Abstract
A novel Leuconostoc mesenteroides NRRL B-1299 dextransucrase gene, dsrE, was isolated, sequenced, and cloned in Escherichia coli, and the recombinant enzyme was shown to be an original glucansucrase which catalyses the synthesis of α-1,6 and α-1,2 linkages. The nucleotide sequence of the dsrE gene consists of an open reading frame of 8,508 bp coding for a 2,835-amino-acid protein with a molecular mass of 313,267 Da. This is twice the average mass of the glucosyltransferases (GTFs) known so far, which is consistent with the presence of an additional catalytic domain located at the carboxy terminus of the protein and of a central glucan-binding domain, which is also significantly longer than in other glucansucrases. From sequence comparison with family 70 and α-amylase enzymes, crucial amino acids involved in the catalytic mechanism were identified, and several original sequences located at some highly conserved regions in GTFs were observed in the second catalytic domain.
Glucansucrase enzymes from oral streptococci, Leuconostoc mesenteroides strains, and some Lactobacillus and Neisseria sp. catalyze the transfer of glucosyl residues from sucrose to synthesize α-d-glucopyranosyl homopolymers and oligomers. When sucrose is the sole substrate, high-molecular-weight polymers are obtained. Depending on the glucansucrase-producing strain, the synthesized glucans differ in size and structure. When efficient acceptors, such as maltose or isomaltose, are added to the reaction medium, glucansucrases catalyze the synthesis of low-molecular-weight oligosaccharides and the regiospecificity of several dextransucrases (type of linkages) from the Leuconostoc genus is conserved in oligosaccharide synthesis (8, 13, 41, 45).
To date, 17 glucosyltransferase (GTF)-encoding genes from Streptococcus spp., 8 glucansucrase-encoding genes from L. mesenteroides, and 1 gene from Lactobacillus reuteri have been cloned (for reviews, see references 3, 16, 32, and 59). Sequence information shows that they are closely related and share a common structure. These genes code for large enzymes with an average molecular mass of 160,000 Da composed of two main domains: an N-terminal conserved catalytic core of about 900 amino acids and a C-terminal domain covering 300 to 400 residues thought to be responsible for glucan binding and constituted by a series of repeated units (32). In addition, biochemical studies revealed that glucansucrases share many mechanistic features with amylolytic enzymes (15). Structural homologies were later confirmed by amino acid sequence comparison with glucoside hydrolases from family 13 (20). In family 13, the catalytic domain is formed of eight β-sheets alternating with eight α-helices, conferring a (β/α)8 barrel structure (55). Two structure predictions (9, 26) concluded that glucansucrase enzymes also possess a catalytic (β/α)8 barrel domain. However, MacGregor et al. (26) observed that well-recognized sequence segments appear in a different order, which tends to show that the β/α barrel elements are circularly permutated. This led to a new classification of the glucansucrase enzymes in family 70 of the glycoside hydrolases on the CAZy website (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html). The only exception is amylosucrase from Neisseria polysaccharea, which belongs to family 13 since its structure is not circularly permutated and is closely related to the α-amylase structure with a much lower molecular size than that of the glucansucrase enzymes (40, 52).
Throughout family 13, sequence alignment and site-directed mutagenesis enabled the location of several residues known to be involved in the formation of the glucosyl enzyme intermediates and structural data confirmed the involvement of these amino acids in the α-retaining catalytic process of glycoside hydrolases (57).
Nevertheless, key elements involved in the selectivity towards an acceptor molecule or in the enzyme regiospecificity still remain unclear. Thus, cloning and sequencing of new and original dextransucrase genes, as well as studies of the reaction products and catalytic parameters of the enzymes expressed, are necessary to improve the understanding of the various specificities.
Among glucansucrases, L. mesenteroides NRRL B-1299 is known to produce a highly branched dextran, which is very unusual since it contains 27 to 35% α-1,2 branch linkages as well as a limited amount of α-1,3 branch linkages (24, 48). The dextransucrase activity of L. mesenteroides NRRL B-1299 was also used in acceptor reactions with maltose in order to obtain oligosaccharides with α-1,2 osidic bonds (39, 42). These α-1,2 gluco-oligosaccharides resist hydrolysis by digestive enzymes in animals and humans because of the configuration of their osidic bonds and selectively stimulate intestinal microflora such as Bifidobacterium sp., Lactobacillus sp., or Bacteroides sp. (10, 58). Thus, these molecules correspond to the definition of prebiotic agents which are food ingredients that are potentially beneficial to the health of consumers (44). This prebiotic effect is already exploited in animal and human nutrition as well as in dermocosmetic applications (37).
According to previous observations describing the existence of more than one glucansucrase in L. mesenteroides NRRL B-1299 (12, 23, 24), two dextransucrase genes from L. mesenteroides NRRL B-1299 have already been cloned. The dsrA gene encodes an intracellular dextransucrase DSR-A, which synthesizes a dextran containing 87% α-1,6 linkages and 13% α-1,3 linkages (29). The second gene, dsrB, corresponds to an enzyme responsible for α-1,6 bond synthesis only (31). However, strategies based on the design of degenerate oligonucleotides from highly conserved sequences in GTF-encoding genes did not permit the gene encoding the α-1,2 synthesizing dextransucrase to be cloned. Screening of the proteins can be successfully applied to isolate new original dextransucrase genes, as it was shown for the alternansucrase of L. mesenteroides NRRL B-1355 (3).
Using this strategy, we report here the cloning and sequencing of dsrE, a new dextransucrase gene encoding an enzyme catalyzing the synthesis of dextran and oligosaccharides containing α-1,2 linkages. The original structure of DSR-E, exhibiting two catalytical domains, is discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains were stored at −80°C in 15% glycerol.
L. mesenteroides NRRL B-1299 (National Center for Agricultural Utilization Research, Peoria, Ill.) was grown in a rotary shaker at 27°C at 200 rpm in standard medium (40 g of sucrose per liter, 20 g of potassium hydrogen phosphate per liter, 20 g of yeast extract per liter, 0.2 g of MgSO4 · 7H2O per liter, 0.01 g of MnSO4 · H2O per liter, 0.01 g of NaCl per liter, 0.02 g of CaCl2 per liter, 0.01 g of FeSO4 · 7H2O per liter) with an adjusted pH of 6.9.
Escherichia coli strains DH5α and JM109 were grown in Luria-Bertani (LB) medium. Selection of strains with cloned inserts in pUC18 or pGEM-T Easy was done on agar plates with 100 μg of ampicillin per ml, 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml.
Plasmid pBAD/TOPO Thiofusion (Invitrogen) was used for the cloning and expression of dsrE in E. coli TOP10 cells grown in LB medium supplemented with 50 μg of ampicillin per ml.
Digested and dephosphorylated pUC18 plasmid was purchased from Amersham Biosciences and used for standard cloning. PCR product cloning requires the pGEM-T Easy plasmid (Promega) for DNA fragments less than 2 kbp and the TOPO-XL plasmid (Invitrogen) for larger fragments.
Gel electrophoresis and enzyme detection.
After a 7-h L. mesenteroides NRRL B-1299 culture, the broth was centrifuged (4,000 × g, 4°C, 30 min). Extracellular insoluble dextransucrase activity probably bound to the cell wall was recovered in the cell pellet (11), concentrated 10-fold in sodium acetate buffer (20 mM, pH 5.4), and heated for 5 min at 95°C with the loading buffer (62.5 mM Tris HCl, 4% sodium dodecyl sulfate [SDS], 6 M urea, 0.01% bromophenol blue, and 200 mM β-mercaptoethanol). Three hundred microliters of the mixture was loaded onto a 7% (mass/vol) polyacrylamide gel. After separation, total proteins were stained with amido black, and dextransucrase enzymes, which remain active after a short renaturation in 20 mM sodium acetate buffer, pH 5.4, were detected in situ following dextran synthesis and a polymer stain reaction with periodic acid-Schiff base (15). Bands corresponding to active dextransucrase were excised and incubated separately in a 2-ml reaction mixture containing 100 g of sucrose per liter, 50 g of maltose per liter, and 20 mM sodium acetate buffer. After a 24-h reaction corresponding to sucrose exhaustion, the reaction was stopped by heating the mixture at 95°C for 5 min and the medium was centrifuged for 5 min at 15,000 × g to eliminate the insoluble dextran. Reverse-phase chromatography (C18 column, Ultrasep 100, 6-μm particle size, 5 by 300 mm; Bishoff Chromatography) was carried out with ultrapure water as the eluant at a constant flow rate of 0.5 ml · min−1. Each sample was analyzed within 30 min at room temperature, with oligosaccharides detected by refractometry. Protein sequencing on the selected band was performed by the Laboratoire de Microséquençage, Institut Pasteur, Paris, France.
Nucleic acid isolation and manipulation.
E. coli plasmid isolation and L. mesenteroides genomic DNA purification were done with the QiaPrep Spin Plasmid kit and the Blood and Cell Culture DNA Maxi kit (Qiagen), respectively. DNA manipulation used standard methods (28). Restriction and modification enzymes were purchased from New England Biolabs or Gibco BRL and used according to the manufacturer's recommendations.
PCR amplification of homologous probe.
Following protein sequencing, two selected peptides (29-FYFESGK and 24-FESQNNNP) were used to synthesize degenerate oligonucleotides (Isoprim) (Table 1).
TABLE 1.
Oligoprimers used for PCR amplification
| Designation | Descriptiona | Sequence (5′-3′)b |
|---|---|---|
| 29-dir | FYFESGK | TT(C/T)TA(C/T)TT(C/T)GA(A/G)TCAGG(C/G)AA(A/G) |
| 24-inv | FESQNNNP | (T/G)GG(G/A)TT(G/A)TT(G/A)TTTTGTGA(T/C)TCAAA |
| IPCR-rev | nt 6334-6363 | CCCTTTACAAGCTGATTTTGCTTATCTGCG |
| IPCR-dir | nt 8876-8907 | GGGTCAAATCCTTACTATACATTGTCACACGG |
| pBAD-PS/ZV-dir | nt 677-706 | GCCATGGCAAATACGATTGCAGTTGACACG |
| pBAD-ent-rev | nt 8482-8505 | AATTTGAGGTAATGTTGATTTATC |
nt, nucleotides.
Primer sequences are designed from the noncoding strand (IPCR-dir or ECHO-dir) or from the complementary strand (IPCR-rev or ECHO-inv).
A 666-bp fragment was generated by PCR with a Perkin-Elmer thermal cycler model 2400 and 50 ng of genomic DNA, 10 μM of primer 29-dir, 10 μM of 24-inv, 250 μM concentrations of each deoxynucleoside triphosphate, and Taq polymerase (Sigma) (25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 5 min).
Southern hybridization and genomic libraries of L. mesenteroides NRRL B-1299.
Chromosomal DNA from L. mesenteroides NRRL B-1299 was digested to completion with several restriction enzymes and separated on agarose gels, and genomic libraries were transferred to Hybond N+ nylon membranes (Amersham Biosciences). Hybridization was performed with the radiolabeled ([α-32P]dATP; Amersham Biosciences) 666-bp PCR fragment. The labeling reaction was performed with the Mega-Prime DNA labeling system kit (Amersham Biosciences) followed by purification of the probe on MicroSpin columns S-200 HR (Amersham Biosciences). Prehybridization and hybridization were performed with high stringency conditions (65°C, overnight) according to standard methods (28).
Inverse PCR.
L. mesenteroides NRRL B-1299 genomic DNA was digested by BclI or EcoRV under the conditions recommended by the supplier and, after recircularization, used as a template in an inverse PCR (Extrapol II DNA polymerase [Eurobio]) (25 cycles of 94°C for 30 s, 51°C for 30 s, and 72°C for 3 min). The two primers were designed according to the pSB2 insert sequence (Table 1).
DNA sequencing and sequence analysis.
DNA fragment sequencing was carried out by Genome Express in both directions. Nucleotide sequence analyses were performed by using the Open Reading Frame (ORF) Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), Blast (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) (2), ClustalW (http://www2.ebi.ac.uk/clustalw) (56), PRODOM (http://protein.toulouse.inra.fr/prodom.html) (7), PFAM (http://pfam.wustl.edu/hmmsearch.shtml) (5), and SAPS (http://bioweb.pasteur.fr/seqanal/interfaces/saps.html) (6) internet programs.
Protein expression with the pBAD cloning system (Invitrogen).
An 8,385-bp fragment, containing the dsrE gene deleted from the sequence encoding the signal peptide, was generated by PCR with primers pBAD-PS/ZV-dir and pBAD-rev (Table 1). After purification, the PCR product was directly ligated into the pBAD/TOPO Thiofusion vector. Transformed E. coli TOP10 clones integrating a plasmid with the dsrE gene cloned into the proper orientation, that is, downstream of the arabinose promoter and in frame with the thioredoxin tag (N-terminal) and the V5 epitope and histidine tag (C-terminal), were selected. Correct construction of the plasmid containing the dsrE gene was confirmed by sequence analysis, showing that a single mutation occurred, which resulted in a D10R mutation in the variable domain.
Cells of E. coli TOP10 harboring pBAD-dsrE were grown in LB medium for 4 h after induction with 0.002% (mass/vol) arabinose. The pellet was resuspended at a final optical density at 600 nm (OD600) of 80 in 20 mM sodium acetate buffer (pH 5.4) and 1% (vol/vol) Triton X-100 with 1 mM phenylmethylsulfonyl fluoride to prevent proteolysis in the cell extract after cell disruption by sonication.
Enzyme assays.
Enzyme reactions were assayed under standard conditions at 30°C, in 20 mM sodium acetate buffer (pH 5.4), 0.05 g of CaCl2 per liter, 1 g of NaN3 per liter, and 100 g of sucrose per liter.
The DSR-E activity was determined by measuring the release of reducing sugars with the di-nitro-salicylic (DNS) assay (54), with 1 U defined as the amount of enzyme which catalyzes the formation of 1 μmol of fructose · min−1 under standard conditions.
Oligosaccharides were synthesized in reaction medium containing 100 g of maltose per liter, 200 g of sucrose per liter, and either 0.5 U of recombinant DSR-E from raw cellular extract per liter or 0.5 U of wild-type extracellular cell wall-associated glucansucrase from L. mesenteroides NRRL B-1299 per liter.
Nucleotide sequence accession number.
The nucleotide and deduced amino acid sequences of dsrE have been submitted to the EMBL nucleotide sequence database under accession number AJ430204.
RESULTS
Isolation, micropurification, and peptide microsequencing of an active α-1,2 linkage-synthesizing dextransucrase.
SDS-polyacrylamide gel electrophoresis (PAGE) analysis of cell wall-associated proteins of L. mesenteroides NRRL B-1299 revealed the presence of four main bands exhibiting GTF activity with molecular masses between 195 and 283 kDa.
Oligosaccharide syntheses were then performed with the excised active bands, and with the tested enzymes, all exhibited the ability to synthesize α-1,2 linkages. In order to ensure no contamination with forms that could be degraded by proteolysis, the upper band corresponding to the protein at 283 kDa was selected.
This band was then dried and used for protein trypsin digestion and peptide microsequencing.
Cloning and sequencing of the dsrE gene.
Following peptide sequencing, degenerate primers (Table 1), designed according to the codon frequency table of dsr genes from L. mesenteroides NRRL B-1299, were synthesized. They allowed the amplification of a 666-bp DNA fragment named probe 1 (Fig. 1). After DNA sequencing, the corresponding 222-amino-acid sequence was shown to belong to a new glucansucrase, the peptide sharing 35% identity with DSR-S from L. mesenteroides NRRL B-512F and 36% identity with DSR-B from L. mesenteroides NRRL B-1299. Sequence comparison located the peptide at the junction between the highly variable region and the catalytic domain of glucansucrases.
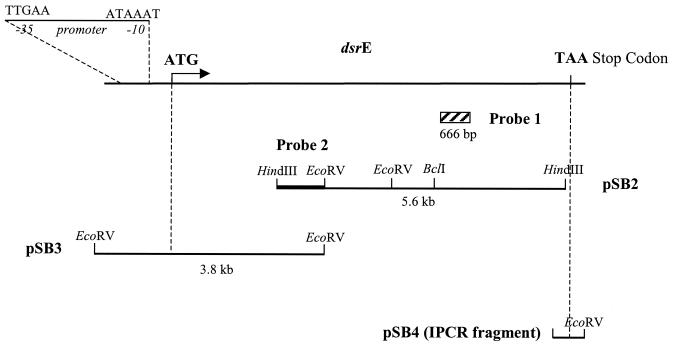
FIG. 1.
Cloning of the dsrE gene from L. mesenteroides NRRL B-1299. Plasmids pSB2 and pSB3 were isolated by screening of genomic libraries with probes 1 and 2, respectively. Plasmid pSB4 contains the inverted PCR fragment obtained with primers IPCR-rev and IPCR-dir.
A first HindIII genomic library was therefore screened with probe 1, and one recombinant plasmid, pSB2, containing a 5.6-kbp insert was purified. Sequence analysis of the HindIII insert revealed an ORF covering the whole fragment.
Then an EcoRV genomic library was screened with a HindIII-EcoRV probe (probe 2), which was isolated from the N terminus of the 5.6-kbp pSB2 insert (Fig. 1). The recombinant plasmid pSB3, positively tested by dot blot, carried a 3.8-kbp insert which, after sequencing, was shown to contain the initiation codon and the promoter region of the new dextransucrase gene, designated dsrE.
Inverse PCR was performed with divergent oligonucleotide primers designed from the previously described sequence of the pSB2 insert on self-ligated L. mesenteroides NRRL B-1299 genomic DNA digested either by BclI or by EcoRV. A unique expected 1-kbp fragment was amplified on the EcoRV recircularized libraries and was then cloned in pGEM-T Easy, leading to pSB4, prior to sequencing. The amplified sequence located downstream of the HindIII site was 221 bases long and contained the termination codon of the dsrE ORF situated 30 nucleotides downstream of the HindIII restriction site. Each insert was sequenced in both directions.
Nucleotide sequence analysis.
The combined nucleotide sequence of the different inserts (pSB2 and pSB3) and the inverted PCR product stretch over 9,264 bp. The 8,508-nucleotide ORF starts with an ATG codon at position 566 and terminates with a TAA stop codon at position 9071.
By analogy with promoter sequences from GTFs (34), the putative −35 TTGAAT and −10 ATAAAT sequences are located at positions 415 and 433. They share 67 and 50% identity, respectively, with the −35 TTGACA and −10 TATAAT consensus sequences from E. coli (25). A putative ribosome-binding site is located 9 bp upstream of the start codon, with a hexanucleotide sequence AGGAGC that is 84% identical to the Shine-Dalgarno proposed consensus sequence AGGAGG (50). No putative transcription termination sequences can be identified downstream of the stop codon.
The ORF search on the entire nucleotide sequence reveals several other ORFs, the most interesting being situated upstream of dsrE on the opposite strand. This ORF is preceded by a putative ribosome-binding site (GGAGAC), and the corresponding translated 111-amino-acid peptide was submitted to a Blastp search and shown to share homologies with the transcription regulator PlcR from Streptococcus pneumoniae, Bacillus anthracis, and Bacillus thuringiensis and a bacteriocin gene (mesF) from L. mesenteroides Y105 (19).
Amino acid sequence analysis.
The 8,508-nucleotide sequence of dsrE encodes a 2,835-amino-acid protein with a predicted pI of 4.88 and a theoretical molecular mass of 313,267 Da. Despite strong homologies with already known dextransucrases, DSR-E is characterized by a unique and very unusual structure involving the presence of two catalytic domains.
Indeed, alignment of the deduced amino acid sequence of DSR-E with available GTF and DSR sequences shows the presence of (i) a signal sequence, (ii) a poorly conserved region, (iii) a highly conserved catalytic domain, and (iv) a glucan-binding domain (GBD). Following this domain, the analysis reveals the presence of an extra catalytic domain at the carboxy terminus of DSR-E, as confirmed by PRODOM and Blast analysis (Fig. 2).
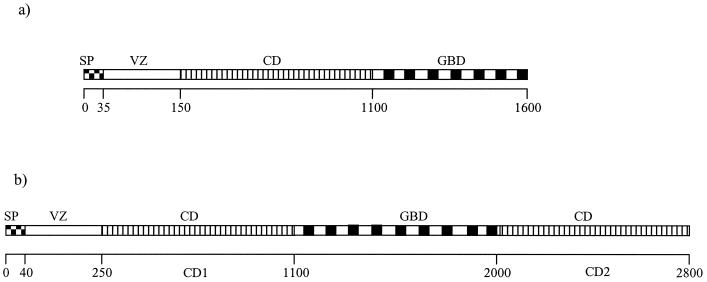
FIG. 2.
Schematic general structure of GTFs and DSRs (a) and DSR-E (b). Abbreviations: SP, N-terminal signal sequence; VZ, variable region; CD, catalytic domain. Amino acid numbering from the N-terminal end is shown.
Signal peptide.
Consistent with the extracellular location of the protein, the sequence encodes a typical gram-positive 40-amino-acid signal peptide, highly conserved with other signal peptides from DSRs of L. mesenteroides. It consists of a positively charged N-terminal part followed by a hydrophobic central region and a more-polar C-terminal region (60). The predicted cleavage site is located between amino acids 40 and 41 according to the algorithm of Nielsen et al. (38).
Variable region.
Then, following the signal peptide, DSR-E displays a 207-amino-acid highly variable domain. Sequence analysis revealed the presence of a new 14-amino-acid repeated motif named S, which is rich in alanine, threonine, and aspartic acid residues and is found very conserved seven times and more-diverging four times (Fig. 3). Its consensus sequence shares some homology with a repeated unit that we also observed in the sequence of DSR-T from L. mesenteroides B-512F (16) and that we designated motif T (Table 2).
FIG. 3.
Alignment of the seven most-conserved S repeats found in the DSR-E variable domain.
TABLE 2.
Sequence comparison of motif S from L. mesenteroides NRRL B-1229 DSR-E and motif T from L. mesenteroides NRRL B-512F DSR-T
| L. mesenteroides strain | Enzyme | Motif | Consensus sequence of repeated motifa |
|---|---|---|---|
| NRRL B-512F | DSR-T | T | -TDDKAA-TTAATS |
| T D | |||
| NRRL B-1299 | DSR-E | S | PAADKAVDTTPAT |
| T T |
Boldface type indicates conserved amino acids.
Catalytic domains.
The first catalytic domain (CD1) ranges from amino acids 248 to 1141, whereas the second one (CD2) is located at the C terminus of the protein, between amino acids 1980 and 2835. CD1 and CD2 have 45% identity and 65% similarity with each other. Both domains contain the conserved amino acids already identified in GTFs or DSRs as being essential for enzymatic activity (Fig. 4).
FIG. 4.
Alignment of highly conserved sequences in glucansucrase catalytic domains. AS, N. polysaccharea (40); GTF-A, L. reuteri (59); GTF-B, Streptococcus mutans GS5 (51); GTF-I, S. downei Mfe28 (15); GTF-S, S. downei Mfe28 (18); DSR-S, L. mesenteroides NRRL B-512F (61); DSR-A, L. mesenteroides NRRL B-1299 (29); DSR-B, L. mesenteroides NRRL B-1299 (31); ASR, L. mesenteroides NRRL B-1355 (3); CD1 and CD2, catalytic domains of DSR-E, L. mesenteroides NRRL B-1299 (this study); —, gap in the sequence; ▪, key amino acid residues of the N-terminal end of the catalytic domain (35); ⇓, putative acid catalyst (9, 26); ➣ putative nucleophile (9, 26); •, putative calcium binding site (9); ⧫, putative residues stabilizing the transition state (26, 35); Δ, residue involved in glucan structure determination (34, 43, 49). Underlined sequences in italics are DSR-E sequences which diverge from consensus sequences.
By analogy with α-amylase family enzymes (9, 24), Asp527 (CD1) and Asp2210 (CD2) can be proposed to play the role of nucleophile. Glu565 (CD1) and Glu2248 (CD2) would be the general acid-base catalysts. Finally, Asp638 (CD1) and Asp2322 (CD2) would correspond to the second aspartic acid of the catalytic triad, always conserved in the active site of α-amylase. Transition state stabilization also requires the interaction with two histidines in α-amylases. These residues correspond to a His and a Gln in glucansucrases of family 70 (30, 36, 47) and are conserved in both catalytic domains of DSR-E: it concerns His637 and Gln1019 for CD1 and His2321 and Gln2694 for CD2. In addition, three conserved amino acid residues essential for the function of Streptococcus downei GTF-I (Trp344, Glu349, and His355) also occur in the one-third of the core region assumed to be outside the (β/α)8 barrel (35). Among them, Trp and Glu residues which appear to be involved in the reaction mechanism are present in both catalytic domains, at positions 425 and 430 for CD1 and positions 2122 and 2127 for CD2. However, in the same block, only the CD1 His436 is conserved when compared to GTF-I His355, whereas Trp2135 is found at this position in the second catalytic domain.
Nevertheless, though there is an overall good conservation of key amino acids, some stretches of sequence located in usually conserved regions of GTFs and DSRs differ clearly in CD2. Thus, as shown in Fig. 4, 2214FIHNDTI and 2323KGVQEKV diverged from the corresponding consensus sequences, NVDADLL and SEVQTVI , respectively, in DSRs.
GBD.
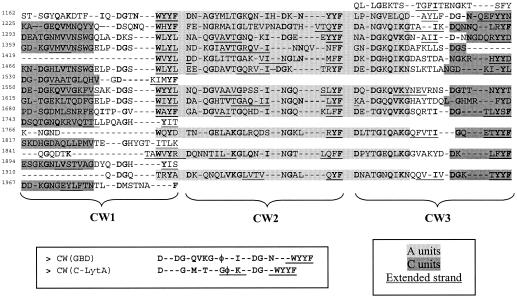
Finally, between the two catalytic domains, the GBD can be found. Usually around 500 amino acids long in GTFs and DSRs, it covers 839 amino acids in DSR-E. Many studies have shown that the GBD of glucansucrases is composed of a series of repeated units divided into 4 classes: A, B, C, and D repeats. According to the consensus sequences proposed by Russell (46), regularly alternating A and C repeats are found in the DSR-E GBD in which three putative cell wall binding domains (CW units) can be localized (Fig. 5). These triplets show homology with the two CW unit triplets forming the C-terminal choline binding domain of the autolysin LytA from S. pneumoniae (C-LytA). The three-dimensional structure of this domain was recently solved (14). It revealed a new solenoid fold formed by 6 CW units. Each CW unit constitutes an independent β-hairpin consisting of two antiparallel β-strands connected by a short internal loop region. Three consecutive hairpins (i.e., 3 CW units) form a complete turn of a left-handed superhelix, conferring the solenoid fold. In DSR-E GBD, two β-strands can also be predicted in the CW units identified by using the secondary structure prediction tool Predator (Fig. 5). This new alignment enables three more-divergent new C repeats to be isolated. One A and one C repeat did not match the newly proposed tandem model.
FIG. 5.
Alignments of CW unit triplets found in the DSR-E GBD. Also shown is the repetitive presence of consecutive A and C motifs, as well as putative extended strands, analogous to those participating in the solenoid fold of C-LytA (14). Strongly conserved residues are printed in boldface type.
Expression of dsrE in E. coli.
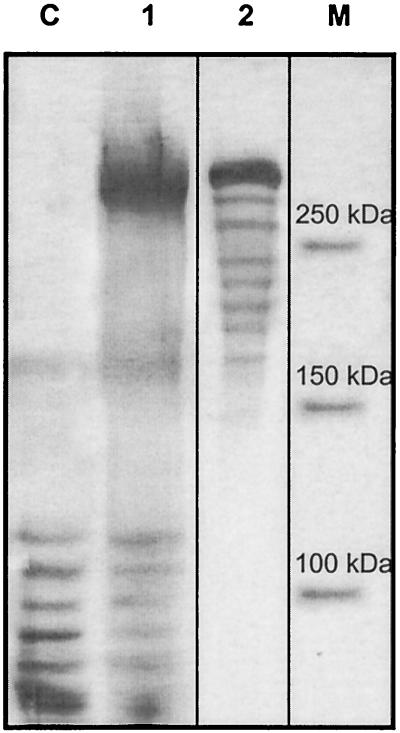
The DSR-E-encoding gene was cloned in an E. coli expression vector under the control of the araBAD promoter (PBAD), leading to plasmid pBAD-dsrE. After optimization of the expression conditions, the highest dextransucrase activity shown by DNS measurements was obtained when E. coli TOP10(pBAD-dsrE) cells were incubated for 4 h with 0.002% arabinose (mass/vol), added at an OD600 of 0.5. Using these conditions, the activity reached 1,063 U/liter of culture. As shown in Fig. 6, SDS-PAGE gels of crude cellular extracts of recombinant cultures revealed that the protein undergoes proteolytic degradation. However, the main band at 320 kDa corresponds to the deduced molecular mass of DSR-E.
FIG. 6.
SDS-PAGE profiles and zymogram of recombinant DSR-E produced by E. coli TOP10. Lanes: C, cellular extract, pBAD/TOPO Thiofusion, and Coomassie staining (negative control); 1, cellular extract, pBAD-dsrE, and Coomassie staining; 2, cellular extract, pBAD-dsrE, and periodic acid-Schiff staining (loading 5 mU of activity); M, broad-range prestained precision protein standard (Bio-Rad).
Characterization of enzyme activity.
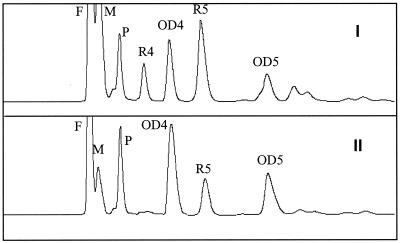
DSR-E was characterized by oligosaccharide synthesis with maltose as the acceptor molecule (Fig. 7). Comparisons of the chromatograms of the DSR-E reaction products with those produced by the native enzymes from L. mesenteroides NRRL B-1299 revealed the presence of (i) oligosaccharides from the OD series and (ii) oligosaccharide R5, which was previously shown to be an α-d-glucopyranosyl-(1 → 2)-α-d-glucopyranosyl-(1 → 6)-α-d-glucopyranosyl-(1 → 6)-α-d-glucopyranosyl-(1 → 4)-α-d-glucopyranose (13). The presence of R5 clearly shows that DSR-E is the enzyme responsible for α-1,2 linkage synthesis.
FIG. 7.
Characterization of the products synthesized by the recombinant DSR-E. High-performance liquid chromatography analysis of gluco-oligosaccharides obtained with native DSRs from L. mesenteroides NRRL B-1299 (I) and the recombinant DSR-E (II). Peak identification: F, fructose; M, maltose; P, panose.
DISCUSSION
The data reported here describe the cloning of a new gene, named dsrE, obtained from L. mesenteroides strain NRRL B-1299. As confirmed by glucan synthesis and periodic acid-Schiff activity staining, cultures of E. coli clones harboring the plasmid carrying the dsrE gene produced an active dextransucrase. Recombinant DSR-E was demonstrated to produce, with maltose as the acceptor, ODi and Ri oligosaccharides which contain α-1,2 linkages.
In addition to its unique regiospecificity, DSR-E possesses a very original structure, never observed before, characterized by the presence of an additional catalytic domain at the carboxy terminus. With a calculated molecular mass of 313,267 Da, DSR-E has twice the average mass of GTFs and DSRs (32).
The role and significance of the variable nonconserved region located downstream of the signal peptide remain unclear. Several studies showed that its deletion does not affect the enzyme activity (1, 33). However, in DSR-E we observed the presence of a 14-amino-acid repeated unit never identified before. That repeat, named S, could thus play a possible role in the enzymatic activity and/or specificity of DSR-E. Similarly, some glucansucrases with unusual specificities, for example, alternansucrase and DSR-T, also possess interesting repeats in the variable domain. So, the influence of this particular repeat on the catalytic properties of DSR-E must be evaluated with deletion experiments.
DSR-E is also remarkable because of the presence of a long GBD, ensuring the junction between the two catalytic domains CD1 and CD2. Along the GBD sequence, regularly alternating A and C repeats are found in which three CW units can be localized. From these observations, it can be suggested that the structure of GBD resembles that of the C-terminal choline binding domain of the autolysin LytA from S. pneumoniae. Thus, A-C tandem repeats could be due to the recurrence of specific duplication events of an ancestral CW unit triplet, corresponding to a complete turn of an original superhelix. This hypothesis corroborates (i) the initial suggestions of Giffard and Jacques (17), who proposed a definition of a fundamental repeating unit from which all classes of repeats (A, B, C, and D) are derived, and (ii) several studies that describe the presence of tandem repeats (4, 18, 22, 46).
Attention has been focused recently on repeated elements in the variable one-third of glucansucrases from L. mesenteroides (21). Unlike DSR-S, DSR-B, and ASR (alternansucrase), no A and C motifs can be found in this region. However, DSR-E is the very first and sole glucansucrase in which a catalytic domain is located after the GBD. Such a structure can be related to the presence of repeated units upstream of the catalytic domain in other DSRs. Either DSR-E might be the product of gene fusion caused by the recombination of two dsr genes, or recombination events between two dsrE genes have led to the presence of repeated elements usually found in the GBD in the N terminus of glucansucrases from L. mesenteroides.
The third and fifth domains correspond to two potential catalytic domains (CD1 and CD2), conferring to DSR-E a unique structure never observed before by analogy to glycoside hydrolase family 70 enzymes. Both domains cover about 900 amino acids, as in other GTFs (32), and thus, because of the presence of all the amino acids thought to play key roles in catalysis, DSR-E seems to possess a double catalysis system. CD1 and CD2 share 44% identity with each other and an average identity with other GTF and DSR catalytic domains of 53 and 44% for CD1 and CD2, respectively. The lower similarity of CD2 can be explained by several regions that diverge from consensus sequences. A tryptophan residue at position 2135 stands for a usually conserved histidine residue. This amino acid is thought to play a role in glucan and oligosaccharide binding (35). Peptide 2210DAVDFIHNDTIQR in block C, the block containing the putative nucleophile, is very different from the highly conserved DAVDNVDADLLQI peptide found in all GTFs and DSRs. The usually conserved residues located just downstream of the first catalytic Asp in glucansucrases could constitute part of the subsite +1, which is involved in the acceptor binding (27). The structure of this site determines the positioning of the acceptor molecule and thus the type of glucosidic bond formed. All glucansucrase enzymes from family 70 have an Asn residue at a position equivalent to N555 of DSR-S. In CD2, the corresponding dipeptide NV is replaced with 2214FI, which can also be found in the amylosucrase sequence. Structural data obtained for this enzyme (52) suggest that the Phe residue could be engaged in the specificity towards the fructo-furanosyl ring of sucrose (53). Moreover, the Ile residue at position 2215 is also found in GTF-A from L. reuteri (59) and is strongly conserved in the α-amylase family.
Likewise, in block E, the 2315KGVQEKV peptide from CD2, following the second Asp of the catalytic triad, differs from the consensus sequence SEVQTVI found in most GTFs and DSRs. Besides, Arguello-Morales et al. (3) also observed in the ASR sequence a specific tripeptide located at the same position, and it can also be noticed that GTF-A from L. reuteri exhibits an original tripeptide just downstream of the third carboxylic acid of the triad (59). In this conserved area, Glu2327 can be found in a position usually involved in the glucan structure determination. Indeed, the corresponding mutant T667R in DSR-S was found to synthesize a glucan with 13% α-1,3 linkages compared to less than 5% for the wild-type enzyme (43). Consistent with this result, it appears that the presence of a carboxylic acid instead of a neutral amino acid (threonine) at the corresponding position of GTF-S increased the synthesis of α-1,3 glucosidic bonds by 30% (49). In addition, concerning GTF-I from S. downei, Monchois et al. (34) also attribute an influence of D569 on oligosaccharide synthesis trough interaction with the acceptor molecule. Thus, DSR-E possesses, because of its two catalytic domains, both a neutral residue in CD1, Thr643, and an acidic residue in CD2, Glu2327.
Moreover, when aligning the DSR-E catalytic domain sequences with other central regions of glucansucrases, stretches of sequence are significantly longer, for example, between the general acid catalyst and the second aspartic acid residue of the catalytic triad in CD2, or shorter, as shown by a 16-amino-acid gap located upstream of the Ca2+ binding site of CD2 (data not shown).
In addition to the attentive study of both catalytic domain sequences, the question is, as no glucansucrase with two catalytic domains has been previously observed, how does such a molecule work? DSR-E possesses two nucleophiles (D527 and D2210) and all the conserved residues required for the formation of two glucosyl enzyme complexes. Thus, DSR-E seems to possess two fully active catalytic domains, and we can assume that the specificity in synthesizing α-1,2 linkages is related to one of the two domains—most probably CD2, which presents a distinctive stretch of sequence compared to CD1. One domain (CD1) would catalyze the transfer of a glucose moiety from sucrose to a maltose residue to give panose, which in turn would be glucosylated, either in CD1, resulting in the addition of a new α-1,6 glucosidic bond, or in CD2, specific for the α-1,2 linkage. It can be suggested that the presence of the GBD enables CD1 and CD2 to be maintained in close proximity for an optimal branching of the polymer.
To determine whether both domains are active, deletion studies and site-directed mutagenesis must be performed to evaluate the influence of each catalytic domain on the activity and specificity of DSR-E.
REFERENCES
- 1.Abo, H., T. Matsumura, T. Kodama, H. Ohta, K. Fukui, K. Kato, and H. Kagawa. 1991. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase). J. Bacteriol. 173:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arguello-Morales, M. A., M. Remaud-Simeon, S. Pizzut, P. Sarçabal, R. M. Willemot, and P. Monsan. 2000. Sequence analysis of the gene encoding alternansucrase, a sucrose glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. FEMS Microbiol. Lett. 182:81-85. [DOI] [PubMed] [Google Scholar]
- 4.Banas, J., L. Simon, J. Williams, J. Ferretti, and R. R. B. Russell. 1994. Analysis of primer-independent GTF-I from Streptococcus salivarius. FEMS Microbiol. Lett. 123:349-354. [DOI] [PubMed] [Google Scholar]
- 5.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brendel, V., P. Bucher, I. Nourbakhsh, B. E. Blaisdell, and S. Karlin. 1992. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl. Acad. Sci. USA 89:2002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpet, F., F. Servant, J. Gouzy, and D. Kahn. 2000. ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 28:267-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Côté, G. L., and J. F. Robyt. 1982. Isolation and partial purification of an extracellular glucansucrase from Leuconostoc mesenteroides NRRL B-1355 that synthesises an alternating (1-6), (1-3) α-D-glucan. Carbohydr. Res. 127:95-107. [DOI] [PubMed] [Google Scholar]
- 9.Devulapalle, K. S., S. Goodman, Q. Gao, A. Hemsley, and G. Mooser. 1997. Knowledge-based model of a glucosyltransferase from oral bacterial group of mutans streptococci. Protein Sci. 6:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djouzy, Z., C. Andrieux, V. Pelenc, S. Somarriba, F. Popot, F. Paul, P. Monsan, and O. Szylit. 1995. Degradation and fermentation of α-gluco-oligosaccharides by bacterial strains from human colon: in vitro and in vivo studies in gnotobiotic rats. J. Appl. Bacteriol. 79:117-127. [DOI] [PubMed] [Google Scholar]
- 11.Dols, M., M. Remaud-Simeon, M. Vignon, R. M. Willemot, and P. F. Monsan. 1997. Properties of soluble and insoluble dextransucrases from Leuconostoc mesenteroides NRRL B-1299. Appl. Biochem. Biotechnol. 62:47-52. [Google Scholar]
- 12.Dols, M., M. Remaud-Simeon, R. M. Willemot, M. R. Vignon, and P. F. Monsan. 1998. Characterization of the different dextransucrase activities excreted in glucose, fructose, or sucrose medium by Leuconostoc mesenteroides NRRL B-1299. Appl. Environ. Microbiol. 64:1298-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dols, M., M. Remaud-Simeon, R. M. Willemot, M. R. Vignon, and P. F. Monsan. 1998. Structural characterisation of the maltose acceptor-products synthesised by Leuconostoc mesenteroides NRRL B-1299 dextransucrase. Carbohydr. Res. 305:549-559. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Tornero, C., R. Lopez, E. Garcia, G. Gimenez-Gallego, and A. Romero. 2001. A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 8:1020-1024. [DOI] [PubMed] [Google Scholar]
- 15.Ferreti, J. J., M. L. Gilpin, and R. R. B. Russell. 1987. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus Mfe28. J. Bacteriol. 169:4271-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funane, K., K. Mizuno, H. Takahara, and M. Kobayashi. 2000. Gene encoding a dextransucrase-like protein in Leuconostoc mesenteroides NRRL B-512F. Biosci. Biotechnol. Biochem. 64:29-38. [DOI] [PubMed] [Google Scholar]
- 17.Giffard, P. M., and N. A. Jacques. 1994. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 73:1133-1141. [DOI] [PubMed] [Google Scholar]
- 18.Gilmore, K. S., R. R. B. Russell, and J. J. Ferreti. 1990. Analysis of the Streptococcus downei gtfS gene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect. Immun. 58:2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hechard, Y., J. M. Berjeaud, and Y. Cenatiempo. 1999. Characterisation of the mesB gene and expression of bacteriocins by Leuconostoc mesenteroides Y105. Curr. Microbiol. 39:265-269. [DOI] [PubMed] [Google Scholar]
- 20.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janecek, S., B. Svensson, and R. R. B. Russell. 2000. Location of repeat elements in glucansucrases of Leuconostoc and Streptococcus species. FEMS Microbiol. Lett. 192:53-57. [DOI] [PubMed] [Google Scholar]
- 22.Kato, C., and H. K. Kuramitsu. 1990. Carboxyl-terminal deletion analysis of the Streptococcus mutans glucosyltransferase-I enzyme. FEMS Microbiol. Lett. 72:299-302. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D., and J. F. Robyt. 1996. Dextransucrase constitutive mutants of Leuconostoc mesenteroides NRRL B-1299. Enzyme Microb. Technol. 17:1050-1056. [Google Scholar]
- 24.Kobayashi, M., and K. Matsuda. 1977. Structural characteristics of dextran synthesised by dextransucrases from Leuconostoc mesenteroides NRRL B-1299. Agric. Biol. Chem. 41:1931-1937. [Google Scholar]
- 25.Lewin, B. 1997. Genes. Oxford University Press, Inc., New York, N.Y.
- 26.MacGregor, E. A., H. M. Jespersen, and B. Svensson. 1996. A circularly permuted α-amylase-type α/β barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 378:263-266. [DOI] [PubMed] [Google Scholar]
- 27.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Monchois, V., R.-M. Willemot, M. Remaud-Simeon, C. Croux, and P. Monsan. 1996. Cloning and sequencing of a gene coding for a novel dextransucrase from Leuconostoc mesenteroides NRRL B-1299 synthesising only α-1,6 and α-1,3 linkages. Gene 182:23-32. [DOI] [PubMed] [Google Scholar]
- 30.Monchois, V., M. Remaud-Simeon, R. R. B. Russell, P. Monsan, and R.-M. Willemot. 1997. Characterisation of Leuconostoc mesenteroides NRRL B-512F dextransucrase (DSRS) and identification of amino-acid residues playing a key role in enzyme activity. Appl. Microbiol. Biotechnol. 48:465-472. [DOI] [PubMed] [Google Scholar]
- 31.Monchois, V., M. Remaud-Simeon, P. Monsan, and R.-M. Willemot. 1998. Cloning and sequencing of a gene coding for an extracellular dextransucrase (DSRB) from Leuconostoc mesenteroides NRRL B-1299 synthesising only a α-1,6 glucan. FEMS Microbiol. Lett. 159:307-315. [DOI] [PubMed] [Google Scholar]
- 32.Monchois, V., R. M. Willemot, and P. Monsan. 1999. Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol. Rev. 23:131-151. [DOI] [PubMed] [Google Scholar]
- 33.Monchois, V., M. A. Argüello-Morales, and R. R. B. Russell. 1999. Isolation of an active catalytic core of Streptococcus downei Mfe28 GTF-I glucosyltransferase. J. Bacteriol. 181:2290-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monchois, V., M. Vignon, and R. R. B. Russell. 1999. Mutagenesis of Asp-569 of glucosyltransferase I glucansucrase modulates glucan and oligosaccharide synthesis. Appl. Environ. Microbiol. 66:1923-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monchois, V., M. Vignon, and R. R. B. Russell. 1999. Isolation of key amino-acid residues at the N-terminal end of the core region of Streptococcus downei glucansucrase GTF-I. Appl. Microbiol. Biotechnol. 52:660-665. [DOI] [PubMed] [Google Scholar]
- 36.Monchois, V., M. Vignon, P. C. Escalier, B. Svensson, and R. R. B. Russell. 2000. Involvement of Gln 937 of Streptococcus downei GTF-I glucansucrase in transition-state stabilisation. Eur. J. Biochem. 267:4127-4136. [DOI] [PubMed] [Google Scholar]
- 37.Monsan, P., and F. Paul. 1995. Enzymatic synthesis of oligosaccharides. FEMS Microbiol. Rev. 16:187-192. [Google Scholar]
- 38.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 39.Paul, F., A. Lopez-Munguia, M. Remaud, V. Pelenc, and P. Monsan. August 1992. Method of the production of α-1,2 oligodextrans using Leuconostoc mesenteroides NRRL B-1299. U.S. patent 5,141,858.
- 40.Potocki de Montalk, G., M. Remaud-Simeon, R. M. Willemot, V. Planchot, and P. Monsan. 1999. Sequence analysis of the gene encoding amylosucrase from Neisseria polysaccharea and characterization of the recombinant enzyme. J. Bacteriol. 181:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remaud, M., F. Paul, P. Monsan, A. Lopez-Mungia, and M. Vignon. 1992. Characterisation of α-1,3 branched oligosaccharides synthesised by acceptor reaction with the extracellular glucosyltransferases from Leuconostoc mesenteroides NRRL B-742. J. Carbohydr. Chem. 11:359-378. [Google Scholar]
- 42.Remaud-Simeon, M., A. Lopez-Munguia, V. Pelenc, F. Paul, and P. Monsan. 1994. Production and use of glucosyltransferases from Leuconostoc mesenteroides NRRL B-1299 for the synthesis of oligosaccharides containing α-1,2 linkages. Appl. Biochem. Biotechnol. 44:101-107. [DOI] [PubMed] [Google Scholar]
- 43.Remaud-Simeon, M., R. M. Willemot, P. Sarçabal, G. Potocki de Montalk, and P. Monsan. 2000. Glucansucrases: molecular engineering and oligosaccharide synthesis. J. Mol. Catal. B 10:117-128. [Google Scholar]
- 44.Roberfroid, M. B. 1997. Health benefits of non-digestible oligosaccharides. Adv. Exp. Med. Biol. 427:211-219. [DOI] [PubMed] [Google Scholar]
- 45.Robyt, J. F., and T. F. Walseth. 1978. The mechanism of acceptor reactions of Leuconostoc mesenteroides NRRL B-512F dextransucrase. Carbohydr. Res. 61:433-435. [DOI] [PubMed] [Google Scholar]
- 46.Russell, R. R. B. 1990. Molecular genetics of glucan metabolism in oral streptococci. Arch. Oral Biol. 35:53S-58S. [DOI] [PubMed] [Google Scholar]
- 47.Sarçabal, P., M. Remaud-Simeon, R. M. Willemot, G. Potocki de Montalk, B. Svensson, and P. Monsan. 2000. Identification of key amino-acid residues in Neisseria polysaccharea amylosucrase. FEBS Lett. 474:33-37. [DOI] [PubMed] [Google Scholar]
- 48.Seymour, F. R., E. C. M. Chen, and S. H. Bishop. 1979. Methylation structural analysis of unusual dextrans by combined gas-liquid chromatography and mass spectrometry. Carbohydr. Res. 68:113-121. [Google Scholar]
- 49.Shimamura, A., Y. J. Nakano, H. Musaka, and H. K. Kuramitsu. 1994. Identification of amino-acid residues in Streptococcus mutans glucosyltransferases influencing the structure of the glucan product. J. Bacteriol. 176:4845-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shine, J., and L. Dalgarno. 1974. The 3′ sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiroza, T., S. Ueda, and H. K. Kuramitsu. 1987. Sequence analysis of the gtfB gene from Streptococcus mutans. J. Bacteriol. 169:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skov, L. K., O. Mirza, A. Henriksen, G. Potocki de Montalk, M. Remaud-Simeon, P. Sarçabal, R. M. Willemot, P. Monsan, and M. Gajhede. 2000. Crystallisation and preliminary X-ray studies of recombinant amylosucrase from Neisseria polysaccharea. Acta Crystallogr. Sect. D 56:203-205. [DOI] [PubMed] [Google Scholar]
- 53.Skov, L. K., O. Mirza, A. Henriksen, G. Potocki de Montalk, M. Remaud-Simeon, P. Sarçabal, R. M. Willemot, P. Monsan, and M. Gajhede. 2001. Amylosucrase, a glucan-synthesising enzyme from the α-amylase family. J. Biol. Chem. 276:25273-25278. [DOI] [PubMed] [Google Scholar]
- 54.Sumner, J., and S. Howell. 1935. A method for the determination of invertase activity. J. Biol. Chem. 108:51-54. [Google Scholar]
- 55.Svensson, B. 1994. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity and stability. Plant Mol. Biol. 25:141-157. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting-position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uitdehaag, J. C. M., R. Mosi, K. H. Kalk, B. A. van der Veen, L. Dijkhuizen, S. G. Withers, and B. W. Dijkstra. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Biol. 6:432-436. [DOI] [PubMed] [Google Scholar]
- 58.Valette, P., V. Pelenc, Z. Djouzy, C. Andrieux, F. Paul, P. Monsan, and O. Szylit. 1993. Bioavailibility of new synthesised glucooligosaccharides in the intestinal tract of gnotobiotic rats. J. Sci. Food Agric. 62:121-127. [Google Scholar]
- 59.Van Geel-Schutten, G. H. 2000. Exopolysaccharide synthesis by Lactobacillus reuteri. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 60.Von Heijne, G. 1990. The signal peptide. J. Membr. Biol. 115:195-201. [DOI] [PubMed] [Google Scholar]
- 61.Wilke-Douglas, M., J. T. Perchorowicz, C. M. Houck, and B. R. Thomas. June 1989. Methods and compositions for altering physical characteristics of fruit and fruit products. Patent Cooperation Treaty patent WO 89/12386.