Abstract
Two-component systems are signal transduction systems which enable bacteria to regulate cellular functions in response to changing environmental conditions. In most cases regulation is accomplished on the transcriptional level by a response regulator protein, which, according to the phosphorylation state of its receiver domain, displays different affinities for its target promoters. Here we describe identification of genes regulated by the two-component system HP166-HP165 of Helicobacter pylori and characterization of the corresponding target promoters. We demonstrated that expression of the HP166-HP165 two-component system is negatively autoregulated under conditions favoring autophosphorylation of the histidine kinase. Furthermore, we found that response regulator HP166 activates transcription of genes encoding a protein family with an unknown function present in H. pylori 26695, as well as an operon composed of five H. pylori-specific genes. While open reading frame HP166 is an essential gene, the target genes of the response regulator are not required for growth under in vitro culture conditions.
The microaerophilic, gram-negative bacterium Helicobacter pylori is the causative agent of chronic type B gastritis and peptic ulcer disease in humans and is also considered a risk factor for the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (16, 31). Infection with this pathogen is widespread and affects approximately 50% of the world’s population.
Several factors associated with the pathogenesis of H. pylori have been identified, including the essential colonization factors urease (11) and flagella (43), adhesins (21, 29), and proteins which contribute to the mucosal damage caused by infection (i.e., the vacuolating cytotoxin VacA [34, 44], the neutrophil-activating protein NAP [14, 36], and the cytotoxin-associated antigen CagA [10]). Recently, it has been demonstrated that CagA is directly injected into host cells via a type IV secretion system encoded by the cag pathogenicity island (30, 42), whose presence is a genetic marker of the so-called type I strains exhibiting increased virulence.
Very little information is available regarding the H. pylori molecular mechanisms that regulate expression of both housekeeping genes and genes assumed to contribute to virulence. Although several genes whose expression is induced at low pH have been identified by microarray and differential display technology (2, 13), the mechanisms of transcriptional regulation underlying these processes remain unclear. Sequence analysis of the genomes of two unrelated H. pylori isolates, isolates 26695 and J99 (1, 45), revealed the presence of few regulatory genes. Two transcriptional repressor proteins have been characterized in some detail. The H. pylori Fur protein is involved in iron-dependent regulation of transcription (6, 7), and the HspR repressor negatively regulates expression of the operons encoding the major chaperones of H. pylori (41).
Two-component systems are widespread prokaryotic signal transduction systems which allow regulation of cellular functions in response to changing environmental conditions. Each of these systems is composed of a sensor protein, which perceives environmental stimuli with its N-terminal input domain, and a cognate response regulator, which elicits a cellular response via its C-terminal output domain, frequently by acting as a transcriptional activator. Communication between the sensor protein and the response regulator is achieved by phosphotransfer from a highly conserved histidine residue in the sensor’s transmitter domain, which is autophosphorylated in the presence of the appropriate stimulus, to an aspartic acid residue in the N-terminal receiver domain of the regulator protein. Phosphorylation of the receiver domain then triggers a conformational change of the response regulator, which activates its C-terminal output domain. The H. pylori genome contains only four open reading frames (ORFs) with homology to two-component histidine kinases and six genes encoding response regulators. Based on structural and functional homologies, one sensor-response regulator pair has been identified as the H. pylori CheA-CheY two-component system regulating chemotaxis (4, 45), while the remaining two-component proteins probably are involved in transcriptional regulation. So far, information about the regulons controlled by the two-component systems of H. pylori is available only for histidine kinase HP244 and the NtrC-like response regulator HP703, which positively regulate the transcription of genes encoding components of the flagellar basal body and hook and the minor flagellin gene flaB (5, 40). Here we found that a two-component system composed of histidine kinase HP165 and response regulator HP166 (5), which belongs to the OmpR family of two-component regulators, negatively autoregulates its own expression and that HP166 acts as a transcriptional activator of several H. pylori-specific genes encoding proteins whose functions are not known.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strains G27, G25, G46, and G50 are clinical isolates (50). H. pylori P76 is a spontaneous streptomycin-resistant derivative of mouse-adapted strain P49 (15). H. pylori G27/HP165::km has been described previously (5). When recovered from frozen stocks, the H. pylori strains were grown under microaerophilic conditions (Oxoid) on Columbia agar plates containing 5% horse blood, 0.2% cyclodextrin, and Dent’s or Skirrow’s antibiotic supplement at 37°C for 2 or 3 days. After passage on fresh plates the bacteria were cultured in a 5% CO2-95% air atmosphere at 37°C. Liquid cultures of H. pylori were grown in Brucella broth containing Dent’s or Skirrow’s antibiotic supplement and 5% fetal calf serum. When required, blood agar plates or broth was supplemented with kanamycin or chloramphenicol at a final concentration of 20 μg/ml. The Escherichia coli strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in Luria-Bertani broth. When necessary, antibiotics were added at the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 30 μg/ml.
TABLE 1.
Strains and plasmids
| Bacterial strain or plasmid | Relevant feature(s) | Reference or source | |||
|---|---|---|---|---|---|
| H. pylori strains | |||||
| G27 | Clinical isolate | 50 | |||
| G25 | Clinical isolate | 50 | G46 | Clinical isolate | 50 |
| G50 | Clinical isolate | 50 | |||
| P76 | Spontaneous streptomycin-resistant derivative of mouse-adapted strain P49 | 15 | G27/HP165::km | ||
| G27 with a kanamycin resistance cassette replacing ORF HP165 | 5 | ||||
| G27/P1408-gfp | G27 with a fusion of the promoter region of ORF HP1408 to a promoterless gfp gene flanked by a kanamycin resistance cassette and replacing ORF HP1408 | This study | |||
| G27/P1408-gfpΔ165 | G27/HP165::km with a fusion of the promoter region of ORF HP1408 to a promoterless gfp gene flanked by a chloramphenicol resistance cassette and replacing ORF HP1408 | This study | |||
| G27/P166-gfp | G27 with a fusion of the promoter region of ORF HP166 to a promoterless gfp gene flanked by a kanamycin resistance cassette and replacing ORF HP1408 | This study | |||
| G27/P166Δ-gfp | G27 with a fusion of the 3′ truncated promoter region of ORF HP166 to a promoterless gfp gene flanked by a kanamycin resistance cassette and replacing ORF HP1408 | This study | |||
| G27/P166-gfpΔ165 | G27/HP165::km with a fusion of the promoter region of ORF HP166 to a promoterless gfp gene flanked by a chloramphenicol resistance cassette and replacing ORF HP1408 | This study | |||
| G27/P166Δ-gfpΔ165 | G27/HP165::km with a fusion of the 3′ truncated promoter region of ORF HP166 to a promoterless gfp gene flanked by a chloramphenicol resistance cassette and replacing ORF HP1408 | This study | |||
| G27/pHel-P120-gfp | G27 harboring pHel2 carrying a fusion of the promoter region of ORF HP120 to a promoterless gfp gene | This study | |||
| P76/Δ1408 | P76 with the regions located between the 23S rDNA loci and ORFs HP1413 and HP422 replaced by kanamycin and chloramphenicol resistance cassettes, respectively | This study | |||
| P76/Δ119 | P76 with the regions located between ORFs HP1186 to HP1189 and ORFs HP117 to HP121 replaced by kanamycin and chloramphenicol resistance cassettes, respectively | This study | |||
| E. coli strains | |||||
| DH5α | Strain used for high-efficiency transformation | Gibco | |||
| XL1-Blue | Strain used for high-efficiency transformation | Stratagene | |||
| M15 | Strain used for overproducing His6-HP166 | Qiagen | |||
| Plasmids | |||||
| pBluescript SK | Cloning vector | Stratagene | |||
| pSL1180 | Cloning vector | Pharmacia | |||
| pILL600 | Plasmid containing the kanamycin resistance cassette from C. coli | 24 | |||
| pDT2548 | Plasmid containing the chloramphenicol resistance gene from C. coli | 48 | |||
| pHel2 | Shuttle vector for H. pylori | 18 | |||
| pQE-166 | pQE30 expressing the His6-tagged response regulator HP166 | 5 | |||
| pLSV16gfp | Listeria-E. coli shuttle vector used as source of the promoterless gfp-mut2 gene | 8 | |||
| pGG1 | pBluescript SK carrying a 284-bp DraI fragment from H. pylori G27 with homology to the upstream region and part of the coding region of ORF HP1408 in H. pylori 26695 | This study | |||
| pGG2 | pBluescript SK carrying a 183-bp DraI fragment from H. pylori G27 with homology to the upstream region of ORF HP119 in H. pylori 26695 | This study | |||
| pGG3 | pBluescript SK carrying a 183-bp DraI fragment from H. pylori G27 with homology to the upstream region of ORF HP119 in H. pylori 26695 | This study | |||
| pSL-gfp1409 | pSL1180 containing the promoterless gfp-mut2 gene and a 504-bp DNA fragment derived from ORF HP1409 | This study | |||
| pSL-P1408-gfp/km | pSL1180 carrying the kanamycin resistance cassette flanked by a fusion of a 287-bp DNA fragment derived from the promoter region of ORF HP1408 to the gfp-mut2 gene and a 504-bp DNA fragment derived from ORF HP1409 | This study | |||
| pSL-P1408-gfp/cm | pSL1180 carrying the chloramphenicol resistance cassette flanked by a fusion of a 287-bp DNA fragment derived from the promoter region of ORF HP1408 to the gfp-mut2 gene and a 504-bp DNA fragment derived from ORF HP1409 | This study | |||
| pSL-P166-gfp/km | pSL1180 containing a 543-bp DNA fragment from the 23S rDNA locus, a 153-bp DNA fragment comprising the promoter region of ORF HP166 which is fused to the gfp-mut2 gene, the kanamycin resistance cassette, and a 504-bp DNA fragment derived from ORF HP1409 | This study | |||
| pSL-P166Δ-gfp/km | Same as pSL-166-gfp/km except that there is a 36-bp deletion in the 3′ part of the promoter region of ORF HP166 | This study | |||
| pSL-P166-gfp/cm | pSL1180 containing a 543-bp DNA fragment from the 23S rDNA locus, a 153-bp DNA fragment comprising the promoter region of ORF HP166 which is fused to the gfp-mut2 gene, the chloramphenicol resistance cassette, and a 504-bp DNA fragment derived from ORF HP1409 | This study | |||
| pSL-P166Δ-gfp/cm | Same as pSL-166-gfp/cm except that there is a 36-bp deletion in the 3′ part of the promoter region of ORF HP166 | This study | |||
| pHel-P120-gfp | pHel2 carrying a 612-bp DNA fragment derived from the upstream region of ORF HP120 and fused to the gfp-mut2 gene | This study | |||
| pSL-1413:km:23S | pSL1180 containing a 506-bp DNA fragment derived from ORF HP1413 and a 543-bp DNA fragment derived from the 23S rDNA locus flanking the kanamycin resistance cassette | This study | |||
| pSL-422:cm:23S | pSL1180 containing a 539-bp DNA fragment derived from ORF HP422 and a 543-bp DNA fragment derived from the 23S rDNA locus flanking the chloramphenicol resistance cassette | This study | |||
| Bacterial strain or plasmid | Relevant feature(s) | Reference or source | |||
| pSL-1186:km:1189 | pSL1180 containing a 595-bp DNA fragment derived from ORF HP1186 and a 521-bp DNA fragment derived from ORF HP1189 flanking the kanamycin resistance cassette | This study | |||
| pSL-117:cm:121 | pSL1180 containing a 494-bp DNA fragment derived from ORF HP117 and a 499-bp DNA fragment derived from ORF HP121 flanking the chloramphenicol resistance cassette | This study | |||
| pSL-166PE | pSL1180 containing a 549-bp DNA fragment from H. pylori G27 comprising the promoter region and part of the coding DNA of ORF HP166 | This study | |||
| pSL-119PE1 | pSL1180 containing a 783-bp DNA fragment from H. pylori G27 with similarity to the promoter and part of the coding region of ORF HP119 in H. pylori 26695 | This study | |||
| pSL-119PE2 | pSL1180 containing a 700-bp DNA fragment from H. pylori G27 with similarity to the promoter and part of the coding region of ORF HP119 in H. pylori 26695 | This study | |||
| pSL-1408f | pSL1180 containing a 360-bp BamHI-EcoRI fragment derived from the P1408-gfp fusion in pSL-1408-gfp/km | This study | |||
| pSL-119f | pSL1180 containing a 298-bp BamHI-EcoRI fragment derived from the promoter region of ORF HP119 cloned in pSL-119PE1 | This study | |||
| pSL-166f | pSL1180 containing a 308-bp BamHI-EcoRI fragment comprising the promoter region and part of the coding DNA of ORF HP166 | This study |
General techniques.
DNA manipulation, cloning procedures, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblot analysis were carried out by using standard procedures. PCR amplifications were performed with a model 60 thermocycler obtained from Biomed by using Deep Vent DNA polymerase (New England Biolabs). All cloned PCR products were subjected to automated sequencing (Big Dye kit; Perkin-Elmer) to ensure proper amplification. Site-directed mutagenesis was performed with a QuickChange site-directed mutagenesis kit (Stratagene). Reverse transcription (RT)-PCR with RNA extracted from H. pylori G27 was performed by using a ProStar First Strand RT-PCR kit (Stratagene) according to the manufacturer’s instructions.
Purification of His6-HP166.
Recombinant His6-tagged response regulator His6-HP166 encoded on plasmid pQE-166 (5) was overexpressed in E. coli, and the protein was purified by affinity chromatography on Ni2+-nitrilotriacetic acid (NTA) agarose essentially as described by Perraud et al. (32).
In vitro phosphorylation of His6-HP166.
His6-HP166 was phosphorylated in vitro by incubating the protein in phosphorylation buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol) in the presence of either 50 mM acetylphosphate, 10 mM phosphoramidate, or 10 mM carbamylphosphate for 30 or 60 min at 25°C. Phosphoramidate was prepared by the method described by Sheridan et al. (37).
Solid-phase DNA binding assay.
A 50-μl portion of an Ni2+-NTA magnetic agarose bead suspension (Qiagen) equilibrated with binding buffer (50 mM NaH2PO4, 50 mM NaCl, 20 mM imidazole; pH 8.0) was incubated with 18 μg of purified response regulator His6-HP166 in 500 μl (final volume) of binding buffer for 1 h at 4°C with constant shaking. After the test tube was placed on a suitable magnetic separator, the binding buffer was removed, and the magnetic beads were washed with 500 μl of interaction buffer (20 mM Tris-HCl, 10 mM MgCl2, 100 mM KCl, 20 mM imidazole; pH 7.5). Subsequently, 50 μg of DraI-digested chromosomal DNA of H. pylori G27 in 500 μl (final volume) of interaction buffer was added to the magnetic beads, and the mixture was incubated for 20 min at room temperature with constant shaking. To allow phosphorylation of response regulator His6-HP166, acetylphosphate was added to the binding and interaction buffers at a final concentration of 50 mM. After the beads were washed three times with 500 μl of interaction buffer, the bound DNA was eluted with 50 μl of 1.25 M NaCl, and the eluate was analyzed by agarose gel electrophoresis. To clone the DNA fragments bound to His6-HP166, the eluates from three individual binding assays were pooled and subsequently separated on a 2% agarose gel. The DNA fragments were eluted from the gel and cloned into the SmaI site of pBluescript SK. The resulting plasmids were digested with EcoRI and BamHI, and the restriction hydrolysates were subjected to binding assays to confirm the interaction of the cloned insert with His6-HP166. Plasmids with DNA inserts capable of binding to His6-HP166 were characterized by sequence analysis.
Construction of H. pylori strains containing promoter-gfp fusions.
To construct H. pylori strain G27/P1408-gfp, a 287-bp DNA fragment comprising the promoter region of ORF HP1408 was amplified from chromosomal DNA of H. pylori G25 by using primer pair Prom5-Prom3 providing BamHI and XbaI restriction sites at the 5′ and 3′ termini. This DNA fragment was cloned into pSL-gfp1409 carrying an XbaI-PstI fragment encoding the promoterless gfp-mut2 gene (9) and a 504-bp PstI-SacI fragment derived from ORF HP1409 which was obtained by PCR performed with primer pair HP1409-5-HP1409-3 on chromosomal DNA of H. pylori G27. The resulting plasmid was linearized with PstI, and a kanamycin resistance cassette and a chloramphenicol resistance cassette from Campylobacter coli were inserted to obtain plasmids pSL-P1408-gfp/km and pSL-P1408-gfp/cm, respectively, which were subsequently used for transformation of H. pylori G27; this resulted in chromosomal integration of the PHP1408-gfp fusion. To construct G27/P166-gfp and G27/P166Δ-gfp, a 543-bp EcoRI-BamHI DNA fragment comprising part of the 23S ribosomal DNA (rDNA) locus and the intergenic region located upstream of ORF HP1408 was PCR amplified with primer pair 23S-5–23S-3 and was cloned into pSL-gfp1409 together with a 153-bp BamHI-XbaI DNA fragment derived from the promoter region of ORF HP166 in H. pylori G27 (primer pair 166gfp-5-166gfp-3) or a 117-bp BamHI-XbaI DNA fragment from the promoter region of ORF HP166 which lacked the binding site for the response regulator HP166 (primer pair 166gfp-5-166Δgfp-3). The resulting plasmids were linearized with PstI, the kanamycin resistance cassette was inserted to obtain plasmids pSL-P166-gfp/km and pSL-P166Δ-gfp/km, and these plasmids were used for transformation of H. pylori G27. Allelic exchange reactions resulted in replacement of ORF HP1408 with the PHP166-gfp and PHP166Δ-gfp promoter fusions. Plasmids pSL-P166-gfp/cm and pSL-P166Δ-gfp/cm differ from pSL-P166-gfp/km and pSL-P166Δ-gfp/km only by the replacement of the kanamycin resistance cassette with a chloramphenicol resistance cassette and were used for transformation of G27/HP165::km to obtain G27/P166-gfpΔ165 and G27/P166Δ-gfpΔ165. pHel-P120-gfp was constructed by cloning a 612-bp BamHI-XbaI DNA fragment containing the upstream region of ORF HP120 which was PCR amplified from chromosomal DNA of H. pylori G27 with primer pair HP120-5–HP120-3 and the XbaI-PstI fragment encoding the promoterless gfp-mut2 gene into pSL1180. The resulting PHP120-gfp promoter fusion was subsequently excised with BamHI and ClaI and was ligated into pHel2; this yielded the shuttle plasmid pHel-P120-gfp, which was used for transformation of H. pylori G27.
Construction of H. pylori deletion mutants P76/Δ1408 and P76/Δ119.
To construct the double-deletion mutant P76/Δ1408, H. pylori P76 was first transformed with the suicide plasmid pSL-1413:km:23S. pSL-1413:km:23S was obtained by cloning a 506-bp PstI-SacI fragment derived from ORF HP1413 (PCR amplified with primer pair 1413-5–1413-3 from chromosomal DNA of H. pylori G27) and a 543-bp EcoRI-PstI fragment derived from the 23S rDNA (PCR amplified with primer pair 23S-5–23S-3) into pSL1180 and then inserting the C. coli kanamycin resistance cassette into the PstI site. H. pylori transformants harboring the correctly integrated kanamycin cassette, which was verified by PCR analysis performed with chromosomal DNA and primers complementary to sequences flanking the integration site, were subsequently transformed with plasmid pSL-422:cm:23S. pSL-422:cm:23S contained 539-bp PstI-SacI and 543-bp EcoRI-PstI fragments flanking the chloramphenicol resistance cassette from C. coli, which were derived from ORF HP422 and the 23S rDNA and were PCR amplified by using primer pairs 422-5–422-3 and 23S-5–23S-3, respectively. Correct integration of the chloramphenicol resistance cassette into the kanamycin- and chloramphenicol-resistant transformants was determined by PCR analysis.
Similarly, P76/Δ119 was constructed by performing allelic exchange reactions mediated by subsequent transformation of H. pylori P76 with the suicide plasmids pSL-1186:km:1189 and pSL-117:cm:121. To construct pSL-1186:km:1189, a 595-bp BamHI-PstI fragment derived from ORF HP1186 (PCR amplified with primer pair 1186-5–1186-3) and a 521-bp PstI-SacI fragment derived from ORF HP1189 (PCR amplified with primer pair 1189-5–1189-3) were cloned into pSL1180, and the kanamycin resistance cassette was subsequently inserted into the PstI site. pSL-117:cm:121 harbored 494-bp EcoRI-PstI and 499-bp PstI-SacI fragments derived from ORFs HP117 (PCR amplified with primer pair 117-5–117-3) and HP121 (PCR amplified with primer pair 121-5–121-3), respectively, flanking the chloramphenicol resistance cassette.
To confirm the deletions in mutants P76/Δ119 and P76/Δ1408, Southern blot analysis was performed with 569- and 504-bp DNA probes derived from ORFs HP1187 (PCR amplified with primer pair 1187-5–1187-3) and HP1409 (PCR amplified with primer pair 1409-5–1409-3), respectively.
RNA isolation.
RNA was prepared as described previously (4).
Primer extension and 5′ RACE analysis.
Primer extension experiments were carried out as previously described (4). Sequencing reaction mixtures containing plasmids pSL-166PE, pSL-1408-gfp/km, pSL-119PE1, and pSL-119PE2 and analyzed on 6% urea-polyacrylamide gels were used as standards for determinations of the transcription initiation sites. pSL-166PE, pSL-119PE1, and pSL-119PE2 contained DNA fragments comprising the promoter regions of ORFs HP166, HP119, and HP119, respectively, which were PCR amplified with primer pairs HP166-5–HP166-3, JHP1114-1113-5–JHP1114-1113-3, and HP119-1188-5–JHP1114-1113-3, respectively, from chromosomal DNA of H. pylori G27. The sequences of oligonucleotides gfpPE, HP166PE, and HP119PE, which were used as primers for cDNA synthesis, are shown in Table 2. 5′ Rapid amplification of cDNA ends (RACE) experiments were performed by using the 5′ RACE system, version 2.0 (Gibco-BRL), according to the manufacturer’s instructions.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′)a | Siteb | Strand | Position (nucleotides)c |
|---|---|---|---|---|
| Prom-5 | aatgtaggatccGACATGCAATGCTTAAAGTTTGC | BamHI | + | 1477176–1477198 |
| Prom-3 | gggaaatctagaAAAAAAGATGACATGGGG | XbaI | − | 1477538–1477555 |
| HP1409-5 | gcaatgctgcagCAATCAGTGGGGCTTACAG | PstI | + | 1477889–1477907 |
| HP1409-3 | gcaatggagctcGCGTTACGCAAATAAGTG | SacI | − | 1478412–1478429 |
| 23S-5 | catgtagaattcAGCTCCTTGACAGCTCCGC | EcoRI | + | 1476708–1476726 |
| 23S-3 | atcgtaggatccTTTATCCATTTCTTTCAAACTC | BamHI | − | 1477230–1477251 |
| 166gfp-5 | agtctaggatccTATTCCATGAAAACAAAGCC | BamHI | − | 174616–174635 |
| 166gfp-3 | atgctatctagaCTGCAACTTTAAAAACGATTAAC | XbaI | + | 174482–174504 |
| 166Δgfp-3 | tcgtactctagaTAAGGGACTAATTCTAGC | XbaI | + | 174518–174536 |
| HP166-5 | caagctgaattcGAAAAGGAAGCCATGAAGCG | EcoRI | − | 174779–174798 |
| HP166-3 | tctttg ctgcagCGCCTACACACTTCAAGCC | PstI | + | 174259–174277 |
| HP120-5 | gtctgatctagaCCAACCTTTTTTTAAAAAACAATC | XbaI | − | 132845–132865 |
| HP120-3 | ctaagaggatccTAGTGAGGGAAGAAATGG | BamHI | + | 132252–132275 |
| JHP1114-1113-5 | caagctgaattcGTTTGATAAGAAAACCTTGCACG | EcoRI | 1239384–1239406 | |
| JHP1114-1113-3 | agagtgctgcagCACCTATTGCTGAATTAAACC | PstI | 1238624–1238644 | |
| HP119-1188-5 | caagctgaattcGCTCTAAAGCTCAAGCTAAAATGC | EcoRI | − | 1258324–1258347 |
| 1413-5 | tatcatctgcagCTCCCCCTAATCTGTCCG | PstI | + | 1483967–1483984 |
| 1413-3 | catgctgagctcGACCCCTGAACTAAACCTC | SacI | − | 1484455–1484473 |
| 422-5 | tagcacgagctcGTGGGAGCGTATCAAGAAG | SacI | + | 437839–437859 |
| 422-3 | tatcactgcagCTCCTGGCGTTTTAGCCG | PstI | − | 438361–438378 |
| 121-5 | tacgatctgcagAACGCGCAGTTAAGCCATGGTC | PstI | + | 132348–132369 |
| 121-3 | tacgatgagctcGGGCTTAACGAATATGAAAG | SacI | − | 132828–132847 |
| 117-5 | tcatgcgaattcGAAAGAGTGGATAAACCCTATTC | EcoRI | + | 127293–127315 |
| 117-3 | tacgacctgcagTCAAAACGCGCAATAAAACTCC | PstI | − | 127766–127787 |
| 1186-5 | tcaagcggatccGCCAATCGCCCATCAACATTG | BamHI | + | 1255920–1255940 |
| 1186-3 | agctatctgcagTTAGCGGGTCTCAGCTGAGC | PstI | − | 1256359–1256380 |
| 1189-5 | cagtgcctgcagTCAAGCGTTCTTAATGTAATGC | PstI | + | 1259619–1259640 |
| 1189-3 | cagtgcgagctcCCGCTTTAGAGTGTTTGG | SacI | − | 1260113–1260130 |
| 1187-5 | agctcaggatccCAGAAAGATTTCATTAAATATG | BamHI | − | 1257294–1257315 |
| 1187-3 | actcatctgcagCTACATCCTTTTACTATAACC | PstI | + | 1256746–1256766 |
| gfp75 | tgaccagaattcCAAGAATTGGGACAACTC | EcoRI | ||
| R4-2 | ctatcaggatccCTTCTATCATACTTCAATTAACTTC | BamHI | + | 174446–174470 |
| 166f-5 | gtcatggaattcGCAATACCAATTGAGCACGC | EcoRI | − | 174735–174754 |
| 119f-5 | aatgtaggatccGATAAAGGCTTACACTCAAGC | BamHI | 1239097–1239118 | |
| 119f-3 | tgaccagaattcTCATCATCATTGTTGCAAGC | EcoRI | + | 130723–130742 |
| HP166PE | GCTAATTCTATATCATCTTC | + | 174414–174437 | |
| HP119PE | TCATCATCATTGTTGCAAGC | + | 130723–130742 | |
| gfpPE | GCCCATTAACATCACCATC | |||
| FBA1408-1 | AATTAAAACGCATCATTAACCATTGATTAAATA | + | 1477490–1477522 | |
| FBA1408-2 | GTGGTATTTAATCAATGGTTAATGATGCGTTTT | − | 1477494–1477526 |
Uppercase letters indicate nucleotides derived from the genome sequences of H. pylori 26695 (45) or H. pylori J99 (JHP1114-1113-5, JHP1114-1113-3, and 119f-5 [1]) and the gfp-mut2 gene (gfpPE and gfp75 [9]). Lowercase letters indicate nucleotides introduced for cloning purposes. Underlining indicates restriction recognition sequences.
Restriction recognition sites.
DNase I footprinting.
DNase I footprint experiments were performed essentially as described by Dickneite et al. (12). Promoter DNA fragments derived from plasmids pSL-166f, pSL-119f, and pSL-1408f were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase at one extremity and gel purified, and approximately 100,000 cpm of each probe was used for footprint experiments. pSL-166f and pSL-119f contained 308- and 298-bp DNA fragments comprising the promoter regions of HP166 and HP119, respectively, which were obtained by PCR performed with chromosomal DNA of H. pylori G27 and primer pairs 166f-5–R4-2 and 119f-5–119f-3. pSL-1408f contained a 360-bp DNA fragment derived from the PHP1408-gfp promoter fusion in plasmid pSL-P1408-gfp/cm, which was obtained by PCR performed with primer pair Prom5-gfp75.
DNA filter binding assays.
The 37-bp DNA probe used for filter binding comprised nucleotides −56 to −19 with respect to the transcriptional start site of ORF HP1408 and was obtained by annealing oligonucleotides FBA1408-1 and FBA1408-2. The annealing reaction was performed by incubating the oligonucleotide solution for 5 min at 94°C and cooling the reaction mixture stepwise with subsequent 10-min incubation periods at 50, 37, 30, 20, and 4°C. The DNA probe was labeled by filling in the 3′ ends using the DNA polymerase I Klenow fragment and [α-32P]dATP, [α-32P]dTTP, and [α-32P]dCTP. Various amounts of His6-HP166 that had been phosphorylated in vitro with acetylphosphate, phosphoramidate, and carbamylphosphate were incubated with 32P-labeled DNA (final concentration, <0.5 nM) in assay buffer (20 mM HEPES [pH 7.9], 50 mM KCl, 5 mM CaCl2, 2 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 50% glycerol) containing 50 mM acetylphosphate, 10 mM phosphoramidate, and 10 mM carbamylphosphate, respectively. When binding of the unphosphorylated response regulator was analyzed, the low-molecular-weight phosphate donor in the assay buffer was replaced by 35 mM KH2PO4/K2HPO4 buffer (pH 7.9) in order to keep the ionic strength of the assay buffer constant. After incubation at room temperature for 25 min, 135-μl aliquots were filtered through nitrocellulose filters (NC 45; Schleicher & Schuell). The filters were immediately washed with 250 μl of assay buffer with 10% glycerol and without bovine serum albumin, dried, and analyzed by liquid scintillation counting. The binding isotherms were calculated as described by Weiss et al. (49). As we were not able to predict the number of binding sites present in the 37-bp DNA probe, the data points were fitted for linear binding to one or two binding sites, as well as for cooperative binding to two binding sites (49).
RESULTS
Response regulator HP166 negatively autoregulates expression of the HP166-HP165 two-component system.
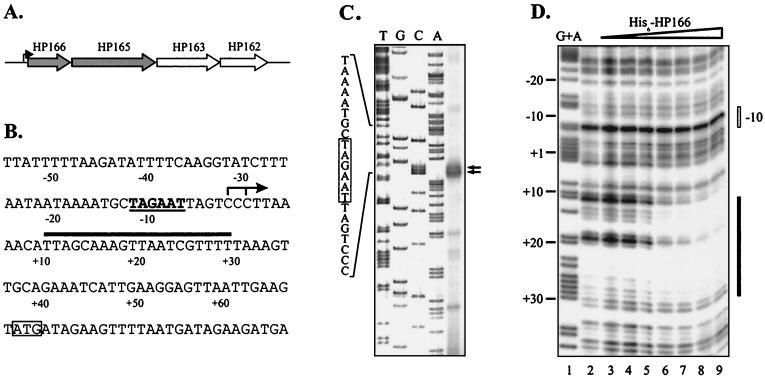
The genes encoding response regulator HP166 and histidine kinase HP165 are located adjacent to each other and, as demonstrated by RT-PCR (data not shown), are organized in an operon together with the gene coding for delta-aminolevulinic acid dehydratase (HP163) and an ORF encoding a conserved hypothetical protein (HP162) (Fig. 1A). Using the results of 5′ RACE experiments performed with RNA extracted from H. pylori G27, we mapped the transcription initiation site at position −64 with respect to the translational start codon of HP166. As shown in Fig. 1C, primer extension analysis yielded similar transcriptional start sites at positions −66 and −68 corresponding to a promoter composed of a TAGAAT −10 element with no further similarity to the −35 element of the E. coli promoter consensus sequence or the recently postulated H. pylori-specific upstream promoter element TTAAGC (47). However, a TG dinucleotide motif was located one nucleotide upstream of the −10 box, which is characteristic of the E. coli −10 extended promoters (23). To analyze the putative autoregulatory role of response regulator HP166, we performed DNase I footprint experiments with the recombinant His6-tagged HP166 protein using a 309-bp DNA fragment harboring the HP166 promoter. As shown in Fig. 1D, addition of 4 μg of His6-HP166 incubated with acetylphosphate prior to the binding reaction resulted in protection of a region ranging from position +10 to position +29 with respect to the transcriptional start site, suggesting that HP166 acts as a transcriptional repressor of its own promoter. At slightly higher protein concentrations unphosphorylated His6-HP166 produced a very similar footprint. The relatively small effect of in vitro phosphorylation on the binding affinity of His6-HP166 for the PHP166 promoter might have been due to the low phosphorylation efficiency obtained when low-molecular-weight phosphate donors were used (see below).
FIG. 1.
(A) Schematic representation of the operon encoding response regulator HP166 and the cognate histidine kinase HP165. In the genome sequence of H. pylori 26695 the histidine kinase gene is annotated as two separate ORFs, HP165 and HP164, probably due to a sequencing error. The arrow indicates the PHP166 promoter. (B) Schematic representation of the PHP166 promoter. The transcriptional start point is indicated by an arrow, and the −10 promoter element is underlined. The translational start codon is enclosed in a box. The binding site of response regulator HP166 is indicated by a solid bar. The numbers below the sequence indicate positions relative to the transcriptional start site. (C) Determination of the transcriptional start site of ORF HP166 by primer extension analysis. Total RNA extracted from H. pylori G27 was hybridized to radiolabeled oligonucleotide HP166PE. Transcriptional start points are indicated by arrows. A part of the promoter sequence is shown on the left. The −10 promoter element is enclosed in a box. The sequence reaction (lanes A, C, G, and T) was performed by using oligonucleotide HP166PE and plasmid pSL-166PE as the template. (D) DNase I footprint analysis of His6-HP166 on the PHP166 promoter. A specific 308-bp end-labeled BamHI-EcoRI fragment was incubated with different amounts of His6-HP166. In lanes 2 to 9 0, 1, 2, 4, 6, 8, 10, and 12 μg of His6-HP166 incubated with acetylphosphate were added. The numbers on the left indicate nucleotide positions with respect to the transcriptional start site. The solid bar on the right indicates the area of DNase I protection. The open bar indicates the position of the −10 promoter element. Lane 1 contained a G+A sequence reaction mixture with the DNA probe used as a size marker (27).
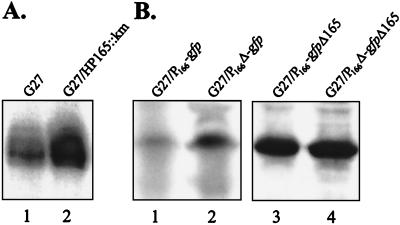
In order to investigate the in vivo significance of the in vitro binding of His6-HP166 to its own promoter, we studied expression of HP166 by performing primer extension analysis with equal amounts of RNA extracted from H. pylori wild-type strain G27 and isogenic mutant G27/HP165::km, which carries a kanamycin resistance cassette instead of the gene encoding histidine kinase HP165 (5). An H. pylori mutant with a deletion of the gene coding for response regulator HP166 could not be analyzed because this gene could not be inactivated in H. pylori G27 (5). As shown in Fig. 2A, inactivation of the corresponding histidine kinase HP165 resulted in clear derepression of the PHP166 promoter, suggesting that phosphorylation of the response regulator is a prerequisite for repressor function. To confirm the effect of the binding of HP166 to the sequence motif identified in DNase I footprint experiments, the PHP166 promoter and a truncated derivative lacking the putative binding site for HP166 were fused to a promoterless reporter gene (gfp) encoding green fluorescent protein (GFP). The gfp fusions were integrated into the chromosomes of H. pylori G27 and G27/HP165::km by allelic exchange by replacing ORF HP1408 and using antibiotic resistance cassettes (Kmr and Cmr, respectively) cloned downstream of the gfp gene. Protein lysates prepared from liquid cultures of the corresponding strains grown to an optical density at 600 nm of 1.0 were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently analyzed by Western blot hybridization using a polyclonal antibody directed against GFP. In the genetic background of H. pylori G27 deletion of the putative repressor binding site in the PHP166 promoter resulted in a significant increase in gfp expression compared to the expression of the wild-type PHP166 promoter fusion (Fig. 2B), while in G27/HP165::km expression of gfp remained unchanged regardless of whether the repressor binding site was deleted from the PHP166 promoter fusion (Fig. 2B).
FIG. 2.
(A) Primer extension analysis of equal amounts of RNA extracted from H. pylori G27 (lane 1) and G27/HP165::km (lane 2) with oligonucleotide HP166PE specific for ORF HP166. (B) Western blot analysis of protein lysates of H. pylori G27/P166-gfp (lane 1), G27/P166Δ-gfp (lane 2), G27/P166-gfpΔ165 (lane 3), and G27/P166Δ-gfpΔ165 (lane 4). The membrane was hybridized with a polyclonal antiserum directed against GFP.
Identification of target genes of response regulator HP166.
In order to identify promoter sequences which are recognized by response regulator HP166, the interaction of HP166 with DNA was analyzed by a magnetocapture assay based on Ni2+-NTA magnetic agarose beads. His6-HP166 was bound to the magnetic beads and then incubated with chromosomal DNA of H. pylori G27 which was digested with DraI to obtain an average fragment length of 500 bp. Interestingly, binding of DNA fragments to His6-HP166 was observed only when the protein was incubated with acetylphosphate prior to the addition of DNA. Analysis of the eluted DNA by agarose gel electrophoresis revealed four bands corresponding to DNA fragments at approximately 400 to 500, 300, 210, and 180 bp (data not shown). These fragments were isolated from the agarose gel after electrophoresis of the pooled eluates from three individual binding assays. Probably due to the small amount of DNA recovered from the gel, we could not clone the 400- to 500-bp and 210-bp fragments. Ligation of the 300- and 180-bp fragments into pBluescript SK yielded plasmids pGG1, pGG2, and pGG3.
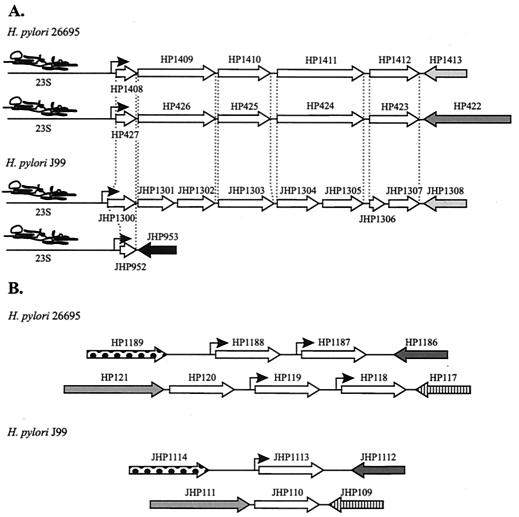
Sequence analysis of the insert of pGG1 and comparison to the genome sequence of H. pylori 26695 demonstrated that the 284-bp DraI fragment comprised the upstream region, as well as part of the coding sequence of ORF HP1408 (82.7% identity in a 309-bp overlap, positions 1477446 to 1477772 in the H. pylori 26695 genome). As determined by RT-PCR experiments (data not shown), ORF HP1408 is the first gene of an operon comprising ORFs HP1408, HP1409, HP1410, HP1411, and HP1412 (Fig. 3), all of which encode H. pylori-specific proteins whose functions are not known. Interestingly, two copies of this operon are present in the genome of H. pylori 26695 (HP1408 to HP1412 and HP427 to HP423), and both copies are located adjacent to a 23S rDNA locus. The second completely sequenced H. pylori strain, strain J99, contained an operon encoding orthologs of HP1408 to HP1412 (i.e., ORFs JHP1300 to JHP1307) (Fig. 3). Furthermore, an ortholog of HP427 (JHP952) is annotated in J99 and is preceded by an upstream region identical to the region upstream of JHP1300 to JHP1307. Digestion of the DNA insert in pGG1 with NspI, which cleaved the fragment at the position of the annotated translational start codon of HP1408, demonstrated that in the magnetocapture assay binding to His6-HP166 was conferred by the 73-bp subfragment comprising the upstream region of ORF HP1408 (data not shown).
FIG. 3.
Schematic representation of the genomic organization of the target genes whose promoter region is bound by HP166. (A) The genome of H. pylori 26695 contains two operons comprising the paralogous ORFs HP1408 to HP1412 and HP427 to HP423. The H. pylori J99 operon JHP1300 to JHP1307 encodes orthologs of HP1408 to HP1412; however, the J99 orthologs of HP1409, HP1411, and HP1412 are split into two separate ORFs (JHP1301 and JHP1302, JHP1304 and JHP1305, and JHP1306 and JHP1307, respectively). The ORFs adjacent to the operons HP1408 to HP1412 and JHP1300 to JHP1307 (i.e., HP1413 and JHP1308) are orthologs. ORF JHP925 is an ortholog of HP1408 and HP427 and is preceded by a promoter region identical to the promoter region that precedes JHP1300. Paralogous and orthologous ORFs in the operons are indicated by dotted lines. (B) Members of the HP119 gene family (HP118, HP119, HP120, HP1187, and HP1188) are clustered at two loci in the genome of H. pylori 26695. The J99 orthologs (JHP113 and JHP110) are located at the corresponding loci in the J99 genome; i.e., HP1189 and JHP1114, HP1186 and JHP1112, HP121 and JHP111, and HP117 and JHP109 are orthologous genes. Promoters regulated by HP166 are indicated by arrows. Orthologous ORFs flanking the HP166 target operons and genes are indicated by identical shading. The organization of operons encoding ORFs orthologous to HP1408 to HP1412 in H. pylori G27 and P76 is similar to the organization in strain 26695. The precise numbers of copies and organization of orthologs of HP119 in H. pylori G27 and P76 are not known.
pGG2 and pGG3 comprised 183-bp DNA inserts that exhibited 93.4% identity to each other and were homologous to the DNA region located immediately upstream of the translational start codon of ORF HP119 of H. pylori 26695 (98.3% identity in a 183-bp overlap with respect to the insert of pGG2, positions 130771 to 130953 in the H. pylori 26695 genome). HP119 belongs to a protein family which in H. pylori 26695 comprises five members (HP118, HP119, HP120, HP1187, and HP1188), while H. pylori J99 contains only two orthologous genes (ORFs JHP1113 and JHP110). This protein family is characterized by the presence of two different types of N-terminal sequences (one present in HP119 and JHP1113 and one present in HP118, HP120, HP1187, HP1188, and JHP110), a variable middle region composed of repeated sequence motifs with a predicted coiled coil structure, and an approximately 200-amino-acid C-terminal region which is almost the same in all members of the family. The functions of these H. pylori-specific proteins are not known. In the chromosome of H. pylori 26695 the paralogous ORFs are clustered at two different loci (HP118 to HP120 and HP1187 to HP1188) (Fig. 3) and, with the exception of ORF HP120, have almost identical upstream sequences, with similarity ranging up to position −160 with respect to the translational start codon of ORF HP1187.
Characterization of target promoters activated by response regulator HP166.
To identify the promoters of the putative target genes of HP166, primer extension analysis was performed. As unequivocal annealing of a primer oligonucleotide to ORF HP1408 was not feasible due to the presence of repeated sequence motifs, we decided to construct a transcriptional fusion with a promoterless gfp reporter gene. To do this, the upstream region of ORF HP1408 was amplified from chromosomal DNA of H. pylori strains G27, G25, G46, and G50, and the sequences of the putative promoter regions were compared. Binding of the 73-bp subfragment of the insert in pGG1 by His6-HP166 indicated that the promoter of ORF HP1408 is located in this sequence. Because the hexanucleotide motif with the highest level of similarity to a −10 promoter element in the 73-bp subfragment exhibited two deviations from the −10 box consensus sequence in the sequence derived from strain G27 but only one mismatch in the case of strains G25, G46, and G50, the 287-bp promoter fragment amplified from H. pylori G25 was chosen to construct the gfp fusion. Subsequently, in H. pylori G27 ORF1408 was replaced by the promoter-gfp fusion by allelic exchange. Primer extension analysis with a gfp-specific oligonucleotide was performed with RNA extracted from H. pylori G27/P1408-gfp, and this yielded a transcriptional start site at position +2 with respect to the annotated translational start codon of ORF HP1408 in the genome sequence of H. pylori 26695, thus placing the start codon of this ORF 31 bp farther downstream (Fig. 4A). The transcription initiation site corresponded to a −10 promoter element with the sequence TACAAT in H. pylori G25, which was not preceded by additional promoter elements with similarities to either the E. coli −35 sequence or the proposed H. pylori consensus sequence.
FIG. 4.
Determination of transcriptional start sites of ORFs HP1408 and HP119 by primer extension analysis. Total RNA extracted from H. pylori G27/P1408-gfp (A) and G27 (B) were hybridized to radiolabeled oligonucleotides gfpPE and HP119PE, respectively. Transcriptional start points are indicated by arrows on the right. Parts of the respective promoter sequences are shown on the left. The −10 promoter elements are enclosed in boxes. Sequencing reactions (lanes A, C, G, and T) were performed by using the oligonucleotides used in the primer extension experiments and plasmid pSL-P1408-gfp/km (A) or pSL-119PE2 (B) as the template.
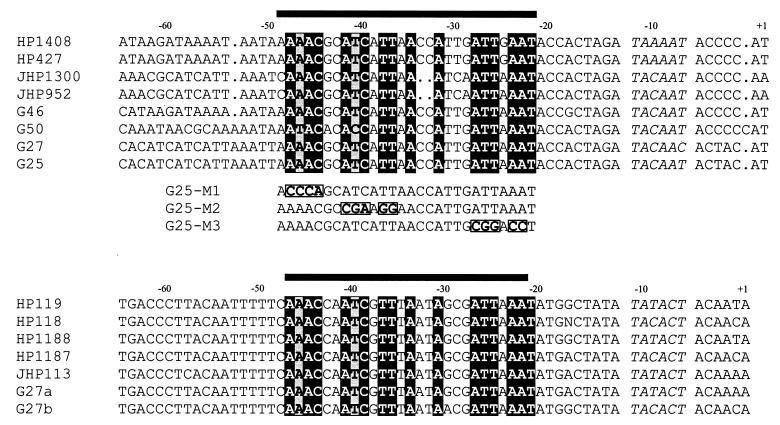
The promoter of ORF HP119 was mapped by primer extension analysis performed with RNA extracted from H. pylori G27 and was found to be located at positions −42 and −43 with respect to the translational start codon (Fig. 4B). PCR fragments that were 783 and 700 bp long and harbored the promoter regions of G27 genes orthologous to HP119 were obtained by using primer oligonucleotides derived from the sequence encoding the N terminus of HP119 and either ORF HP1189 or the sequence encoding the C-terminal part of HP120. The promoter sequences in the 783- and 700-bp PCR fragments were identical to the corresponding sequences in plasmids pGG2 and pGG3, respectively, and revealed −10 promoter elements of the sequences TATACT and TACACT, respectively (G27a and G27b in Fig. 5).
FIG. 5.
Alignment of the promoter sequences of ORFs positively regulated by response regulator HP166. The alignment shows the promoter sequences of ORFs HP1408 (upper part) and HP119 (lower part) and their orthologs derived from different H. pylori strains. The HP and JHP designations indicate sequences derived from the previously published genomes of H. pylori 26695 and J99, respectively. The promoter sequences of the HP1408 orthologs in strains G46, G50, G27, and G25 were obtained by performing PCR with primer pair Prom5-Prom3. The promoter sequences of the HP119 orthologs in H. pylori G27 were derived from the DNA inserts in plasmids pGG2 (G27a) and pGG3 (G27b). The numbers above the sequences indicate positions relative to the transcriptional start point. The −10 promoter elements are italicized. The bars indicate the regions which are protected from DNase I digestion in footprint experiments with His6-HP166 and the PHP1408 and PHP119 promoter probes. Nucleotides in the response regulator binding site which are identical in all of the sequences aligned are indicated by black shading, and nucleotides which are different in a single promoter sequence are indicated by gray shading. The sequences below the upper part of the promoter alignment (G25-M1, G25-M2, and G25-M3) represent the mutated response regulator binding sites in the PHP1408 promoter which were analyzed in DNase I footprint experiments (see text for details).
To investigate whether ORF HP120, which was under control of a different promoter, was transcribed, a 612-bp fragment comprising the upstream region of HP120 was fused to the promoterless gfp gene cloned into pHel2, and the resulting plasmid was transformed into H. pylori G27. Subsequently, 5′ RACE experiments performed with gfp-specific primers and RNA extracted from the transformed strain revealed a transcription initiation site at position −40 with respect to the translational start codon of ORF HP120. This transcriptional start site corresponded to a consensus −10 promoter element (TATAAT).
A comparison of the DNA sequences located immediately upstream of the −10 promoter elements of ORFs HP1408 and HP119 and their orthologs in several H. pylori strains revealed significant similarities in a 28-bp DNA segment which overlapped the −35 region (Fig. 5), indicating that this segment was a putative binding site of response regulator HP166. However, the conserved DNA segment did not contain obvious sequence motifs of dyad symmetry which are frequently involved in the binding of regulatory proteins.
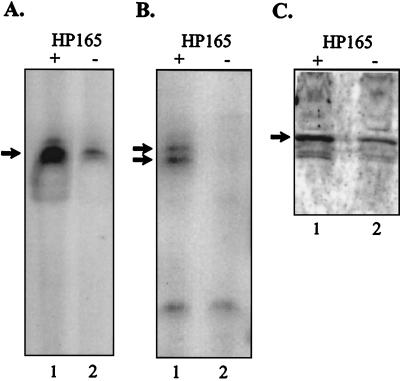
To study the effect of the HP166-HP165 two-component system on expression of ORFs HP1408 and HP119, the PHP1408-gfp fusion was introduced into the histidine kinase mutant G27/HP165::km, and primer extension experiments with the oligonucleotide primers complementary to gfp and ORF HP119 were performed by using RNA extracted from the resulting strains (G27/P1408-gfpΔ165 and G27/HP165::km, respectively). As shown in Fig. 6 transcription from the PHP1408 promoter was clearly reduced in the histidine kinase mutant (Fig. 6A), while an HP119-specific transcript was not detected in G27/HP165::km (Fig. 6B), suggesting that phosphorylated response regulator HP166 had a positive regulatory effect. Additionally, protein lysates were prepared from liquid cultures of strains G27/P1408-gfp and G27/P1408-gfpΔ165 grown to an optical density at 600 nm of 1.0 and were subjected to immunoblot analysis performed with a polyclonal antibody directed against GFP; the results confirmed that gfp expression in histidine kinase mutant G27/P1408-gfpΔ165 was reduced and therefore suggested that phosphorylated HP166 had a positive regulatory effect (Fig. 6C).
FIG. 6.
Analysis of expression of ORFs HP1408 and HP119. (A) Primer extension analysis of equal amounts of RNA extracted from H. pylori G27/P1408-gfp (lane 1) and G27/P1408-gfpΔ165 (lane 2) with oligonucleotide gfpPE. (B) Primer extension analysis of equal amounts of RNA extracted from H. pylori G27 (lane 1) and G27/HP165::km (lane 2) with oligonucleotide 119PE. (C) Western blot analysis of protein lysates prepared from H. pylori G27/P1408-gfp (lane 1) and G27/P1408-gfpΔ165 (lane 2). The membrane was probed with a polyclonal antiserum raised against GFP. The arrows indicate the positions of specific cDNA (A and B) and the GFP (C).
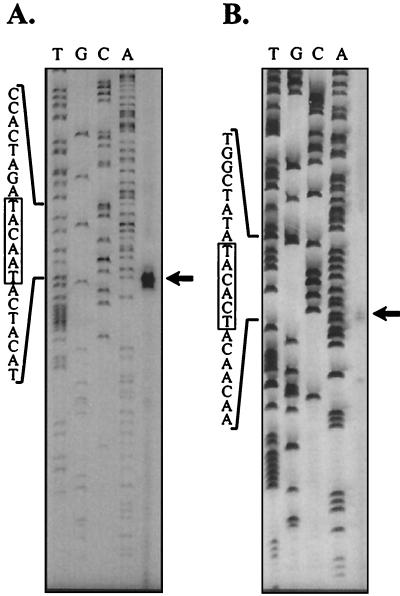
Response regulator HP166 binds to a 26-bp sequence motif in the promoters of ORFs HP1408 and HP119.
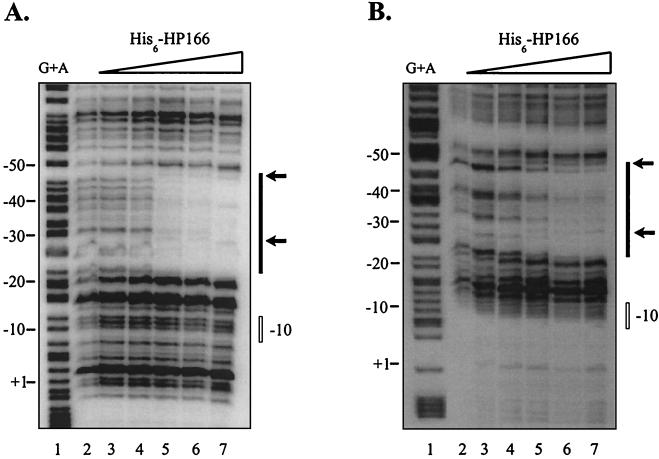
In order to define the binding site of response regulator HP166 in the promoter regions of ORFs HP1408 and HP119, DNase I footprint experiments were performed. Figure 7A shows the results of a footprint experiment carried out with a 360-bp DNA fragment derived from the PHP1408-gfp fusion in plasmid pSL-P1408-gfp/cm. Addition of 2 μg of His6-HP166 phosphorylated in vitro with acetylphosphate resulted in a region protected from DNase I digestion which extended from position −22 to position −49 with respect to the transcriptional start site and the appearance of two bands with enhanced DNase I sensitivity at sites within and delimiting the protected region. When the same experiment was performed with unphosphorylated His6-HP166, a similar footprint was obtained. However, the same degree of protection could be detected only after addition of 6 μg of protein (data not shown). When 2 μg of His6-HP166 treated with acetylphosphate was added to a 298-bp DNA fragment comprising the promoter region of ORF HP119, bands with enhanced DNase I sensitivity were detected, while a clear protected area ranging from position −22 to position −47 with respect to the transcriptional start site of ORF HP119 appeared when 6 μg of His6-HP166 was added (Fig. 7B). Addition of 6 μg of unphosphorylated His6-HP166 resulted in a similar footprint; however, DNase I-hypersensitive sites were not detected (data not shown). Again, the observed similarity of the binding affinities of His6-HP166 treated or not treated with acetylphosphate for the PHP1408 and PHP119 promoters might be attributable to the low degree of in vitro phosphorylation obtained upon incubation of His6-HP166 with acetylphosphate (see below). With both promoter probes the region protected from DNase I digestion was located at the same distance with respect to the −10 promoter elements and corresponded to the DNA segment showing pronounced sequence similarity in PHP1408 and PHP119 (Fig. 5), indicating that this DNA segment comprised the sequence motif required for binding of response regulator HP166.
FIG. 7.
DNase I footprint analysis of His6-HP166 on the PHP1408 (A) and PHP119 (B) promoters. Specific end-labeled fragments were incubated with different amounts of His6-HP166 that had been incubated with acetylphosphate. In panel A lanes 2 to 7 contained preparations to which 0, 0.5, 1, 2, 4, and 6 μg of His6-HP166, respectively, were added, and in panel B lanes 2 to 7 contained preparations to which 0, 0.5, 2, 4, 6, and 8 μg of His6-HP166, respectively, were added. The numbers on the left indicate nucleotide positions with respect to the transcriptional start site. The solid bars on the right indicate areas of DNase I protection, and the arrows indicate DNase I-hypersensitive sites. The open bars indicate the position of the −10 promoter element. Lane 1 contained a G+A sequence reaction mixture with the DNA probe used as a size marker (27).
As no direct or inverted repeat sequences were present in the sequence motif identified, site-directed mutagenesis of the PHP1408 promoter probe was performed in order to determine the nucleotides required for binding of HP166. As shown in Fig. 5, conserved AT-rich tetra- or hexanucleotide sequences located in the center or at the borders of the protected region in PHP1408 were mutagenized, and DNase I footprint experiments with His6-HP166 were carried out. When the AAAC sequence motif at the 5′ border of the protected region was changed to CCCA, a footprint similar to the footprint obtained with the wild-type PHP1408 promoter probe was observed, while binding of His6-HP166 was completely eliminated when the central ATCATT sequence motif was changed to CGAAGG (data not shown). Changing the 3′ ATTAAA motif to CGGACC led to a shortened footprint covering only 19 bp (data not shown), suggesting that the central region of the sequence motif protected in the wild-type PHP1408 promoter probe is essential for binding of the response regulator.
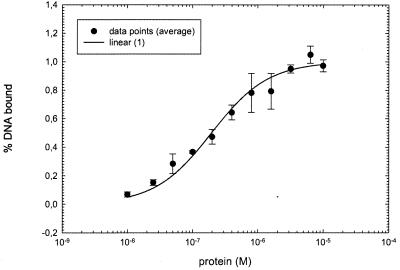
To confirm the binding of response regulator HP166 to the 26-bp sequence motif in the PHP1408 and PHP119 promoters and to investigate the effect of in vitro phosphorylation of His6-HP166 in more detail, DNA filter binding assays were carried out by using a 37-bp DNA probe which contained nucleotides −56 to −19 with respect to the transcriptional start site of ORF HP1408. His6-HP166 incubated with 50 mM acetylphosphate for 30 min bound to the oligonucleotide, and half-maximal binding was observed at a concentration of 200 nM (Fig. 8); the unphosphorylated protein exhibited a lower affinity for the DNA probe and half-maximal binding at a concentration of 780 nM. For both phosphorylated and unphosphorylated His6-HP166 the data points corresponded best to a binding isotherm fitted for linear binding to a single binding site. Quantification of the degree of in vitro phosphorylation by incubating His6-HP166 with radioactively labeled acetylphosphate demonstrated that only 2% of the protein molecules were phosphorylated under the experimental conditions used, suggesting that the rather low affinity of in vitro binding of His6-HP166 to the PHP1408 promoter might be a consequence of inefficient in vitro phosphotransfer. The binding affinity of His6-HP166 for the PHP1408 promoter could not be increased by prolonged incubation with acetylphosphate or treatment of His6-HP166 with phosphoramidate or carbamylphosphate prior to the binding reaction (data not shown).
FIG. 8.
Binding of response regulator His6-HP166 treated with acetylphosphate to a 37-bp oligonucleotide comprising the HP166 binding site of the PHP1408 promoter. Binding was determined on nitrocellulose filters as described in Materials and Methods. The data points were fitted for linear binding to a single binding site. The experiment was carried out three times, and the bars indicate standard deviations.
Deletion of the target genes positively regulated by HP166 has no effect on the growth characteristics of H. pylori P76.
As ORF HP166 proved to be an essential gene, we investigated whether this might be due to the essential functions of its target genes. As shown in Fig. 3, target genes of HP166 under control of PHP1408- and PHP119-like promoters are present at two different loci in the genomes of both H. pylori 26695 and J99. After verifying by PCR analysis that the genome organization in mouse-adapted H. pylori strain P76 is similar to that in H. pylori 26695, we constructed deletion mutants P76/Δ1408 and P76/Δ119. In P76/Δ1408 the regions between the 23S rDNA sequences and the orthologs of ORFs HP1413 and HP422 were replaced by the kanamycin and chloramphenicol resistance cassettes, respectively. P76/Δ119 harbors the kanamycin resistance cassette instead of the DNA located between the orthologs of ORFs HP1189 and HP1186 and the chloramphenicol resistance cassette instead of the DNA located between the orthologs of ORFs HP121 and HP117. Both mutants were indistinguishable in terms of growth characteristics from the P76 wild-type strain. A Southern blot analysis performed with chromosomal DNA extracted from mutants P76/Δ1408 and P76/Δ119 and probes derived from the 5′ end of ORF HP1409 and the 3′ end of ORF HP119, respectively, showed that additional orthologs of the HP1408-HP1412 operon and of ORF HP119 were not present in the genome of H. pylori P76 (data not shown).
DISCUSSION
Sequencing of the H. pylori genome revealed a remarkably low number of transcriptional regulators, which has been interpreted as an adaptation of this microorganism to its only known ecological niche, the human stomach. These regulators include three two-component histidine kinases with cognate response regulators, as well as two orphan response regulators. In this study we identified target genes regulated by the two-component system composed of histidine kinase HP165 and response regulator HP166 and obtained evidence that HP166 acts as both a transcriptional activator and a repressor protein.
Response regulator proteins frequently regulate their own expression, as well as expression of a cotranscribed cognate histidine kinase. Accordingly, binding of His6-HP166 downstream of the PHP166 promoter in vitro (Fig. 1) suggests that there is a negative autoregulatory loop. As transcription of PHP166 is derepressed in a knockout mutant with a mutation of the cognate histidine kinase HP165 (Fig. 2), response regulator HP166 seems to act as a repressor of its own expression in its phosphorylated state. On the other hand, transcription of the PHP1408 and PHP119 promoters, which were identified as target promoters of response regulator HP166 in this study, seems to be enhanced by phosphorylation of the regulator protein, as indicated by the reduced transcription of ORFs HP1408 and HP119 in the histidine kinase mutants G27/P1408-gfpΔ165 and G27/HP165::km (Fig. 6). As the observed reduction in transcription was rather weak when RNA preparations from H. pylori grown under in vitro culture conditions were used, it is possible that histidine kinase HP165 is not fully activated under these environmental conditions, and therefore, more pronounced transcriptional activation of ORFs HP1408 and HP119 would be expected in vivo when the appropriate environmental stimulus is present.
The finding that phosphorylated HP166 had the opposite regulatory effect on its own transcription suggests that tightly balanced synthesis of HP166 is required in response to environmental stimuli perceived by histidine kinase HP165. Furthermore, in this context it should be noted that ORF HP166 proved to be an essential gene which could not be inactivated by knockout mutagenesis unless an additional copy of the gene was present (28), while the cognate histidine kinase HP165 is easily dispensable under in vitro culture conditions (5). A possible explanation for this observation is that the unphosphorylated response regulator HP166 is necessary for transcription of target genes essential for in vitro growth, while an additional set of genes is activated or repressed under environmental conditions which trigger the histidine kinase activity of sensor protein HP165 and accordingly lead to the phosphorylation of HP166. This hypothesis is supported by the finding that a mutated derivative of ORF HP166 encoding a response regulator protein with a D52N substitution lacking the phosphorylation site is able to complement a knockout mutant of wild-type ORF HP166 to cell viability (J. Schär, P. Dietz, and D. Beier, unpublished data).
Therefore, the promoters of the two subsets of target genes should contain response regulator binding sites with different affinities (high-affinity binding sites which are recognized by the unphosphorylated protein and lower-affinity binding sites whose binding requires phosphorylation of HP166). The promoter composition of multiple binding sites that exhibit different affinities for the response regulator protein is a regulatory mechanism encountered in members of the OmpR response regulator subfamily which contain a winged helix-turn-helix motif in the output domain (26). For example, in E. coli OmpR regulates expression of the porins OmpF and OmpC in response to the osmolarity of the growth medium, which governs the phosphorylation state of OmpR by modulating the histidine kinase and/or phosphatase activity of the cognate sensor protein EnvZ (35, 46). This is achieved by cooperative binding of OmpR to multiple OmpR binding sites with different affinities in the ompF and ompC promoters, whose degrees of occupancy depend on the concentration of phosphorylated OmpR and consequently lead either to expression of ompF or to repression of ompF and expression of ompC (20, 33). In case of the TorRS two-component system of E. coli (22, 38), the TorR response regulator represses its own expression by binding to a high-affinity TorR binding site located immediately downstream of the −10 box of the TorR promoter (3). Binding to this site is independent of TorR phosphorylation, as repression of torR is also observed in a torS mutant and in the absence of trimethylamine N-oxide, which is the stimulus that triggers TorS kinase activity (3). Transcription of the torCAD operon, which is divergently transcribed from torR, is achieved by the subsequent binding of the phosphorylated response regulator to two additional tor boxes with low binding affinity located in the short intergenic region between torR and torCAD (3, 39). As repression of ORF HP166 seems to be enhanced by phosphorylation of the response regulator, we propose that this negative feedback loop might ensure the short-term expression of target genes requiring phosphorylated HP166 for transcription. Simultaneously, this mechanism may eliminate putatively harmful overexpression of target genes essential for in vitro growth by limiting the total amount of response regulator protein as a rapid response to potentially short-term environmental changes.
The binding sites for response regulator HP166 which were identified in the PHP1408 and PHP119 promoters and downstream of the PHP166 promoter by DNase I footprint analysis are highly asymmetric. While the 26-bp motif which is protected in the PHP1408 and PHP119 promoters is well conserved (the residues at 14 of 26 positions are identical) in the paralogous loci of the same H. pylori strain and also in orthologous loci of different strains (Fig. 5), the 20-bp repressor binding site located downstream of the PHP166 promoter (Fig. 1A) exhibits only a limited degree of similarity to the activator binding sites; an AARYNNATCRTT motif is conserved in the three HP166 binding sites identified. In fact, mutagenesis of the ATCRTT motif interfered with binding of the response regulator protein to the PHP1408 promoter in DNase I footprint experiments, demonstrating that this motif is essential for the interaction of HP166 with its binding site. However, this core motif seems not to be sufficient for binding of HP166 since the upstream region of ORF HP1408 contains several repeats of an ATCAT(A/T) motif, which are not protected in DNase I footprint experiments even in the presence of elevated amounts of protein. Most of the members of the OmpR response regulator subfamily for which DNA recognition sites have been determined appear to bind direct repeat DNA sequences. However, while the pho box representing the binding site of two molecules of the E. coli PhoB response regulator is composed of a perfect direct repeat sequence (25), OmpR binding sites exhibit only limited sequence similarity. Huang and Igo (19) demonstrated that a single OmpR binding site spans approximately 18 bp and is composed of two asymmetric half sites, each of which contains a highly conserved G · C base pair. The ompF promoter contains three adjacent OmpR binding sites (sites F1 to F3), and it has been shown that OmpR binds to the F1 site as a tandemly arranged dimer (17). The asymmetry of the binding sites of response regulator HP166 is reminiscent of the rather degenerate structure of the OmpR binding sites.
The binding isotherm for binding of HP166 to the 26-bp motif in the PHP1408 promoter which is protected from DNase I digestion (Fig. 8) revealed the rather weak DNA binding affinities of HP166 under the experimental conditions used in vitro; the binding affinities of His6-HP166 treated with acetylphosphate and of the untreated protein differed approximately fourfold. This observation might have been due to the fact that incubation of His6-HP166 with acetylphosphate resulted in phosphorylation of only 2% of the response regulator molecules (data not shown). However, the finding that in spite of the low efficiency of in vitro phosphorylation treatment of His6-HP166 with acetylphosphate reproducibly resulted in an increase in DNA binding affinity, together with the observation that expression of target genes HP1408 and HP119 was reduced in H. pylori strains carrying a knockout mutation of the cognate histidine kinase HP165, argues in favor of the hypothesis that HP1408 and HP119 are positively regulated by the phosphorylated response regulator.
The genes identified so far that appear to be activated by response regulator HP166 encode H. pylori-specific proteins whose functions are not known. Interestingly, several copies of both sets of genes (i.e., the HP1408-HP1412 operon and the members of the HP119 gene family) are present in the chromosome of H. pylori 26695, and the numbers of copies of the genes in H. pylori strains 26695 and J99 are different (1, 45). H. pylori 26695, J99, and G27 all contain a gene belonging to the HP119 family (i.e., HP120 and JHP110) whose expression seems not to be regulated by the HP166-HP165 two-component system, as the PHP120 promoter does not contain sequence motifs resembling the binding site of HP166 which was identified in the PHP1408 and PHP119 promoters. However, since the PHP120 promoter was mapped by 5′ RACE analysis and the amount of transcript was not quantified in the G27 wild-type strain and the histidine kinase mutant G27/HP165::km, convincing evidence for constitutive expression of the PHP120 promoter is missing. Interestingly, in H. pylori G27 the gene corresponding to ORF HP120 contains a frameshift mutation (Dietz and Beier, unpublished observations). Knockout mutagenesis of the genes orthologous to HP1408 to HP1412 and HP119 could easily be performed in H. pylori P76 without changing its growth behavior, demonstrating that these genes do not contribute to the lethal effect observed upon inactivation of ORF HP166. Analysis of the mutants P76/Δ1408 and P76/Δ119 in the mouse model should provide information regarding a putative role of the target genes of HP166 in the infectious process.
Acknowledgments
We thank R. Gross for critically reading the manuscript and V. Weiss and A. Bock for helpful discussions.
This work was supported by grants Be 1543/2-1 and Be 1543/2-3 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S., C.-Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J.-T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansaldi, M., G. Simon, M. Lepelletier, and V. Mejean. 2000. The TorR high-affinity binding site plays a key role in both torR autoregulation and torCAD operon expression in Escherichia coli. J. Bacteriol. 182:961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179:4676–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bereswill, S., F. Lichte, S. Greiner, B. Waidner, F. Fassbinder, and M. Kist. 1999. The ferric uptake regulator (Fur) homologue of Helicobacter pylori: functional analysis of the coding gene and controlled production of the recombinant protein in Escherichia coli. Mol. Microbiol. Immunol. 188:31–40. [DOI] [PubMed] [Google Scholar]
- 7.Bereswill, S., S. Greiner, A. H. M. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubert, A., Z. Sokolovic, S.-K. Chun, L. Papatheodorou, A. Simm, and W. Goebel. 1999. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261:323–336. [DOI] [PubMed] [Google Scholar]
- 9.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38. [DOI] [PubMed] [Google Scholar]
- 10.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, and R. Rappuoli. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174:2466–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickneite, C., R. Böckmann, A. Spory, W. Goebel, and Z. Sokolovic. 1998. Differential interaction of the transcription factor PrfA and the PrfA-activating factor (Paf) of Listeria monocytogenes with target sequences. Mol. Microbiol. 27:915–928. [DOI] [PubMed] [Google Scholar]
- 13.Dong, Q., D. Hyde, C. Herra, C. Kean, P. Murphy, C. O. O’Morain, and M. Bucklcy. 2001. Identification of genes regulated by prolonged acid exposure in Helicobacter pylori. FEMS Microbiol. Lett. 196:245–249. [DOI] [PubMed] [Google Scholar]
- 14.Evans, D. J., Jr., D. G. Evans, T. Takemura, H. Nakano, H. C. Lampert, D. Y. Graham, D. N. Granger, and P. R. Kvietys. 1995. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 63:2213–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez-Duarte, O. G., B. Lucas, Z.-X. Yan, K. Panthel, R. Haas, and T. F. Meyer. 1998. Protection of mice against gastric colonization by Helicobacter pylori by single oral dose immunization with attenuated Salmonella typhimurium producing urease subunits A and B. Vaccine 16:460–471. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin, C. S. 1997. Helicobacter pylori gastritis, peptic ulcer, and gastric cancer: clinical and molecular aspects. Clin. Infect. Dis. 25:1017–1019. [DOI] [PubMed] [Google Scholar]
- 17.Harrison-McMonagle, P., N. Denissova, E. Martinez-Hackert, R. H. Ebright, and A. M. Stock. 1999. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J. Mol. Biol. 285:555–566. [DOI] [PubMed] [Google Scholar]
- 18.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519–528. [DOI] [PubMed] [Google Scholar]
- 19.Huang, K.-J., and M. M. Igo. 1996. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J. Mol. Biol. 262:615–628. [DOI] [PubMed] [Google Scholar]
- 20.Huang, K.-J., C.-Y. Lan, and M. M. Igo. 1997. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. USA 94:2828–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilver, D., A. Arnqvist, J. Ögren, I.-M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373–377. [DOI] [PubMed] [Google Scholar]
- 22.Jourlin, C., A. Bengrine, M. Chippaux, and V. Mejean. 1996. An unorthodox sensor protein (TorS) mediates the induction of the tor structural genes in response to trimethylamine N-oxide in Escherichia coli. Mol. Microbiol. 20:1297–1306. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35 recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 232:406–418. [DOI] [PubMed] [Google Scholar]
- 24.Labigne-Roussel, A., P. Coutcoux, and L. Tompkins. 1988. Gene disruption and replacement as feasible approach for mutagenesis of Campylobacter jejuni. J. Bacteriol. 170:1704–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino, K., M. Amemura, T. Kawamoto, S. Kimura, H. Shinagawa, A. Nakata, and M. Suzuki. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259:15–26. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301–312. [DOI] [PubMed] [Google Scholar]
- 27.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel, T. K., K. C. DeWalt, N. R. Salama, and S. Falkow. 2001. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter 6:15–23. [DOI] [PubMed] [Google Scholar]
- 29.Odenbreit, S., M. Till, D. Hofreuter, G. Faller, and R. Haas. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537–1548. [DOI] [PubMed] [Google Scholar]
- 30.Odenbreit, S., J. Püls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497–1500. [DOI] [PubMed] [Google Scholar]
- 31.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267–1271. [DOI] [PubMed] [Google Scholar]
- 32.Perraud, A.-L., B. Kimmel, V. Weiss, and R. Gross. 1998. Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol. Microbiol. 27:875–887. [DOI] [PubMed] [Google Scholar]
- 33.Pratt, L. A., and T. J. Silhavy. 1995. Porin regulon of Escherichia coli, p. 105–127. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 34.Reyrat, J.-M., V. Pelicic, E. Papini, C. Montecucco, R. Rappuoli, and J. L. Telford. 1999. Towards deciphering the Helicobacter pylori cytotoxin. Mol. Microbiol. 34:197–204. [DOI] [PubMed] [Google Scholar]
- 35.Russo, F. D., and T. J. Silhavy. 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222:567–580. [DOI] [PubMed] [Google Scholar]
- 36.Satin, B., G. Del Giudice, V. Della Bianca, S. Dusi, C. Laudanna, F. Tonello, D. Kelleher, R. Rappuoli, C. Montecucco, and F. Rossi. 2000. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 191:1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheridan, R. C., J. F. McCullough, and Z. T. Wakefield. 1971. Phosphoramidic acid and its salts. Inorg. Synth. 13:23–26. [Google Scholar]
- 38.Simon, G., V. Mejean, C. Jourlin, M. Chippaux, and M. C. Pascal. 1994. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of trimethylamine N-oxide reductase. J. Bacteriol. 176:5601–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon, G., C. Jourlin, M. Ansaldi, M.-C. Pascal, M. Chippaux, and V. Mejean. 1995. Binding of the TorR regulator to cis-acting direct repeats activates tor operon expression. Mol. Microbiol. 17:971–980. [DOI] [PubMed] [Google Scholar]
- 40.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spohn, G., and V. Scarlato. 1999. The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol. Microbiol. 34:663–674. [DOI] [PubMed] [Google Scholar]
- 42.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA 97:1263–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telford, J. L., P. Ghiara, M. Dell’Orco, M. Comanducci, D. Burroni, M. Bugnoli, M. F. Tecce, S. Censini, A. Covacci, Z. Xiang, E. Papini, C. Montecucco, L. Parente, and R. Rappuoli. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its role in gastric disease. J. Exp. Med. 179:1653–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. [DOI] [PubMed] [Google Scholar]
- 46.van Alphen, W., and B. Lugtenberg. 1977. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J. Bacteriol. 131:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanet, A., L. Marsan, A. Labigne, and M.-F. Sagot. 2000. Inferring regulatory elements from a whole genome. An analysis of Helicobacter pylori ς80 family of promoter signals. J. Mol. Biol. 297:335–353. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23–28. [DOI] [PubMed] [Google Scholar]
- 49.Weiss, V., F. Claverie-Martin, and B. Magasanik. 1992. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc. Natl. Acad. Sci. USA 89:5088–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang, Z., S. Censini, P. F. Bayelli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]