Abstract
Bacteria need dedicated systems that allow appropriate adaptation to the perpetual changes in their environments. In Bacillus subtilis, two HtrA-like proteases, HtrA and HtrB, play critical roles in the cellular response to secretion and heat stresses. Transcription of these genes is induced by the high-level production of a secreted protein or by a temperature upshift. The CssR-CssS two-component regulatory system plays an essential role in this transcriptional activation. Transcription of the cssRS operon is autoregulated and can be induced by secretion stress, by the absence of either HtrA or HtrB, and by heat stress in a HtrA null mutant strain. Two start sites are used for cssRS transcription, only one of which is responsive to heat and secretion stress. The divergently transcribed htrB and cssRS genes share a regulatory region through which their secretion and heat stress-induced expression is linked. This study shows that CssRS-regulated genes represent a novel class of heat-inducible genes, which is referred to as class V and currently includes two genes: htrA and htrB.
Since the conditions in natural environments are highly variable and unpredictable, eubacteria need systems that support their adaptation to perpetual changes. For this purpose, relevant stimuli must be sensed and identified by the cells and subsequently the resulting information must be transformed into appropriate transcriptional or behavioral responses.
In many cases, two-component systems are used for signal transduction (18, 24). The presence of multiple two-component systems is a prerequisite to adequately respond to diverse stimuli received by the cells. Such systems consist of a sensor histidine kinase and a cognate response regulator. In many cases, the sensor kinase is located in the cytoplasmic membrane. It is responsible for sensing environmental or nutritional stimuli and transferring this information to the second protein of the system through autophosphorylation and phosphotransfer reactions. When the cognate response regulator is phosphorylated, it either activates or represses the transcription of specific genes, thereby eliciting a cellular response appropriate to the original stimuli. The genome of the gram-positive bacterium Bacillus subtilis encodes 34 two-component systems (8). The functions of most of them have not yet been determined. However, one of these two-component systems, CssR-CssS (for “control of secretion stress regulator and sensor”) (11), has recently been reported to respond to secretion stress generated by overproduction of the α-amylase AmyQ of Bacillus amyloliquefaciens. The cssR gene and the downstream cssS gene form a bicistronic operon, but little is known about its regulation.
In B. subtilis, heat-inducible genes can be divided into four different classes on the basis of their regulatory mechanisms (5). The transcription of class I genes, which code for classical chaperones, is σA dependent and regulated by the HrcA repressor (14, 29, 31). The class II genes respond not only to heat but also to other stresses such as exposure to ethanol or salt, starvation for glucose or phosphate, or growth under anaerobic conditions. Transcription of these genes is regulated by σB (3, 10, 19). The class III genes also respond to general stress, but they are regulated by the CtsR transcriptional repressor (5, 6). All the heat-responsive genes that do not belong to one of these three classes have been collectively termed class IV genes (ahpC, clpX, ftsH, htpG, lonB, ykdA, yvtA, and trxA) (6).
The B. subtilis genome contains three genes encoding putative membrane-bound HtrA-like proteases: YkdA, YvtA, and YyxA (16, 25). Transcription of ykdA and yvtA is inducible by heat stress or α-amylase overproduction, whereas yyxA is insensitive to these stimuli (15, 16). Furthermore, transcription of the ykdA and yvtA genes is negatively auto- and cross-regulated. Because of their similarity to the HtrA protein of Escherichia coli and their induction upon heat stress, YkdA and YvtA are referred to as HtrA and HtrB, respectively (for “high-temperature requirement”). At present, htrA and htrB are listed as class IV heat stress responsive genes since their respective promoters have −10 regions (but not −35 regions) typical of σA-type promoters and lack both σB-type promoters and the consensus binding sites for HrcA or CtsR. Instead, the control regions of htrA and htrB have a fourfold-repeated octameric consensus sequence positioned in the vicinity of the −35 regions, suggesting a novel regulatory mechanism of expression (15). Interestingly, it has been established that the htrA gene is one of the targets of the CssRS two-component system (11). However, a comparison of the phenotypes of cssS and htrA mutant strains indicated that at least one other protease is controlled by CssRS.
The present studies were aimed at the identification of additional CssRS-controlled genes and the establishment of a possible dual role of CssRS in heat and secretion stress responses. The results show that expression of htrA, htrB, cssR, and cssS is responsive to secretion stress in a manner dependent on the CssRS two-component system. In addition, the induction of htrA and htrB expression by heat stress in a wild-type background is CssRS dependent, signifying that this is a new class of heat-inducible genes, termed class V.
MATERIALS AND METHODS
Plasmids, bacterial strains, and media.
Tables 1 and 2 list the bacterial strains and plasmids, respectively, used in this study. Bacteria were grown in TY medium (1% tryptone, 0.5% yeast extract, 1.0% NaCl). Minimal medium was prepared as described by Leskela et al. (13). Antibiotics were used in the following concentrations: ampicillin, 50 μg/ml; erythromycin, 1 μg/ml; kanamycin, 10 μg/ml; spectinomycin, 100 μg/ml; chloramphenicol, 5 μg/ml. To visualize α-amylase activity, TY plates were supplemented with 1% starch.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | 1 |
| BFA3041 | 168 htrB(yvtA)::pMutin4 | This study |
| BFA2461 | 168 cssR(yvqA)::pMutin4 | 11 |
| BV2001 | 168 cssS::Sp | 11 |
| BV2006 | 168 cssS::pMutin2 | 11 |
| BV2015 | 168 cssS::Sp htrB(yvtA)::pMutin4 | This study |
| BV2016 | 168 cssS::Sp amyE::P cssRS(yvqAB)-bgaB | This study |
| BV2017 | 168 htrA(ykdA)Δ439 amyE::P cssRS(yvqAB)-bgaB | This study |
| BV2018 | 168 cssS::Sp htrA(ykdA)Δ439 amyE::P cssRS(yvqAB)-bgaB | This study |
| BV2019 | 168 htrB(yvtA)::Km amyE::P cssRS(yvqAB)-bgaB | This study |
| BV2020 | 168 cssS::Sp htrB(yvtA)::Km amyE::P cssRS(yvqAB)-bgaB | This study |
| BV2021 | 168 htrA(ykdA)Δ439 htrB(yvtA)::Km amyE::P cssRS(yvqAB)-bgaB (suppressed) | This study |
| BV2022 | 168 cssS::Sp htrA(ykdA)Δ439 htrB(yvtA)::Km amyE::P cssRS(yvqAB)-bgaB (suppressed) | This study |
| BV2023 | 168 cssS::pMutin2 amyE::XcssS | This study |
| BV2024 | 168 cssS::Sp amyE::P htrA(ykdA)-bgaB | This study |
| BV2025 | 168 cssS::Sp amyE::P htrB(yvtA)-bgaB | This study |
| DN2 | 168 amyE::P htrA(ykdA)-bgaB | 16 |
| DN26 | 168 htrA(ykdA)Δ439 | 16 |
| DN110 | 168 amyE::P htrB(yvtA)-bgaB | 15 |
| DN111 | 168 htrB(yvtA)::Km | 15 |
| DN112 | 168 htrB::kan amyE::P htrB(yvtA)-bgaB | 15 |
| DN113 | 168 htrAΔ439 amyE::P htrB(yvtA)-bgaB | 15 |
| DN119 | 168 htrAΔ439 amyE::P*htrB(yvtA)-bgaB | This study |
| DN120 | 168 htrB::kan amyE::P*htrB(yvtA)-bgaB | This study |
| DN220 | 168 amyE::P cssRS(yvqA/B)-bgaB | This study |
| DN225 | 168 amyE xylR::xylR-amyL P cssRS(yvqAB)::pDN221 | This study |
| DN226 | 168 amyE xylR::xylR-amyL cssR(yvqA):: pMutin4 | This study |
| DN227 | 168 P cssRS(yvqAB)::pDN221 | This study |
| DN228 | 168 amyE xylR::xylR-amyL ∗P cssRS(yvqAB)::pDN222 | This study |
| KS408 | 168 amyE xylR::xylR-amyL | 23 |
| E. coli | ||
| MC1061 | F−araD139 Δ(ara-leu)7696 Δ(lac)X74 galU galK hsdR2 mcrA mcrB1 rspL | 28 |
| TG-1 | supE hsdΔ 5 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 22 |
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristics | Reference |
|---|---|---|
| pUB110 | Kmr | 9 |
| pKTH10 | pUB110 derivative; encodes the α-amylase (AmyQ) of B. amyloliquefaciens; Kmr | 17 |
| pKTH10L | pKTH10 derivative; Kmr | 11 |
| pMutin4 | pBR322-based integration vector for B. subtilis; contains a multiple-cloning site downstream of the Pspac promoter and a promoterless lacZ gene preceded by the RBSa of the spoVG gene; Apr/Emr | 26 |
| pDL | Vector for the integration of transcriptional promoter-bgaB gene fusions in the amyE locus of B. subtilis; Apr Cmr | 30 |
| pDN220 | pDL containing the complete cssR-cssS control region on a PCR-amplified fragment; Apr Cmr | This study |
| pDN221 | pMutin4 containing the complete cssR-cssS control region on a PCR-amplified fragment; Apr Emr | This study |
| pDN222 | pMutin4 containing the complete cssR-cssS control region, with the A → G mutation at position −60 relative to the start codon of cssR; Apr Emr | This study |
| pDN223 | pDL containing the htrB control region with the T → C change at position −218 relative to the start codon of htrB; Apr Cmr | This study |
| pX | Vector for the integration of genes in the amyE locus; the integrated gene will be transcribed from the xylA promotor; carries the xylR gene; Apr Cmr | 12 |
| pXcssS | pX derivative; carries cssS downstream of the xylA promoter; Apr Cmr | This study |
RBS, ribosome-binding site
DNA techniques.
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of competent E. coli cells were carried out as described by Sambrook et al. (22). B. subtilis was transformed as described by Leskela et al. (13). PCR was carried out as described by van Dijl et al. (27) using chromosomal DNA of B. subtilis 168 as a template. The nucleotide sequences of primers used for PCR are listed below; restriction sites used for cloning are underlined. Enzymes were from Roche Molecular Biochemicals (Mannheim, Germany). Constructs were first made in E. coli MC1061 or TG-1 and then introduced into B. subtilis.
To construct the htrB::pMutin4 mutation and the transcriptional htrB-lacZ fusion, an internal fragment of the htrB (yvtA) gene was amplified by PCR with the oligonucleotides yvtAH3 (5′-GGC CAA GCT TCA ACA TCA AAC TGA ACC-3′) and yvtAB1 (5′-GGC CGG ATC CAC AGC CGT TTC TTG C-3′). The amplified fragment (180 nucleotides) was cloned into plasmid pMutin4, using BamHI and HindIII. The B. subtilis 168 htrB::pMutin4 (BFA3041) strain was obtained by Campbell-type (single-crossover) integration of the resulting plasmid into the chromosome of B. subtilis 168. This mutant was verified by Southern hybridization. The B. subtilis 168 cssS::Sp htrB::pMutin4 (BV2015), B. subtilis 168 cssS::Sp amyE::PhtrA-bgaB (BV2024), and B. subtilis 168 cssS::Sp amyE::PhtrB-bgaB (BV2025) strains were, respectively, constructed by transformation of B. subtilis 168 htrB::pMutin4 (BFA3041), B. subtilis 168 amyE::PhtrA-bgaB (DN2), and B. subtilis 168 amyE::PhtrB-bgaB (DN110) with chromosomal DNA of B. subtilis 168 cssS::Sp (BV2001) and selection for spectinomycin resistance.
To construct plasmid pDN220, the cssRS promoter region was amplified by PCR with the oligonucleotides YVQAPF (5′-CCG GAA TTC GTT CTT ACA CTC CTT AAC G-3′) and YVQAPR (5′-CGG GAT CCG CAG TTC ATT CAG GTT ATC C-3′). The amplified fragment (337 nucleotides) was cloned into plasmid pDL, using EcoRI and BamHI. To determine the PcssRS promoter activity in different backgrounds, the strains B. subtilis 168 amyE::PcssRS-bgaB (DN220), B. subtilis 168 htrAΔ439 amyE::PcssRS-bgaB (BV2017), B. subtilis 168 htrB::Km amyE::PcssRS-bgaB (BV2019), and B. subtilis 168 cssS::Sp amyE::PcssRS-bgaB (BV2016) were, respectively, constructed by transformation of B. subtilis 168, B. subtilis 168 htrAΔ439 (DN26), B. subtilis 168 htrB::Km (DN111), and B. subtilis 168 cssS::Sp (BV2001) with plasmid pDN220, selection for chloramphenicol resistance, and screening for an AmyE− phenotype. Two of these strains, BV2017 (htrA) and BV2019 (htrB), were transformed with chromosomal DNA of the BV2001 (cssS) strain; selection for spectinomycin resistance resulted in B. subtilis 168 cssS::Sp htrAΔ439 amyE::PcssRS-bgaB (BV2018) and B. subtilis 168 cssS::Sp htrB::Km amyE::PcssRS-bgaB (BV2020). Strain BV2017 (htrA) was also transformed with chromosomal DNA from the DN111 strain (htrB); selection for kanamycin resistance resulted in the strain B. subtilis 168 htrAΔ439 htrB::Km amyE::PcssRS-bgaB (BV2021; suppressed). Finally, BV2021 (htrA htrB; suppressed) was transformed with chromosomal DNA of the BV2001 (cssS) strain; selection for spectinomycin resistance resulted in B. subtilis 168 cssS::Sp htrAΔ439 htrB::Km amyE::PcssRS-bgaB (BV2022; suppressed).
To construct plasmid pDN221, the cssRS promoter region was amplified by PCR with the oligonucleotides YVQAPF and YVQAPR. The amplified fragment was cloned into plasmid pMutin4, using EcoRI and BamHI. Strain B. subtilis 168 PcssR::pDN221 (DN227) was generated by transforming B. subtilis 168 with plasmid pDN221 and selection for erythromycin resistance. Plasmid pDN222 is identical to plasmid pDN221 except for the introduction of a single PCR-generated A-to-G base change at bp −60 relative to the TTG start codon of cssR. Strain DN228 was constructed by transforming B. subtilis 168 amyE xylR::xylR-amyL (KS408) with plasmid pDN222 and selecting for erythromycin resistance. Plasmid pDN223 was constructed by PCR amplifying the insert of plasmid pDN222 with primers YVTAPF and YVTAPR (15) and cloning the product into pDL as an EcoRI-BamHI fragment. Strains DN119 and DN120 were generated by transforming strains DN26 (htrA) and DN111 (htrB), respectively with plasmid pDN223 with selection for chloramphenicol resistance. Strain B. subtilis 168 amyE xylR::xylR-amyL PcssR::pDN221 (DN225) was created by transformation of KS408 with chromosomal DNA from strain DN227 and selection for erythromycin resistance. Strain B. subtilis 168 amyE xylR::xylR-amyL cssR::pMutin4 (DN226) was generated by transformation of strain KS408 with chromosomal DNA from strain B. subtilis 168 cssR::pMutin4 (BFA2461) and selection for erythromycin resistance.
To construct a strain with a xylose-inducible cssS gene, the complete cssS region was amplified with the oligonucleotides cssS3 (5′-GCT CTA GAA TTG CCG TCT CCT CGT ATC G-3′) and cssS4 (5′-CGC GGA TCC AGC AGA CCT TGT CAG AGA A-3′). The amplified fragment (1,755 nucleotides) was cleaved with XbaI and BamHI and ligated into the SpeI and BamHI sites of plasmid pX, resulting in pXcssS. The mutant B. subtilis 168 cssS::pMutin2 amyE::XcssS (BV2023) was obtained by a double-crossover integration of the XcssS cassette from plasmid pXcssS into the amyE locus of B. subtilis 168 cssS::pMutin2 (BV2006); chloramphenicol-resistant transformants were screened for an AmyE− phenotype.
β-Galactosidase activity assay.
To assay β-galactosidase activities, overnight cultures were diluted in fresh medium and samples were taken at different intervals for optical density readings at 600 nm (OD600) and β-galactosidase activity determinations. For strains containing a transcriptional lacZ fusion, the β-galactosidase assay and the calculation of β-galactosidase units (Miller units: nanomoles per OD600 unit per minute) were performed as described by Hyyryläinen et al. (11). To investigate the effects of heat stress, cells containing a transcriptional bgaB fusion were first grown at 37°C to an OD600 of 0.3 to 0.4 and then divided into two cultures: one remained at 37°C, while the other was transferred to a prewarmed flask and incubated at 48°C. To assay BgaB activity, the LacZ activity assay was used with minor modifications: cell lysis was followed by an incubation at 70°C for 15 min and the subsequent β-galactosidase activity assays were performed at 55°C. Experiments were repeated at least twice, starting with independently obtained transformants. In all experiments, the relevant controls were performed in parallel. Although some differences were observed in the absolute β-galactosidase activities, the ratios between these activities in the various strains tested were largely constant. A ratio of about 1.5 was generally reproducible. Differences in absolute β-galactosidase levels reported in this paper and those reported by Noone et al. (15, 16) are due to different methods for calculating specific activities: in this study β-galactosidase units are calculated as nanomoles per minute per OD600 unit, and in references 15 and 16 they were calculated as as nanomoles per minute per milligram of protein.
Transcriptional analysis.
For the analysis of the effects of heat stress, total RNA was isolated from B. subtilis 168 htrAΔ439 (DN26) at various times before and after heat stress at 48°C. For the analysis of the effects of secretion stress, total RNA isolated at different time points from B. subtilis 168 xylR::xylR-amyL (KS408), grown in TY medium with or without 1% xylose, was used as a template for primer extension. 32P-radiolabeled primers YVQA-RT1 (5′-CTA GAT AAA TGG TGT ATG ACA AGG C-3′) and YVQA-RT2 (5′-GAT GTA ATG TTC CAG CCC-3′) were annealed to 25 μg of total RNA, and primer extension analysis was carried out as previously described (16).
RESULTS
Transcription of htrB is CssS dependent.
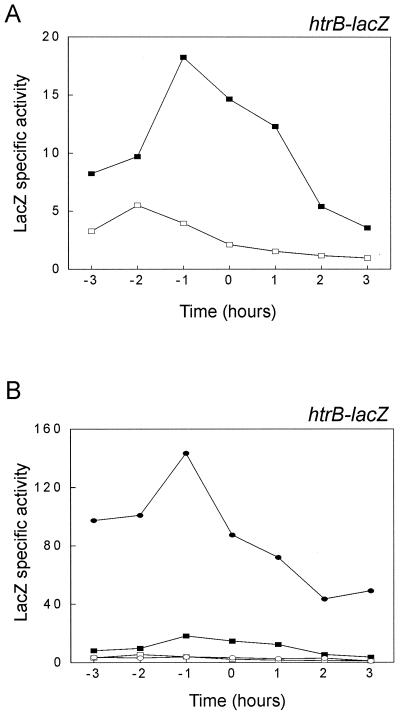
Since the transcription of htrB responds to the same stimuli as the CssRS-dependent transcription of htrA, a transcriptional htrB-lacZ fusion (Fig. 1A) was used to investigate a possible role of CssS in the regulation of htrB. In view of the negative autoregulation of htrB, it is important to note that the chromosomal pMutin4 insertion used to obtain the htrB-lacZ fusion results in the inactivation of this gene. The β-galactosidase activities of B. subtilis 168 htrB::pMutin4 (BFA3041) and the B. subtilis 168 cssS::Sp htrB::pMutin4 strain (BV2015) were measured as a function of growth in TY medium at 37°C. As shown in Fig. 2A, the htrB transcription peaked at about 1 h before the transition (t = 0) between exponential and postexponential growth. The disruption of cssS reduced htrB-lacZ transcription more than threefold and significantly changed the expression profile. To test whether the previously reported induction of htrB by α-amylase overproduction (15) is CssS dependent, plasmid pKTH10L was introduced into the htrB-lacZ strains. This plasmid imposes secretion stress on the cells by overexpression of the α-amylase AmyQ (11). The presence of pKTH10L resulted in a more than eightfold increase in htrB transcription in the cssS+ background without affecting the expression profile (Fig. 2B). In contrast, a strain containing the control vector pUB110 did not affect htrB transcription (data not shown). The disruption of cssS reduced htrB-lacZ transcription in cells subject to AmyQ secretion stress to a level similar to that observed in nonstressed cells (Fig. 2B). Remarkably, the cssS disruption did not reduce htrB-lacZ transcription to background levels. Essentially the same effects were observed when cells were grown in minimal medium (data not shown). Disruption of cssS in the htrB mutant strain containing pKTH10L caused a reduced growth rate during the exponential phase and a reduced cell density in the postexponential phase of growth. For example, cssS htrB mutant cells containing pKTH10L reached an OD600 of about 2, while control cells containing pUB110 reached an OD600 of about 5 (data not shown). These observations demonstrate that both the basal level of transcription and secretion stress induction of htrB are CssS dependent.
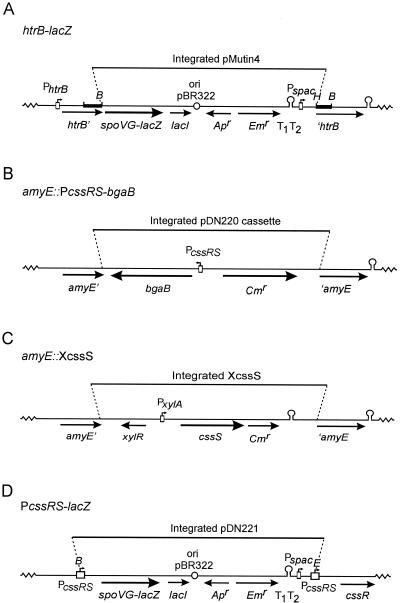
FIG. 1.
Construction of mutant strains. (A) Schematic presentation of the htrB::pMutin4 mutation. The htrB gene was disrupted with pMutin4 by a single-crossover event (Campbell-type integration). Simultaneously, the spoVG-lacZ reporter gene of pMutin4 was placed under the transcriptional control of the htrB promoter region (PhtrB). The chromosomal fragment from the htrB region, which was amplified by PCR and cloned into pMutin4, is indicated by black bars. Only the restriction sites relevant for the construction are shown (B, BamHI; H, HindIII). lacI, E. coli lacI gene; ori pBR322, replication functions of pBR322; Apr, ampicillin resistance marker; Emr, erythromycin resistance marker; T1T2, transcriptional terminators on pMutin4; Pspac,isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent promoter;htrB′, 3′-truncated htrB gene; ′htrB, 5′-truncated htrB gene. (B) Schematic presentation of the amyE region of the chromosome of strains containing an amyE::PcssRS-bgaB mutation. By a double-crossover event, the amyE gene was disrupted with a pDN220-derived cassette containing the bgaB reporter gene placed under the transcriptional control of the cssRS promoter region (PcssRS); Cmr, chloramphenicol resistance marker; amyE′, 3′ truncated amyE gene; ′amyE, 5′ truncated amyE gene. (C) Schematic presentation of the amyE region of the chromosome of strains containing the amyE::XcssS mutation. By a double-crossover event, the amyE gene was disrupted with a pXcssS-derived cassette (XcssS), which contains the cssS gene placed under the transcriptional control of a xylose-inducible promoter (PxylA). Cmr, chloramphenicol resistance marker; xylR, gene specifying the XylR repressor protein; amyE′, 3′ truncated amyE gene; ′amyE, 5′ truncated amyE gene. (D) Schematic presentation of the PcssRS::pDN221 reporter strain. The promoter region of the cssRS genes was duplicated by the insertion of pDN221 into the chromosome via a single-crossover event (Campbell-type integration). Consequently, both the cssRS operon and the spoVG-lacZ reporter gene of pDN221 are placed under the transcriptional control of a cssRS promoter region (PcssRS). Only the restriction sites relevant for the construction are shown (B, BamHI; E, EcoRI). lacI, E. coli lacI gene; ori pBR322, replication functions of pBR322; Apr, ampicillin resistance marker; Emr, erythromycin resistance marker; T1T2, transcriptional terminators on pDN221; Pspac, IPTG-dependent promoter.
FIG. 2.
Analysis of htrB expression in a cssS mutant background. The CssS dependence of htrB-lacZ transcription was analyzed in the absence (A) or presence (B) of pKTH10L-induced secretion stress. The transcriptional htrB-lacZ gene fusion schematically shown in Fig. 1A was used to determine the time courses of htrB expression in cells grown at 37°C in TY medium. The strains used for the analyses were B. subtilis 168 htrB::pMutin4 (BFA3041; solid rectangles), B. subtilis 168 cssS::Sp htrB::pMutin4 (BV2015; open rectangles), B. subtilis 168 htrB::pMutin4 containing pKTH10L (solid ellipses), and B. subtilis 168 cssS::Sp htrB::pMutin4 containing pKTH10L (open ellipses). Samples for the determination of β-galactosidase activities (indicated in nanomoles per minute per OD600 unit) were withdrawn at the times indicated. Zero time (t = 0) indicates the transition point between the exponential and postexponential growth phases. The parental strain B. subtilis 168 was used as a negative control (data not shown).
CssS is essential for the heat stress induction of htrA and htrB.
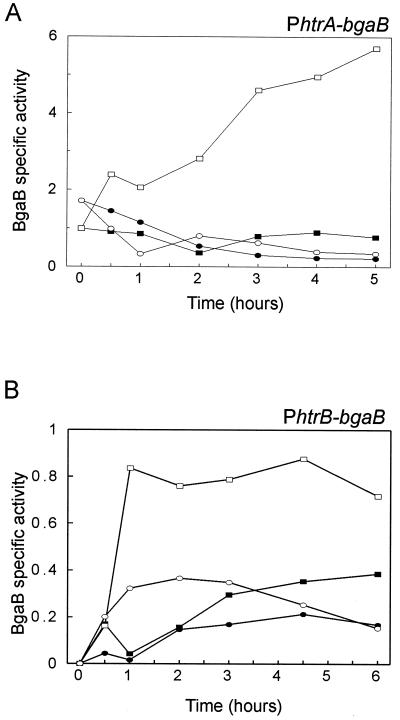
To study whether the CssRS system is involved in the increased transcription of htrA and htrB in response to heat stress, the activities of their respective promoters were monitored using transcriptional fusions with the bgaB reporter gene (which encodes a thermostable β-galactosidase). To avoid interference with normal HtrA and HtrB functions, the promoter-bgaB fusions were positioned at the amyE locus of the B. subtilis chromosome. This resulted in B. subtilis 168 amyE::PhtrA-bgaB (DN2) and B. subtilis 168 amyE::PhtrB-bgaB (DN110). Subsequent disruption of cssS in these strains resulted in B. subtilis BV2024 and B. subtilis BV2025, respectively. To analyze the transcription of htrA-bgaB and htrB-bgaB in response to heat stress, the four strains were first grown in TY medium at 37°C to an OD600 of 0.3 to 0.4. Subsequently, half of the culture was incubated at 48°C and half remained at 37°C. Samples for β-galactosidase measurements were withdrawn from the cultures at specified times after the temperature shift (t = 0). As shown in Figure 3A, htrA-bgaB transcription in the cssS+ background was constant at a low level at 37°C but increased progressively after the temperature upshift to 48°C. When cssS was disrupted, the htrA-bgaB transcription was no longer induced by heat stress (Fig. 3B, 37 and 48°C), showing the critical role of CssS in this process. The htrB-bgaB transcription was low at 37°C (Fig. 3B). In contrast to htrA-bgaB, the htrB-bgaB transcription increased rapidly after heat stress (open rectangles) and then remained constant. Furthermore, when cssS was mutated, the effect of the heat stress on htrB-bgaB transcription was still detectable but was significantly reduced (Fig. 3B, 37 and 48°C). It was established previously that htrA expression is not induced by puromycin (16). Similar experiments have demonstrated that puromycin addition has no effect on htrB expression (data not presented). These findings indicate that heat stress induction of both htrA and htrB is controlled by the CssRS system. The inducing signal for htrA and htrB expression is probably not misfolded cytosolic proteins, since expression of neither gene is induced by puromycin addition.
FIG. 3.
Analysis of htrA and htrB transcription under heat shock conditions. Transcriptional PhtrA-bgaB and PhtrB-bgaB promoter-gene fusions in the amyE locus were used to determine the time courses of htrA and htrB promoter activity in cells grown in TY medium. The cells were first grown at 37°C to an OD600 of 0.3 to 0.4 and then divided into two cultures: one culture remained at 37°C, and the other was transferred to 48°C. Samples for the determination of β-galactosidase (BgaB) activities (indicated in nanomoles per minute per OD600 unit) were withdrawn as a function of time after the temperature shift (t = 0). The parental strain B. subtilis 168 was used as a negative control (data not shown). (A) B. subtilis 168 amyE::PhtrA-bgaB (DN2) at 37°C (solid rectangles) and at 48°C (open rectangles). B. subtilis 168 cssS::Sp amyE::PhtrA-bgaB (BV2024) at 37°C (solid ellipses) and at 48°C (open ellipses). (B) B. subtilis 168 amyE::PhtrB-bgaB (DN110) at 37°C (solid rectangles) and at 48°C (open rectangles). B. subtilis 168 cssS::Sp amyE::PhtrB-bgaB (BV2025) at 37°C (solid ellipses) and at 48°C (open ellipses).
Response of the cssR-cssS operon to heat and secretion stress.
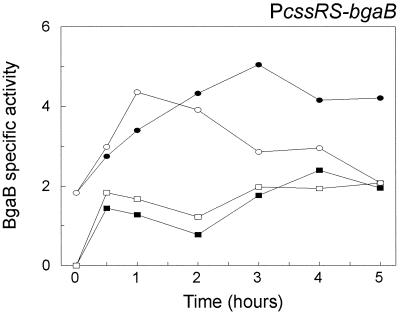
To investigate whether the transcription of the cssRS operon responds to heat stress and secretion stress, a fusion between the promoter region of this operon and the bgaB reporter gene was introduced into the amyE locus of the B. subtilis 168 chromosome, resulting in strain DN220 (Fig. 1B). The cssRS promoter activity was determined in TY medium at 37 and 48°C, as described in the previous section for the PhtrA-bgaB and PhtrB-bgaB constructs. At 37 and 48°C, the activity of the cssRS promoter was low and constant, with little difference in expression between the two temperatures (Fig. 4). Therefore, cssRS expression appeared to be insensitive to heat stress at 48°C. In contrast, secretion stress caused by the overproduction of AmyQ, encoded by plasmid pKTH10, resulted in a more than twofold increase of PcssRS-bgaB transcription at 37°C. An elevated level of PcssRS-bgaB transcription was observed at 48°C in the presence of high α-amylase levels. However, on reaching a maximum at approximately 1 h after the temperature upshift, PcssRS-bgaB transcription started to decrease slowly, reaching the level of the control strain 4 to 5 h after heat induction. Thus, it is apparent that the transcription of cssRS is upregulated in response to secretion stress. In contrast, under the conditions tested, cssRS transcription is not responsive to heat stress in the absence of secretion stress.
FIG. 4.
Analysis of cssRS expression under various stress conditions. A transcriptional bgaB gene fusion in the amyE locus was used to determine the time course of the PcssRS activity in cells grown in TY medium. B. subtilis 168 amyE::PcssRS-bgaB (DN220) was first grown at 37°C to an OD600 of 0.3 to 0.4 and then divided into two cultures: one remained at 37°C (solid rectangles), and the other was transferred to 48°C (open rectangles). Similarly, cultures of this strain containing pKTH10 were incubated at 37°C (solid ellipses) or 48°C (open ellipses). Samples for the determination of β-galactosidase (BgaB) activities (indicated in nanomoles per minute per OD600 unit) were withdrawn as a function of time after the temperature shift (t = 0). The parental strain B. subtilis 168 was used as a negative control (data not shown).
Effect of htrA and htrB mutations on cssRS transcription at 37°C.
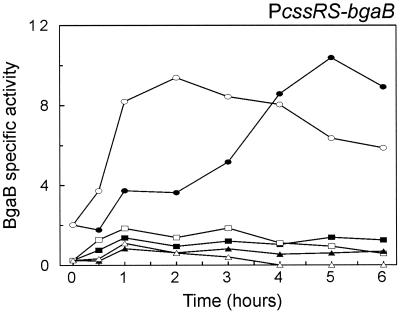
Since HtrA and HtrB are involved in a negative autoregulation and reciprocal cross-regulation of their own genes (15), the possible influences of htrA and htrB mutations on cssRS transcription were investigated. For this purpose, the transcriptional PcssRS-bgaB promoter-gene fusion was introduced into the amyE locus of B. subtilis 168 htrAΔ439 (DN26) and B. subtilis 168 htrB::Km (DN111), resulting in strains BV2017 and BV2019, respectively. In addition, a strain (BV2021) containing mutations in both htrA and htrB and also harboring the PcssRS-bgaB fusion was constructed. Such htrA-htrB double-mutant strains are severely impaired in growth and rapidly accumulate suppressor mutations (15). Therefore, strain BV2021 used in the present studies carried an uncharacterized suppressor mutation that affected growth. The results summarized in Table 3 show the expression levels that persist in each strain at the midexponential phase of the growth cycle in TY medium. Compared to the htrA+ htrB+ cssS+ control strain, the transcription level of the cssRS operon was increased threefold in the htrA single mutant and twofold in the htrB single mutant when cells were grown at 37°C. At this temperature, the cssRS transcription in the htrA htrB double mutant was similar to that in the htrA single mutant, showing that under these conditions, the effects of the mutations were not additive. On closer examination, the transcription of the cssRS operon in the htrA htrB double mutant (BV2021; suppressed) increased during exponential growth and reached its maximum in the postexponential phase (Fig. 5, transition point around 3 h). At that point, the cssRS expression in the htrA htrB double mutant was about fivefold higher than that in the htrA+ htrB+ control strain (DN220). Interestingly, the profile of the cssRS expression in the htrA htrB double mutant was nearly identical to that observed for the htrA single mutant (data not shown). However, in contrast to the midexponential growth phase (Table 3, 37°C), the level of cssRS expression in the postexponential phase was about twofold higher in the double mutant than in the htrA single mutant (data not shown).
TABLE 3.
Expression of transcriptional fusions between the cssR-cssS promoter region and the bgaB reporter gene in various genetics background at 37 and 48°C
| Background strain | Presence of:
|
BgaB activitya at:
|
|||
|---|---|---|---|---|---|
| htrA | htrB | cssS | 37°C | 48°C | |
| DN220 | + | + | + | 1.2 | 1.8 |
| BV2017 | − | + | + | 3.6 | 6.9 |
| BV2019 | + | − | + | 2.6 | 3.9 |
| BV2021; suppressed | − | − | + | 3.7 | 8.2 |
| BV2016 | + | + | − | 0.8 | 1.0 |
| BV2018 | − | + | − | 0.6 | 0.7 |
| BV2020 | + | − | − | 1.6 | 1.5 |
| BV2022; suppressed | − | − | − | 0.8 | 1.1 |
All values represent BgaB activities that were determined 1 h after the cultures were split. One activity unit is defined as 1 nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min per OD600 unit. All values represent the average of two independent experiments.
FIG. 5.
Analysis of cssRS operon expression in the htrA-htrB double mutant. The transcriptional PcssRS-bgaB gene fusion in the amyE locus was used to determine the time courses of the cssR-cssS operon expression in cells grown in TY medium. The cells were first grown at 37°C to an OD600 of 0.3 to 0.4 and then divided into two cultures: one remained at 37°C, and the other was transferred to 48°C. The strains used for the analyses were B. subtilis 168 amyE::PcssRS-bgaB (DN220) at 37°C (solid rectangles) and 48°C (open rectangles), B. subtilis 168 htrAΔ439 htrB::Km amyE::PcssRS-bgaB (BV2021; suppressed) at 37°C (solid ellipses) and 48°C (open ellipses), and B. subtilis 168 cssS::Sp htrAΔ439 htrB::Km amyE::PcssRS-bgaB (BV2022; suppressed) at 37°C (solid triangles) and 48°C (open triangles). β-Galactosidase (BgaB) activities are indicated in nanomoles per minute per OD600 unit. Zero time (t = 0) indicates the time point of the heat shock. The parental strain 168 was used as a negative control (data not shown).
Effect of htrA and htrB mutations on cssRS transcription under heat stress conditions.
Expression of htrA and htrB are both upregulated by heat stress while that of cssRS is elevated in the absence of HtrA and/or HtrB. Since expression of cssRS in the wild-type (htrA+ htrB+) background was insensitive to heat stress, the effects of heat stress on cssRS expression were examined in htrA and htrB mutant backgrounds. Table 3 shows the cssRS transcription levels determined at 37°C and 1 h after a temperature upshift to 48°C. The response of PcssRS-bgaB expression to heat stress differed, depending on the genetic background. In the htrA+ htrB+ cssRS+ control strain (DN220) and in the htrB single mutant (BV2019), there was little difference in cssRS transcription at 37 and 48°C. However, there was an approximately twofold difference in cssRS expression levels in the htrA single mutant (BV2017) and in the htrA htrB double mutant (BV2021; suppressed). Notably, at 48°C the double mutant grew to a lower OD600 (less than 2) than did the single mutants or the parental strain (around 5 [data not shown]). As shown in Fig. 5, the transcription of cssRS in the htrA htrB background increased rapidly for 1 h after heat stress and decreased slowly after 2 h. The maximal levels of cssRS transcription were reached much earlier after heat stress than at 37°C. Since the transcription of cssRS in the htrA or htrB single mutants was lower than that in the double mutant at 48°C, it seems that mutations in both htrA and htrB have an additive effect on cssRS transcription after heat stress (Table 3 and data not shown).
Transcription of cssRS is affected by levels of CssR, CssS, HtrA, HtrB, and secretory proteins.
To investigate whether expression of the cssRS operon is subject to autoregulation, the cssS gene was disrupted in the parental strain and in a series of htrA/B mutants generating the cssS (BV2016) single mutant, the htrA cssS (BV2018) and htrB cssS (BV2020) double mutants, and the htrA htrB cssS (BV2022; suppressed) triple mutant. Expression of the cssRS operon was measured with the bgaB fusion as described in the previous paragraphs. Interestingly, many of these strains did not grow normally at 48°C. The cssS htrA double mutant (BV2018) stopped growing at an OD600 of about 3 instead of 5, the cssS htrB double mutant (BV2020) ceased growth at an OD600 of approximately 2, while the cssS htrA htrB triple mutant (BV2022; suppressed) ceased growth at an OD600 of below 2. Table 3 shows the expression levels of the cssRS operon in these mutant strains at 37°C and after 1 h of heat stress at 48°C. Expression of cssRS was very marginally affected by mutation of cssS at 37°C (1.2 U reduced to 0.8 U) and at 48°C (1.8 U reduced to 1 U). The strongest effect was observed in the htrA mutant background, where the level of cssRS expression decreased approximately 10-fold (6.9 U reduced to 0.7 U) at 48°C and 6-fold (3.6 U reduced to 0.6 U) at 37°C compared with the levels in the cssS+ background. Mutation of cssS in the htrB background had a milder effect (reduced approximately twofold) on PcssRS-bgaB expression. However as shown in Fig. 5, transcription of PcssRS-bgaB in the htrA htrB double mutant dropped to a low level in the absence of cssS.
To investigate whether the response of cssRS transcription to secretion stress is dependent on the CssRS system, a transcriptional cssR-lacZ fusion was constructed by insertion of pMutin4 into the cssR gene of B. subtilis KS408, which contains a xylose-inducible gene for the α-amylase AmyL of Bacillus licheniformis in the xylR locus. The resulting cssR mutant strain DN226 was grown in TY medium at 37°C in the presence or absence of 1% xylose. In contrast to the cssR+ parental strain, the induction of amyL did not result in increased cssR transcription in the cssR mutant background (data not shown). In fact, the cssR transcription was about 10-fold reduced compared to that of the non-secretion-stressed cssR+ control strain (data not shown). Taken together, these data show that the CssRS system regulates its own expression in response to secretion stress. Under conditions of heat stress, CssRS has little effect on cssRS expression in a wild-type background, but this two-component system does regulate its own expression in htrA or htrB single mutants and htrA htrB double-mutant backgrounds.
Increased cssRS transcription on overexpression of CssS.
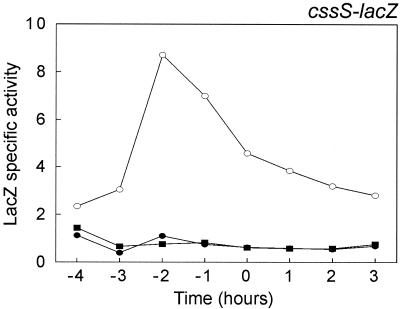
To study the effects of CssS overexpression on the transcription of cssRS, the cssS gene was placed under the control of a xylose-inducible promoter in the amyE locus of a strain containing a cssS-lacZ fusion (BV2006), resulting in strain BV2023 (Fig. 1C). It should be noted that the cssS gene is disrupted by pMutin2 in the latter strain. As shown in Fig. 6, expression of the cssRS operon was up to eightfold higher when cssS was overexpressed after induction with 1% xylose in cells grown at 37°C. In contrast, the transcription of cssS remained low and unaffected in the absence of xylose. As a control, it was established that the presence of xylose had no effect on cssS transcription in the cssS mutant control strain (BV2006) (data not shown). Similar results were observed in minimal medium. These observations imply that the expression of the cssRS operon, encoding the CssRS two-component system, is autoregulated.
FIG. 6.
Analysis of cssS transcription on CssS overexpression. A transcriptional cssS-lacZ gene fusion was used to determine the time courses of cssS expression in cells grown at 37°C in TY medium. The strains used for the analyses were B. subtilis 168 cssS::pMutin2 (BV2006; solid rectangles) and B. subtilis 168 cssS::pMutin2 amyE::XcssS (BV2023) in the absence (solid ellipses) or presence (open ellipses) of 1% xylose. The parental strain 168 was used as a negative control (data not shown). β-Galactosidase activities are indicated in nanomoles per minute per OD600 unit. Zero time (t = 0) indicates the transition point between the exponential and postexponential growth phases.
Mapping of the transcriptional start sites of cssRS.
To determine the transcriptional start points of the cssRS operon by primer extension analysis, two different primers were used. Primer YVQA-RT1 is complementary to a region spanning the putative translational start site of cssR. Primer YVQA-RT2 is located further downstream in the coding sequence of cssR, with its 3′ end being located at bp +63 relative to the putative translational start site (+1).
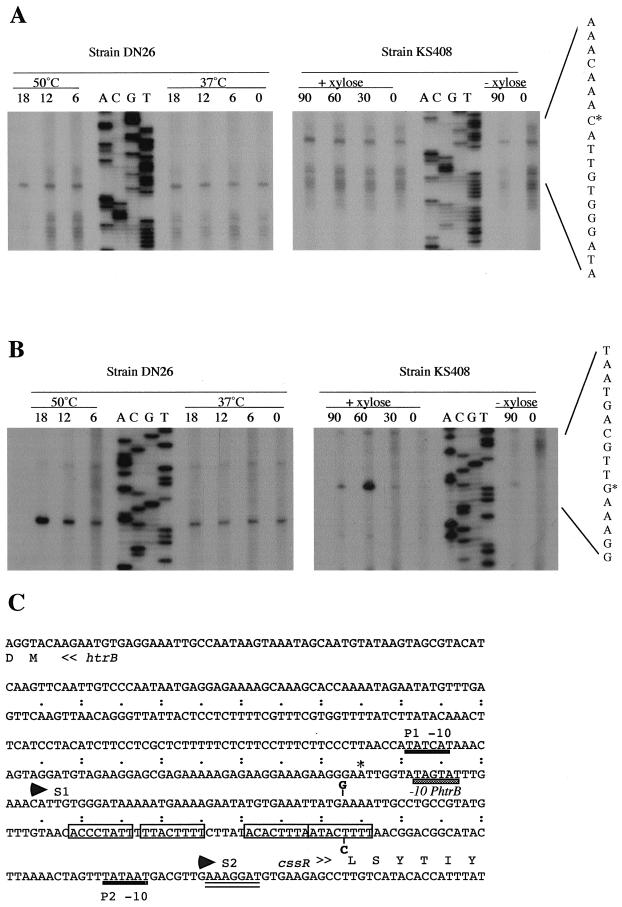
Because the expression levels of cssRS are very low in the parental strain and significantly higher in a htrA mutant background, the effects of heat stress on cssRS transcription were determined with the htrAΔ439 mutant strain (DN26). Using RNA from this latter strain, two transcriptional start points were identified upstream of the cssRS operon. A transcriptional start site designated S1 was identified at a cytosine nucleotide positioned 99 bp upstream of the putative translational start codon. The intensity of this reverse transcript was the same in all RNA samples and did not change in response to heat stress (Fig. 7A, left panel; Fig. 7C). A second transcriptional start site designated S2 was detected at a guanidine nucleotide 18 bp upstream of the putative translational start site (Fig. 7B, left panel; Fig. 7C). In contrast to start site S1, the intensity of the band corresponding to start site S2 was clearly increased in samples from heat-stressed cells. This result using primer extension is consistent with the data presented in Table 3 using a PcssRS-bgaB transcriptional fusion; both results show that the cssRS operon is heat stress inducible in a htrA mutant background. Both transcriptional start sites are located downstream of −10 motifs typical of σA-dependent promoters (Fig. 7C). However, a canonical σA-type −35 motif is not apparent in either promoter.
FIG. 7.
Transcriptional analysis of cssRS. The transcription of cssRS under heat shock and secretion stress conditions was analyzed by primer extension. RNA was isolated from the DN26 mutant (htrAΔ439) under heat shock conditions and the KS408 strain (xylR-amyL) under secretion stress conditions. The time points of sampling are indicated in minutes above each lane. For DN26, zero time represents the time point at which cells grown in TY medium were subject to heat shock. For KS408, zero time represents the beginning of exponential growth (t = 0; OD550 about 0.5) in TY medium with or without 1% xylose. (A) Primer extension of total RNA using primer YVQA-RT1. Equivalent amounts of RNA (12.5 μg) were used from all samples. The sequencing ladders were generated with primer YVQA-RT1. The start point of transcription is indicated (∗). (B) Primer extension of total RNA using primer YVQA-RT2. Equivalent amounts of RNA (12.5 μg) were used from all samples. The sequencing ladders were generated with primer YVQA-RT2. The start point of transcription is indicated (∗). (C) Nucleotide sequence of the intergenic region between the cssRS operon and the divergently transcribed htrB gene. The transcriptional start sites, S1 and S2, are indicated by arrowheads above the sequence. Heavy black bars below the sequence indicate two σA-type −10 motifs, denoted P1 and P2, upstream of the S1 and S2 start sites, respectively. The double-underlined sequence represents the putative ribosome binding site of cssR. The repeated motif (TTTTCACA) common to the inducible promoters of cssRS and htrB are boxed. The grey-shaded box represents the −10 motif of the inducible htrB promoter (−10 PhtrB), and the initiation point of transcription is indicated by an asterisk.
To study the effects of secretion stress on cssRS transcription, the transcription start sites of cssRS were mapped with RNA from strain KS408, which contains a xylose-inducible amyL gene at the xylR locus. Similar to the heat stress experiment, the two transcriptional start sites S1 and S2 were detected with RNA from KS408 with or without xylose induction. While the level of mRNA with the S1 start site was mildly increased in cells grown in the presence of xylose (Fig. 7A, right panel), the level of mRNA with the S2 start site was significantly increased after 60 min of exponential growth under these conditions (Fig. 7B, right panel). In summary, these data indicate that the cssRS operon has two transcriptional start sites: a noninducible S1 site that might correspond to a maintenance promoter and an inducible S2 site for a promoter that is responsive to heat and secretion stress in a htrA mutant background.
The inducible htrB and cssRS promoters share a regulatory sequence.
The htrB and cssRS genes are juxtaposed on the chromosome and are divergently transcribed (Fig. 7C). Furthermore, the inducible promoters of htrB and cssRS have good consensus σA-type −10 regions but do not have σA-type −35 regions. Instead, overlapping with and/or upstream of the −35 regions of both promoters is a 40-bp sequence that contains four copies of the TTTTCACA sequence, shown to be essential for heat-inducible expression of htrA (16). To investigate the possibility that these motifs may participate in inducible expression of both htrB and cssRS, strains were generated in which a single point mutation was introduced into one motif, changing it from TTTTCATA to TTTCCATA at position −60 relative to the cssR start codon (Fig. 7C). To test the effects of this point mutation on the activity of the htrB promoter in response to heat stress, the original or mutant htrB-cssRS regulatory regions were placed in front of the bgaB gene and integrated in the amyE locus of htrA or htrB mutant strains (Table 4). The comparison of the bgaB expression levels directed by the original (PhtrB-bgaB) and the mutant (P*htrB-bgaB) regulatory sequences at 37°C shows that the point mutation reduces the htrB promoter activity in both the htrA (DN113 and DN119) and htrB (DN112 and DN120) mutant backgrounds. More significantly, this mutation also reduces the heat stress-induced activity (1 h at 48°C) of the htrB promoter by 10- to 20-fold in both genetic backgrounds.
TABLE 4.
Comparison of the levels of expression of P htrB-bgaB and of the fusion containing a point mutation P*htrB-bgaB in htrA and htrB null mutant backgrounds at 37°C and after heat stress for 1 h at 48°C
| Background | Strain | BgaB activitya PhtrB-bgaB at:
|
Strain | BgaB activity P*htrB-bgaB at:
|
|||
|---|---|---|---|---|---|---|---|
| htrA | htrB | 37°C | 48°C | 37°C | 48°C | ||
| − | + | DN113 | 2.4 (17) | 40.6 (268) | DN119 | 0.5 (7) | 2.4 (24) |
| + | − | DN112 | 2.5 (12) | 31.7 (184) | DN120 | 0.5 (5) | 1.3 (15) |
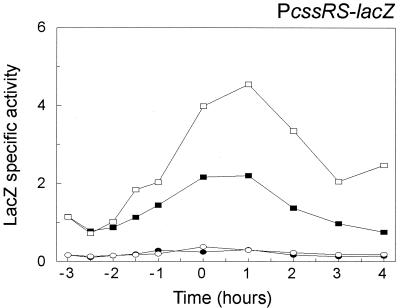
To investigate the effects of the point mutation on the transcription of the cssRS operon in relation to secretion stress, PcssRS-lacZ fusions with and without the mutation (Fig. 1D) were introduced at the cssRS locus of B. subtilis KS408. This strain contains a xylose-inducible amyL gene at the xylR locus. The resulting strains, DN225 and DN228, were grown in TY medium in the presence or absence of 1% xylose, and the β-galactosidase levels were measured throughout the growth cycle (Fig. 8). The promoter activity of the intact PcssRS-lacZ fusion was about twofold increased in postexponential DN225 cells expressing AmyL due to xylose induction. In contrast, the mutated P*cssRS-lacZ displayed significantly reduced levels of promoter activity irrespective of the induction of AmyL synthesis with xylose. It is interesting that a mutation in cssRS or a point mutation in the htrB-cssRS regulatory region reduced the transcription of htrB and cssRS in a similar manner. These data show that through a shared regulatory sequence that participates in heat and secretion stress induction, the expression of htrB and cssRS are linked.
FIG. 8.
Effect of a point mutation in the cssRS promoter on its induction by secretion stress. The transcriptional PcssRS-lacZ fusion in B. subtilis 168 xylR::xylR-amyL PcssRS::pDN221 (DN225), which contains a xylose-inducible amyL gene, was used to determine the time courses of cssRS promoter activity in cells grown at 37°C in TY medium with 1% xylose (open rectangles) or without xylose (solid rectangles). In parallel, the transcriptional P*cssRS-lacZ fusion in B. subtilis 168 xylR::xylR-amyL PcssRS::pDN222 (DN228) was grown in the presence (open ellipses) or absence (solid ellipses) of 1% xylose. β-Galactosidase activities were measured in nanomoles per minute per OD600 unit. Zero time (t = 0) indicates the transition point between the exponential and postexponential growth phases.
DISCUSSION
The synthesis of proteases at elevated levels is one of a variety of cellular responses that counteract the detrimental effects of the presence of misfolded proteins. The present studies show that the CssR-CssS two-component system responds to heat and secretion stress by activating the expression of two HtrA-like proteases encoded by htrA and htrB. Misfolded proteins can accumulate in the cell through thermal denaturation or from a limited availability of appropriate folding catalysts at the extracytoplasmic side of the membrane. The latter mechanism would operate particularly on high-level production of secreted proteins. In this respect, it is important to bear in mind that most proteins of B. subtilis are transported across the membrane in an unfolded conformation via the Sec translocation channel (25). The CssRS two-component regulatory system detects secretion stress by sensing the accumulation of misfolded proteins at the membrane-cell wall interface (11). The CssRS-inducing signal is not cytosolic misfolded proteins, since neither htrA nor htrB expression is induced by puromycin addition. The present observation that the expression of CssRS-controlled genes is, in principle, responsive both to heat and secretion stress indicates that the CssRS system can sense misfolded proteins extracytosolically, irrespective of the cause that leads to their accumulation.
It is interesting that CssRS-dependent heat stress induction of the cssRS operon is observed under these conditions in the absence of htrA and not in a wild-type background. However, cssRS is induced by secretion stress in a CssRS-dependent manner. Therefore, cssR and cssS are both members of the CssRS regulon, but only under secretion stress conditions. Perhaps the level of misfolded proteins generated by heat stress in a wild-type background is sufficient to stimulate the expression of htrA and htrB but insufficient to stimulate the expression of cssRS. The higher level of misfolded protein that persists after heat stress in the absence of HtrA, or overexpression of α-amylase, is necessary to stimulate CssRS-dependent expression of the cssRS operon. This would imply that cssRS is responsive to different levels of misfolded proteins.
These results, together with previous studies, justify the regrouping of heat-inducible genes of B. subtilis into five distinct classes that differ in their regulatory mechanisms. The class I genes are regulated by σA and the HrcA repressor, the class II genes are regulated by σB, and the class III genes are regulated by the CtsR transcription factor. Since the htrA and htrB genes are not regulated by any of the known regulatory systems of the class I, II, or III genes, the present studies indicate that they belong to a separate class of heat stress-inducible genes, which are controlled by the CssRS two-component regulatory system. Since only genes induced in a wild-type background can be considered bona fide members of a class of heat stress genes, it is evident that in sensu strictu, only htrA and htrB are members of this new CssRS-dependent grouping of heat stress genes, termed class V. However, since cssRS is autoregulated under certain genetic and environmental conditions, there are consequently four genes in the CssRS regulon: cssS, cssR, htrA, and htrB
The htrA and htrB genes were previously shown to have one transcriptional start site (15), whereas the cssRS operon has two transcriptional start sites, only one of which is heat and secretion stress responsive. The three stress-inducible promoters have canonical −10 regions typical of σA-type promoters but none have canonical −35 regions. All three stress-inducible promoters have repeated octameric motifs (TTTTCATA) in the vicinity of the −35 regions, and it has been demonstrated that deletion of one copy of this motif from the htrA promoter results in a dramatic (>20-fold) decrease in heat stress induction (16). This study shows that a point mutation in repeat I of the octameric consensus sequence (the point mutation is in the htrB copy corresponding to the one deleted in the htrA promoter [see the discussion of promoter alignment in reference 16] affects heat and secretion stress induction of both the htrB and cssRS genes. These data show that stress-induced expression of htrB and cssRS are linked through this common regulatory sequence, perhaps to make the levels of protease (HtrA and HtrB) and regulator (CssR and CssS) responsive to the prevailing stress conditions. It is tempting to speculate that CssR binds to these repeated motifs to modulate gene expression in response to stress. The identification of a noninducible promoter activity upstream of the cssRS operon suggests the existence of a second mode of regulation for this operon.
The CssRS system bears some resemblance to the CpxA-CpxR two-component system from E. coli. First, CpxA and CssS show amino acid sequence similarities, and the same is true for CpxR and CssR (11). Second, these two systems control the transcription of genes encoding HtrA-like proteases: htrA (degP) of E. coli is regulated by the CpxAR system (4), and htrA and htrB of B. subtilis is regulated by the CssRS system. Finally, like the cpxAR operon (7, 20), the transcription of the cssRS operon is autoregulated. Notwithstanding these similarities, the htrA, htrB, and cssRS promoter regions lack the CpxR consensus binding site. This work unveils an even more remarkable difference, since CpxAR and CssRS seem to respond to different stimuli. While the CssRS two-component system can respond to heat stress, the equivalent task in E. coli is accomplished by σE and not by CpxAR. This could be because the σE-mediated stress response pathway senses misfolded outer membrane or periplasmic proteins whereas the CpxAR pathway senses inner membrane-associated aggregates or misfolded protein (21). In contrast to E. coli, the gram-positive B. subtilis lacks an outer membrane, having a thick matrix of cell wall polymers instead (2). Thus, it seems as if one system might be sufficient to sense and respond to the accumulation of misfolded proteins in the cell envelope of organisms that lack an outer membrane.
The fact that the growth of the htrA-htrB double-mutant strain was severely affected underscores the importance of the proteases encoded by these genes and demonstrates a redundancy in their functions (15). Due to an as yet unidentified suppressor mutation, this strain displayed similar growth characteristics to the parental strain at 37°C. Nevertheless, the suppressor mutation did not completely restore the growth at 48°C. In fact, the growth at 48°C of all strains with a double mutation in cssS, htrA, or htrB was affected. Notably, the strongest effect on growth at 48°C was observed when the two protease genes were mutated, and this was not exacerbated by the introduction of an additional mutation in cssS. This implies that the CssRS system combats the detrimental effects of heat stress primarily by the upregulation of htrA and htrB. Strikingly, the cssS-htrA double mutant grew better at 48°C than did the cssS-htrB double mutant, indicating that HtrA and HtrB have at least partly different functions. The latter view is supported by the observation that htrB transcription showed a faster response to heat stress than did that of htrA. Interestingly, the expression of htrB was still upregulated by heat stress in a cssS mutant background whereas the expression of htrA was no longer heat inducible in the absence of CssS. Thus, it seems that heat-induced expression of htrB may not be exclusively CssS dependent.
In conclusion, this paper documents the existence of a novel class of heat stress-responsive genes in B. subtilis. These class V genes (htrA and htrB) are regulated by the CssRS two-component system. The CssRS two-component system can also be induced by secretion stress, and therefore the regulon comprises four genes (cssR, cssS, htrA, and htrB) under these conditions. Ongoing research is focused on the determination of the precise binding site for CssR and the complete definition of the CssRS regulon.
Acknowledgments
We thank C. Eschevins, M. Hecker, H. L. Hyyryläinen, J. D. H. Jongbloed, V. P. Kontinen, M. Sarvas, W. Schumann, S. Seror, H. Tjalsma, and other members of the Groningen/European Bacillus Secretion Groups for stimulating discussions.
S.B. and J.M.V.D. were supported in part by European Union (EU) “Quality of Life and Management of Living Resources” grants QLK3-CT-1999-00413 and QLK3-CT-1999-00917. A.M. was supported by the CEU projects BIO4-CT95-0278 and QLG2-1999-014555. E.D. was supported by the Ubbo Emmius foundation of the University of Groningen. Work in the K.M.D. laboratory is supported by European Union (EU) “Quality of Life and Management of Living Resources” grant QLG2-1999-01455.
E.D. and D.N. contributed equally to this work.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 3.Benson, A. K., and W. G. Haldenwang. 1993. The sigma B-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J. Bacteriol. 175:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 5.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 6.Derré, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 7.De Wulf, P., O. Kwon, and E. C. Lin. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181:6772-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gryczan, T. J., S. Contente, and D. Dubnau. 1978. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J. Bacteriol. 134:318-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecker, M., and U. Völker. 1990. General stress proteins in Bacillus subtilis. FEMS Microbiol. Ecol. 74:197-214. [Google Scholar]
- 11.Hyyryläinen, H. K., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Prágai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system of Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 12.Kim, L., A. Mogk, and W. Schumann. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71-76. [DOI] [PubMed] [Google Scholar]
- 13.Leskela, S., V. P. Kontinen, and M. Sarvas. 1996. Molecular analysis of an operon in Bacillus subtilis encoding a novel ABC transporter with a role in exoprotein production, sporulation and competence. Microbiology 142:71-77. [DOI] [PubMed] [Google Scholar]
- 14.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noone, D., A. Howell, R. Collery, and K. M. Devine. 2001. YkdA and YvtA, HtrA-like serine proteases in Bacillus subtilis, engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J. Bacteriol. 183:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noone, D., A. Howell, and K. M. Devine. 2000. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J. Bacteriol. 182:1592-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palva, I. 1982. Molecular cloning of alpha-amylase gene from Bacillus amyloliquefaciens and its expression in B. subtilis. Gene 19:81-87. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 19.Price, C. P. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-198. In G. Stortz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 20.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raivio, T. L., and T. J. Silhavy. 1999. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatias. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Stephenson, K., and C. R. Harwood. 1998. Influence of a cell-wall-associated protease on production of alpha-amylase by Bacillus subtilis. Appl. Environ. Microbiol. 64:2875-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 27.van Dijl, J. M., A. de Jong, G. Venema, and S. Bron. 1995. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. Structural and functional similarities with LexA-like proteases. J. Biol. Chem. 270:3611-3618. [DOI] [PubMed] [Google Scholar]
- 28.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 29.Yuan, G., and S. L. Wong. 1995. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177:6462-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan, G., and S. L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the sigma A-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]