored compounds such as indigo, indirubin, and isatin (10, 12).
Screening for improvements in naphthol production and TCE mineralization.
The TOM shuffled library was screened using whole cells in a 96-well plate format as described in Materials and Methods for both naphthol synthesis from naphthalene and chloride ion production from the mineralization of TCE. After screening a few thousand white colonies from our shuffled TOM library, it was found that these colonies were devoid of any TOM activity (83% of the colonies were white). The white colonies were then screened visually and eliminated from further screening in 96-well plates. All white colonies checked by a plasmid DNA miniprep were found to still contain the plasmid pBS(Kan)TOM. After screening colored colonies via 96-well plate assays, many mutants were found that had increased naphthol synthesis and chloride production. All up-mutants were initially confirmed in triplicate via 96-well plates.
Characterization of best mutants.
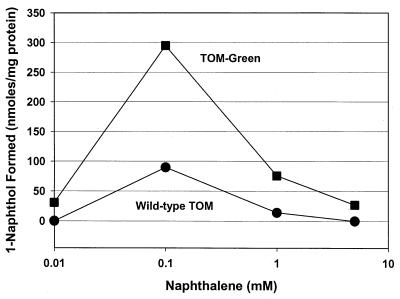
The best mutants initially identified by screening for naphthol synthesis in 96-well plates were further examined in 60-ml vials for naphthol production as a function of naphthalene concentration; the best mutant, TOM-Green (named for the fact that it turns green in LB medium, probably as a result of indigo and isatin formation), was better at 1-naphthol production after 16 h than the wild-type enzyme at all concentrations (Fig. 1). After longer contact (24 h), TOM-Green formed 7.6-fold more naphthol than the wild-type enzyme at the optimal naphthalene concentration of 0.1 mM naphthalene (270 ± 6 versus 36 ± 8 nmol of naphthol/mg of protein) and infinitely more naphthol at 5.0 mM (since the wild-type enzyme had no activity, 94 ± 42 versus 0 nmol of naphthol/mg of protein). Using whole cells contacted with 0.1 mM naphthalene, the naphthol production results were corroborated by determining the initial rates of naphthol synthesis. Cells expressing the mutant TOM-Green enzyme synthesized naphthol 6.4-fold faster than the wild-type TOM-containing strain at 0.1 mM (0.19 ± 0.03 nmol/min/mg of protein versus 0.029 ± 0.004 nmol/min/mg of protein).
FIG. 1.
Effect of naphthalene concentration on enhanced enzyme activity as expressed in TG1. TOM-Green (squares) and wild-type TOM (circles) production of naphthol after 16 h of exposure.
The naphthol products of TOM-Green and wild-type TOM were determined by HPLC analysis to be greater than 97% 1-naphthol, with 2-naphthol as a minor product (Table 1); therefore, DNA shuffling did not significantly alter the regiospecific site of oxidation on naphthalene. These strains containing TOM could be very useful for biocatalytic synthesis of 1-naphthol because of their high yields and their ability to form such a pure product (decreasing the need to perform costly purification).
TABLE 1.
HPLC analysis of naphthol productsa
| Enzyme | % Oxidized naphthalene
|
||
|---|---|---|---|
| 1-Naphthol | 2-Naphthol | Unknown | |
| Wild-type TOM | 98 | 0 | 2 |
| TOM-Green | 97 | 1 | 2 |
| Toluene-o-xylene monooxygenaseb | 74 | 25 | 1 |
| Toluene-3-monooxygenasec | 48 | 49 | 3 |
All plasmids were expressed in E. coli TG1.
From pBZ1260, encoding the touABCDEF gene cluster of Pseudomonas stutzeri OX1 (6).
From pRO1966, encoding tbuA1UBVA2C gene cluster of Pseudomonas pickettii PKO1 (4).
As with 1-naphthol synthesis, from the library of 26,705 mutants, it was found that the best variant for TCE mineralization was TOM-Green. A more detailed analysis was then conducted with this mutant to determine its initial degradation rates on an entire series of chlorinated compounds (Table 2) using gas chromatography. Note that these experiments were conducted so that the substrate was not completely degraded and thus differences between the amounts of chloride generated by TOM-Green and the wild-type enzyme could be discerned. The TCE initial degradation rate was 2-fold faster than that of the wild-type TOM-containing strain, and the degradation rates of both 1,1-DCE and trans-DCE were also increased significantly (Table 2). The degradation rates of cis-DCE and VC were the same as with the wild-type enzyme. These enhanced degradation rates of the chlorinated ethenes by TOM-Green were corroborated by the larger chloride ion concentrations generated from their mineralization (Table 2): 2.8-fold more for TCE, 1.2-fold more for 1,1-DCE, and 1.6-fold more for trans-DCE.
TABLE 2.
Chlorinated ethene degradation and chloride produced by TG1 cells expressing wild-type TOM and the mutant TOM-Greena
| Compound | Initial liquid concn (μM) | Initial degradation rate (nmol/min/mg of protein) | Chloride produced (μM) | ||
|---|---|---|---|---|---|
| Wild-type TOM | TOM- Green | Wild-type TOM | TOM- Green | ||
| TCE | 67 | 1.39 ± 0.05 | 2.5 ± 0.3 | 131 ± 19 | 366 ± 48 |
| 1,1-DCE | 25 | 0.42 ± 0.07 | 1.0 ± 0.2 | 61 ± 7 | 74 ± 1 |
| cis-DCE | 79 | 2.97 | 3.08 | 110 ± 6 | 107 ± 9 |
| trans-DCE | 86 | 1.13 ± 0.06 | 1.60 ± 0.2 | 56.9 ± 0.4 | 92 ± 5 |
| VC | 34 | 2.06 | 2.08 | 86.2 ± 0.6 | 99 ± 4 |
| Toluene | 22 | 0.64 ± 0.04 | 0.44 ± 0.08 | N/Ab | N/A |
Chloride concentrations were determined by quenching reactions so that roughly 50% of each chlorinated ethene was oxidized by TOM-Green. Initial liquid concentrations indicated were calculated based on Henry’s law (2, 25) (200 μM added if all the volatile organic was in the liquid phase). The mean ± 1 standard deviation is shown.
N/A, not applicable.
To confirm that the mutants were not improved because of some artifact of growth or change in expression, growth rates, toluene degradation, and total cellular protein profiles via SDS-PAGE were measured. All of the mutants grew at the same rate as the wild-type TOM-containing strain (1.56 ± 0.05/h in LB) and displayed the same high degree of plasmid stability. The mutant TOM-Green actually had only 70% of the toluene-degrading activity of the wild type (initial degradation rate of 0.44 ± 0.08 nmol/min/mg of protein versus 0.64 ± 0.04 nmol/min/mg of protein at 50 μM [Table 2]). If the increases in chlorinated aliphatic degradation and naphthol synthesis were caused by expression differences, then toluene degradation would also have increased proportionally; instead, there was less activity toward the natural substrate as the enzyme was evolved. Comparison of total cell protein extracts via SDS-PAGE also did not reveal any significant changes in protein expression levels between the mutants and the wild type (data not shown).
To explore the activity of the mutant TOM-Green enzyme further, the oxidation of larger polyaromatic compounds was investigated. TOM-Green was found to have enhanced activity relative to the wild-type enzyme on three-ring compounds, with phenanthrene oxidized 2.1 ± 0.1 times faster at 100 μM and 3.1 ± 0.6 times faster at 200 μM, fluorene oxidized 2.7 ± 0.2 times faster at 100 μM and 2.5 ± 0.2 times faster at 200 μM, and anthracene oxidized 1.3 ± 0.1 times faster at 100 μM and 1.8 ± 0.6 times faster at 200 μM.
Sequence changes.
Using primers generated from the TOM sequence published in U.S. patent 5,543,317 (37), the entire wild-type TOM locus from the plasmid pBS(Kan)TOM, the mutant TOM-Green, and other putative mutants were sequenced. Our wild-type TOM sequence in pBS(Kan)TOM (GenBank accession number AF349675, generated using high-fidelity PCR), which was confirmed by sequencing at least twice in each direction and by sequencing four putative up-mutants in both directions, compared well to the revised TOM sequence in GenBank (AF319657), which differs substantially from that in the patent (37). The only coding difference found for the wild-type TOM used here relative to AF319657 lies in tomA3 (D14N), where a single nucleotide change was found so that our wild-type TOM codes for Asn (AAT) whereas the previously reported sequence codes for Asp (GAT). After sequencing this region of pMS64, it was found that D14N in pBS(Kan)TOM was a result of cloning TOM from pMS64 into pBS(Kan).
The sequence of the mutant TOM-Green had 4 bp changes relative to our wild-type TOM sequence (one silent mutation in tomA1 and two silent mutations and one amino acid change in tomA3), which led to only one amino acid change in the α-subunit of the hydroxylase (valine 106 to alanine 106, t2077c based on AF319657). The wild-type TOM and the TOM-Green mutant sequence retained all the key α-hydroxylase residues of the similar Methylococcus capsulatus (Bath) soluble methane monooxygenase (sMMO) genes, including the diiron ligands (E114, E144, H147, E209, E243, and H246 of sMMO), the catalytic residues (T213 and N214 of sMMO) involved in proton delivery to the active site, the structural residues (D143, R146, S238, D242, and R245 of sMMO) involved in hydrogen bonding between the C and F helices, and the protein docking residues (Y292, W371, and Y376 of sMMO) (7).
The V106A mutation lies in helix B (based on the sMMO crystal structure [11, 32]) of the hydroxylase α-subunit. Interestingly, this amino acid is always a hydrophobic amino acid in all other related nonheme monooxygenases (1, 4, 5, 7, 19, 28, 46). TOM-Green, however, is the first monooxygenase to have an alanine at this position, just three amino acids away from a conserved glutamate involved in binding the diiron catalytic center of the hydroxylase where oxidation occurs.
We postulate that the smaller side chain of the alanine in TOM-Green compared to the valine in wild-type TOM allows greater access of substrate to the catalytic center, based on the crystal structures of the M. capsulatus Bath and the M. trichosporium OB3b nonheme sMMO hydroxylases, which show a clear channel formed by residues in this region (11, 32). Note that the tomA3 gene and mmoX, which encoded the α-subunits, are 36% similar. When the sMMO hydroxylase crystal was solved in two forms, the most prominent difference was an altered side chain conformation on leucine110 in the active-site cavity (31); the leucine 110 in the sMMO hydroxylase is analogous to the valine 106 in TOM.
The leucine 110 of sMMO was postulated to serve as a component in a hydrophobic gate controlling substrate access and product egress from the catalytic diiron active site (31). In the analogous hemerythrin, a leucine gate mutated to alanine exhibited increased autooxidation, while dioxygen association and dissociation constants were dramatically decreased when it was mutated to a tyrosine (31). Our results support these earlier observations in that by creating a smaller gate in TOM-Green, naphthalene, which is larger than the normal substrate toluene, has greater access to the active site. This hypothesis was confirmed by the enhanced oxidation rates by TOM-Green relative to the wild-type enzyme for the even larger substrates phenanthrene, fluorene, and anthracene, all three-ring compounds.
Site-directed mutagenesis of selected active-site residues has been performed on the somewhat related toluene 4-monooxygenase of Pseudomonas mendocina KR1, leading to toluene, m-xylene, and p-xylene products with varied regiospecificity (30). None of the mutations tested was in an analogous position to valine 106 of TOM, and they did not determine if any of their mutants had altered substrate specificity.
Random protein engineering has been used here to evolve five subunits comprising approximately 3.5 kb of the multicomponent biocatalyst TOM for multiple functions. One mutant enzyme, TOM-Green, which had significantly increased synthesis rates and yields of 1-naphthol and which also demonstrated enhanced degradation of recalcitrant chlorinated aliphatics as well as polyaromatic hydrocarbons, was found. The source of this improvement was found to be a key mutation in a probable gate amino acid in the α-subunit of the hydroxylase which appears to allow greater access of substrates into the active site. Currently, molecular breeding techniques (9) are being used with enzymes from the family of nonheme monooxygenases (7) to evolve TOM for further improvements in naphthol synthesis and degradation of individual and mixtures of chlorinated aliphatics.
Acknowledgments
This research was supported by the National Science Foundation (BES-9807146).
We are grateful for the assistance of Doohyun Ryoo (Jeonju University).
REFERENCES
- 1.Bertoni, G., M. Martino, E. Galli, and P. Barbieri. 1998. Analysis of the gene cluster encoding toluene/o-Xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3626–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley, P. M., and F. H. Chapelle. 1998. Effect of contaminant concentration on aerobic microbial mineralization of DCE and VC in stream-bed sediments. Environ. Sci. Technol. 32:553–557. [Google Scholar]
- 3.Bull, A., B. Marrs, and R. Kurane. 1998. Biotechnology for clean industrial products and processes. Towards industrial sustainability. OECD Publications, Paris, France.
- 4.Byrne, A. M., J. J. Kukor, and R. H. Olsen. 1995. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PK01. Gene 154:65–70. [DOI] [PubMed] [Google Scholar]
- 5.Cardy, D. L. N., V. Laidler, G. P. C. Salmond, and J. C. Murrell. 1991. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol. Microbiol. 5:335–342. [DOI] [PubMed] [Google Scholar]
- 6.Chauhan, S., P. Barbieri, and T. K. Wood. 1998. Oxidation of trichloroethylene, 1,1-dichloroethylene, and chloroform by toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3023–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coufal, D. E., J. L. Blazyk, D. A. Whittington, W. W. Wu, A. C. Rosenzweig, and S. J. Lippard. 2000. Sequencing and analysis of the Methylococcus capsulatus (Bath) soluble methane monooxygenase genes. Eur. J. Biochemistry 267:2174–2185. [DOI] [PubMed] [Google Scholar]
- 8.Crameri, A., G. Dawes, E. Rodriguez, Jr., S. Silver, and W. P. C. Stemmer. 1997. Molecular evolution of an arsenate detoxification pathway by DNA shuffling. Nat. Biotechnol. 15:436–438. [DOI] [PubMed] [Google Scholar]
- 9.Crameri, A., S.-A. Raillard, E. Bermudez, and W. P. C. Stemmer. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391:288–291. [DOI] [PubMed] [Google Scholar]
- 10.Eaton, R. W., and P. J. Chapman. 1995. Formation of indigo and related compounds from indolecarboxylic acids by aromatic acid-degrading bacteria: Chromogenic reactions for cloning genes encoding dioxygenases that act on aromatic acids. J. Bacteriol. 177:6983–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elango, N., R. Radhakrishnan, W. A. Froland, B. J. Wallar, C. A. Earhart, J. D. Lipscomb, and D. H. Ohlendorf. 1997. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 6:556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensley, B. D., B. J. Ratzkin, T. D. Osslund, and M. J. Simon. 1983. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science 222:167–169. [DOI] [PubMed] [Google Scholar]
- 13.Frost, J. W., and K. M. Draths. 1995. Biocatalytic syntheses of aromatics from D-glucose: renewable microbial sources of aromatic compounds. Annu. Rev. Microbiol. 49:557–579. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes, K. 1988. The parB (hok/sok) locus of plasmid R1: a general purpose plasmid stabilization system. Bio/Technology 6:1402–1405. [Google Scholar]
- 15.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge University, Cambridge, England.
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98. [Google Scholar]
- 17.Hudlicky, T., D. Gonzalez, and D. T. Gibson. 1999. Enzymatic dihydroxylation of aromatics in enantioselective synthesis: expanding asymmetric methodology. Aldrichim. Acta 32:35–62. [Google Scholar]
- 18.Jahng, D., and T. K. Wood. 1994. Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 60:2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, G. R., and R. H. Olsen. 1995. Nucleotide sequence analysis of genes encoding a toluene/benzene-2-monooxygenase from Pseudomonas sp. strain JS150. Appl. Environ. Microbiol. 61:3336–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumamaru, T., H. Suenaga, M. Mitsuoka, T. Watanabe, and K. Furukawa. 1998. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat. Biotechnol. 16:663–666. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 22.Luu, P. P., C. W. Yung, A. K.-Y. Sun, and T. K. Wood. 1995. Monitoring trichloroethylene mineralization by Pseudomonas cepacia G4 PR1. Appl. Microbiol. Biotechnol. 44:259–264. [Google Scholar]
- 23.McCarty, P. L. 1997. Breathing with chlorinated solvents. Science 276:1521–1522. [DOI] [PubMed] [Google Scholar]
- 24.McClay, K., B. G. Fox, and R. J. Steffan. 2000. Toluene monooxygenase-catalyzed epoxidation of alkenes. Appl. Environ. Microbiol. 66:1877–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercer, J. W., and R. M. Cohen. 1990. A review of immiscible fluids in the subsurface: properties, models, characterization and remediation. J. Contaminant Hydrol. 6:107–163. [Google Scholar]
- 26.Nelson, M. J. K., S. O. Montgomery, E. J. O’Neill, and P. H. Pritchard. 1986. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl. Environ. Microbiol. 52:383–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman, L. M., and L. P. Wackett. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066–14076. [DOI] [PubMed] [Google Scholar]
- 28.Nordlund, I., J. Powlowski, and V. Shingler. 1990. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J. Bacteriol. 172:6826–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelps, P. A., S. K. Agarwal, G. E. Speitel, Jr., and G. Georgiou. 1992. Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl. Environ. Microbiol. 58:3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry 36:9283–9289. [DOI] [PubMed] [Google Scholar]
- 31.Rosenzweig, A. C., H. Brandstetter, D. A. Whittington, P. Nordlund, S. J. Lippard, and C. A. Frederick. 1997. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): Implications for substrate gating and component interactions. Proteins Structure Function Genetics 29:141–152. [PubMed] [Google Scholar]
- 32.Rosenzweig, A. C., C. A. Frederick, S. J. Lippard, and P. Nordlund. 1993. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 366:537–543. [DOI] [PubMed] [Google Scholar]
- 33.Ryoo, D., H. Shim, K. Canada, P. Barbieri, and T. K. Wood. 2000. Aerobic degradation of tetrachloroethylene by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Nat. Biotechnol. 18:775–778. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, P. K., and P. L. McCarty. 1996. Isolation and Characterization of a facultatively aerobic bacterium that reductively dehalogenates tetrachloroethene to cis-1,2-dichloroethene. Appl. Environ. Microbiol. 62:761–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shields, M. S., and S. C. Francesconi. August 1996. Microbial degradation of trichloroethylene, dichloroethylenes, and aromatic pollutants. U.S. patent 5,543,317.
- 38.Shields, M. S., M. J. Reagin, R. R. Gerger, R. Campbell, and C. Somerville. 1995. TOM, a new aromatic degradative plasmid from Burkholderia (Pseudomonas) cepacia G4. Appl. Environ. Microbiol. 61:1352–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shields, M. S., M. J. Reagin, R. R. Gerger, C. Somerville, R. Schaubhut, R. Campbell, and J. Hu-Primmer. 1994. Constitutive degradation of trichloroethylene by an altered bacterium in a gas-phase bioreactor, p. 50–65. In R. E. Hinchee, A. Leeson, L. Semprini, and S. K. Ong (ed.), Bioremediation of chlorinated and polycyclic aromatic hydrocarbon compounds. Lewis Publishers, Boca Raton, Fla.
- 40.Shim, H., S. Chauhan, D. Ryoo, K. Bowers, S. M. Thomas, K. A. Canada, J. G. Burken, and T. K. Wood. 2000. Rhizosphere competitiveness of trichloroethylene-degrading, poplar-colonizing recombinant bacteria. Appl. Environ. Microbiol. 66:4673–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shim, H., and T. K. Wood. 2000. Aerobic degradation of mixtures of chlorinated aliphatics by cloned toluene-o-xylene monooxygenase and toluene o-monooxygenase in resting cells. Biotechnol. Bioeng. 70:693–698. [DOI] [PubMed] [Google Scholar]
- 42.Stemmer, W. P. C. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talukder, M., and C. R. Kates. 1995. Naphthalene derivatives, p. 979–1017. In M. Howe-Grant (ed.), Kirk-Othmer encyclopedia of chemical technology, 4th ed., vol. 16. John Wiley & Sons, Inc., New York, N.Y.
- 44.van Hylckama Vlieg, J. E. T., W. de Koning, and D. B. Janssen. 1996. Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl. Environ. Microbiol. 62:3304–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood, T. K., R. H. Kuhn, and S. W. Peretti. 1990. Enhanced plasmid stability through post-segregational killing of plasmid-free cells. Biotechnol. Tech. 4:36–41. [Google Scholar]
- 46.Yen, K.-M., M. R. Karl, L. M. Blatt, M. J. Simon, R. B. Winter, P. R. Fausset, H. S. Lu, A. A. Harcourt, and K. K. Chen. 1991. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 173:5315–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, H., and F. H. Arnold. 1997. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 25:1307–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]