Abstract
Yersinia enterocolitica O:8 has two contact-dependent type III secretion systems (TTSSs). The Ysa TTSS is encoded by a set of genes located on the chromosome and exports Ysp proteins. The Ysc TTSS and the Yop effector proteins it exports are encoded by genes located on plasmid pYVe8081. In this study, secretion of YspG, YspH, and YspJ by the Ysa TTSS was shown to require pYVe8081. Furthermore, mutations that blocked the function of the Ysc TTSS did not affect YspG, YspH, and YspJ production. This indicated that YspG, YspH, and YspJ are encoded by genes located on pYVe8081 and that they may correspond to Yops. A comparison of Ysps with Yop effectors secreted by Y. enterocolitica indicated that YspG, YspH, and YspJ have apparent molecular masses similar to those of YopN, YopP, and YopE, respectively. Immunoblot analysis demonstrated that antibodies directed against YopN, YopP, and YopE recognized YspG, YspH, and YspJ. Furthermore, mutations in yopN, yopP, and yopE specifically blocked YopN, YopP, and YopE secretion by the Ysc TTSS and YspG, YspH, and YspJ secretion by the Ysa TTSS. These results indicate YspG, YspH, and YspJ are actually YopN, YopP, and YopE. Additional analysis demonstrated that YopP and YspH secretion was restored to yopP mutants by complementation in trans with a wild-type copy of the yopP gene. Examination of Y. enterocolitica-infected J774A.1 macrophages revealed that both the Ysc and Ysa TTSSs contribute to YopP-dependent suppression of tumor necrosis factor alpha production. This indicates that both the Ysa and Ysc TTSSs are capable of targeting YopP and that they influence Y. enterocolitica interactions with macrophages. Taken together, these results suggest that the Ysa and Ysc TTSSs contribute to Y. enterocolitica virulence by exporting both unique and common subsets of effectors.
Yersinia enterocolitica causes a range of gastrointestinal illnesses in humans and is primarily transmitted though the consumption of contaminated food or water (6). After ingestion, this bacterium invades host tissue by traversing M cells to gain access to underlying gastrointestinal system-associated lymphoid tissue called Peyer's patches. Subsequently, Y. enterocolitica disseminates to the mesenteric lymph nodes and occasionally spreads systemically (7, 8). The virulence of Y. enterocolitica is thought to involve the activities of two different contact-dependent type III secretion systems (TTSSs). The Ysc TTSS secretes Yops (Yersinia outer proteins), and the Ysa TTSS secretes Ysps (Yersinia secreted proteins) (10, 17). A third TTSS associated with virulence is an integral part of the flagellum which secretes Fops (flagellar outer proteins) (47). The functions of the Ysc TTSS have been studied extensively. Genes encoding the Ysc secretion apparatus and the secreted Yops are located on a 68-kb plasmid called pYVe8081 in Y. enterocolitica serotype O:8 (42). A similar plasmid-encoded TTSS is also present in Yersinia pestis, Yersinia pseudotuberculosis, and other pathogenic serotypes of Y. enterocolitica (30-32). Studies focusing on the functions of the Ysc TTSS have demonstrated that Yop proteins are secreted in a contact-dependent manner during infection of cultured mammalian cells, with many of the Yops being translocated directly into the cytosol of the cells (33, 43, 44). The Yops perform a number of functions inside eukaryotic cells, leading to inhibition of phagocytosis by macrophages and polymorphonuclear leukocytes, suppression of T- and B-lymphocyte activation, and alteration of cytokine production by T and B lymphocytes (18). Specific activities of Yop effectors include host cell GTPase activation by YopE (1, 45), protein tyrosine phosphatase activity by YopH (16), serine/threonine kinase activity by YopO (YpkA) (14), and down-regulation of mitogen-activated protein (MAP) kinase pathways by YopP (YopJ) (4, 27). Other proteins, including YopB, YopD, YopQ (YopK), YopR, LcrV, YscM1, and YopN, that are secreted by the Ysc TTSS have activities required for coordinating the export and targeting of Yop effectors (2).
The Ysa TTSS, which is encoded by a gene cluster in the chromosome of Y. enterocolitica, was recently described and was shown to be required for the secretion of a number of Ysps (17, 46). The functions of Ysps are not known, but similarities between components that form the secretion apparatus of the Ysa TTSS and other contact-dependent TTSSs indicate that this system plays a role in the pathogenesis of Y. enterocolitica. Consistent with this hypothesis, a strain defective for Ysa TTSS function exhibited reduced virulence when mice were infected orally (17). The flagellar TTSS has been shown to secrete a number of Fops (47). Characterization of one Fop revealed that it was the previously described phospholipase YplA (47). This Fop has been shown to contribute to the survival of Y. enterocolitica in the host and inflammation of the gastrointestinal system-associated lymphoid tissue (39).
Since the apparatus that forms each of these secretion systems has conserved features in Y. enterocolitica, it is possible that some substrates may be recognized by more than one secretion system. Consistent with this hypothesis, an earlier study demonstrated that YplA can be exported by the Ysc, Ysa, and flagellar TTSSs (46). Results from the same study also indicated that secretion of some Ysps by the Ysa TTSS was affected by the loss of pYVe8081. Previously, it was thought that the only type III secretion-related genes carried by pYVe8081 were associated with the Ysc TTSS (46). This led to the hypothesis that some Ysps may be encoded by genes located on pYVe8081 and that these proteins were Yops which are exported by both the Ysc and Ysa TTSSs. To begin to address this hypothesis, we further examined the requirement for pYVe8081 in Ysp secretion.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, Escherichia coli strains were grown at 37°C and Y. enterocolitica strains were grown at 26°C. E. coli S17-1λpir was used to deliver mobilizable plasmids to Y. enterocolitica. For general manipulations, bacteria were cultivated in Luria-Bertani broth (1% tryptone, 0.5% yeast extract, 90 mM NaCl) or on Luria-Bertani agar (Difco). The medium used for the examination of protein secretion by Y. enterocolitica was Luria broth base (L medium; 1% tryptone, 0.5% yeast extract) adjusted to contain NaCl at the final concentrations indicated in the text. When necessary, depletion of calcium from the medium was accomplished by the addition of 20 mM sodium oxalate and 20 mM MgCl2. Antibiotics (in micrograms per milliliter) were used as follows. For Y. enterocolitica, working concentrations were kanamycin, 100; chloramphenicol, 10; tetracycline, 7; nalidixic acid, 20; and gentamicin, 100. For E. coli, working concentrations were kanamycin, 50; chloramphenicol, 25; and tetracycline, 15.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Y. enterocolitica | ||
| JB580v | Serogroup O:8; Nalr ΔyenR (R− M+) | 19 |
| GY4455 | yscK::TnMod-lacZYA-RKm′ | 29 |
| YVM356 | yscR::mini-Tn5 Km2 | 11 |
| YVM351 | yscU::mini-Tn5 Km2 | 11 |
| YVM373 | yscC::mini-Tn5 Km2 | 11 |
| YVM377 | yscL::mini-Tn5 Km2 | 11 |
| GY4478 | pYVe8081− | 46 |
| GY4418 | ysaV::mini-Tn5 Km2 | 46 |
| GY4428 | ysaV::pEP185.2 | This study |
| GY4499 | ysaT::TnMod-RKm′ | 46 |
| GY4618 | yopP::pEP185.2 | This study |
| YVM421 | yopP::mini-Tn5 Km2 | 11 |
| GY1087 | yscR::mini-Tn5 Km2 ysaV::pEP185.2 | This study |
| GY4764 | yopN::pEP185.2 | This study |
| GY4765 | yopE::pEP185.2 | This study |
| E. coli S17-1λpir | recA thi pro hsdRM+ RP4::2-Tc::Mu::Km Tn7 λpir | 41 |
| Plasmids | ||
| pEP185.2 | mob+; pir-dependent oriR6K; Cmr | 19 |
| pTM100 | mob+; derivative of pACYC184; Cmr Tetr | 21 |
| pGY410 | yopOP locus in pCR Blunt II TOPO | This study |
| pGY411 | yopP in pCR Blunt II TOPO | This study |
| pGY415 | yopP in pTM100 | This study |
| pGY423 | yopOP in pTM100 | This study |
| pGY352 | 0.9-kb fragment of ysaV in pEP185.2 | K. G. Venecia and G. M. Younga |
| pGY375 | 0.5-kb fragment of yopP in pCR Blunt II TOPO | This study |
| pGY381 | 0.5-kb fragment of yopP in pEP185.2 | This study |
| pGY449 | 0.47-kb fragment of yopN in pCR Blunt II TOPO | This study |
| pGY450 | 0.6-kb fragment of yopE in pCR Blunt II TOPO | This study |
| pGY451 | 0.47-kb fragment of yopN in pEP185.2 | This study |
| pGY452 | 0.6-kb fragment of yopE in pEP185.2 | This study |
Unpublished data.
The yopP-complementing plasmids were created by cloning DNA fragments generated by PCR using Pfu polymerase (Stratagene). Genomic DNA from wild-type Y. enterocolitica was used as the template for primer YopP3 (5′ AGT GTA TGT CAC TCA GCC 3′), which annealed 3.5 kb upstream of the yopP open reading frame, and primer YopP5 (5′ AGA GCT GA CCG TAT TCC 3′), which annealed just 3′ of the yopP open reading frame. The 4.4-kb fragment, which contains yopO and yopP, was cloned into pCR Blunt II TOPO (Invitrogen) to create pGY410 and then subcloned into pTM100 via EcoRI to create pGY423 (yopOP+). A second yopP-complementing clone, pGY415 (yopP+), was also constructed. PCR was used to amplify a 1.3-kb fragment containing the yopP open reading frame and about 380 bp of upstream sequence. Primers YopP4 (5′GAA TGG ATG TGA CAA GTG 3′) and YopP5 (see above) were used. This fragment was also cloned into pCR Blunt II TOPO to create pGY411 and then subcloned into pTM100 via EcoRI to create pGY415 (yopP+). The orientation of yopP in both pGY423 and pGY415 was checked to ensure that the cloned yopP fragment was in the orientation opposite to that of the vector-derived cat gene. The sequences of the cloned PCR-generated fragments were determined using a DNA-sequencing system and a BigDye terminator cycle-sequencing kit (Applied Biosystems) according to the manufacturer's instructions.
Construction of yopN, yopP, and yopE mutants.
Insertion mutations in yopN, yopP, and yopE were constructed as follows using previously described procedures (19). A fragment of DNA corresponding to an internal region of each open reading frame was amplified by PCR from wild-type Y. enterocolitica genomic DNA. The primers for yopN were YopN1 (5′ TGT AGA TCT GCA CTA AGC GC 3′) and YopN2 (5′ TGC AGA TAG TCA GCG GC 3′), those for yopP were YopP1 (5′ GAT ATA GCG GAT GGA TCC 3′) and YopP2 (5′ CTT ATT GTG GGG TAA AGG 3′), and those for yopE were YopE1(5′ TAT TTC TAC ATC ACT GCC CC 3′) and YopE2 (5′ AAT TGA TGC ATC TGT TGC GC 3′). Each DNA fragment was cloned into pCR Blunt II TOPO to create pGY449, pGY375, and pGY450, respectively (Table 1). The DNA fragments were then subcloned into the suicide vector pEP185.2. The yopN fragment was subcloned by cutting pGY449 with KpnI and XhoI. The corresponding 0.47-kb fragment was then ligated with pEP185.2 that had been digested with KpnI and XhoI to generate pGY451 (Table 1). The yopP fragment was subcloned by cutting pGY375 with BamHI to take advantage of a 5′ BamHI site in yopP and a 3′ BamHI site in the vector. The 0.5-kb BamHI fragment was ligated into the BglII site of pEP185.2 to create pGY381 (Table 1). The yopE fragment was subcloned by cutting pGY450 with KpnI and XhoI. The corresponding 0.6-kb fragment was then ligated with pEP185.2 that had been digested with KpnI and XhoI to generate pGY452 (Table 1). Each of the suicide plasmid derivatives was then mobilized from E. coli S17-1λpir into Y. enterocolitica. Matings were plated on LB medium containing nalidixic acid and chloramphenicol to select for strains that had integrated the plasmid into the targeted locus (Table 1).
Construction of other mutant strains.
To create a yscR ysaV double mutant, pGY352 (Table 1) was transferred into YVM356 as described above. Recombinants were selected as strains resistant to nalidixic acid, kanamycin, and chloramphenicol. One recombinant was saved and designated strain GY1087 (yscR::mini-Tn5 Km2 ysaV::pEP185.2).
Preparation of extracellular proteins, SDS-PAGE, and Western blot analysis.
Extracellular proteins were prepared as described previously (46). Y. enterocolitica strains were grown overnight in Luria broth and subcultured at 1:30 into 5 ml of appropriate medium to induce secretion of Ysps or Yops as indicated in the text. Cultures were grown at 26 or 37°C for 6 h and then used to isolate secreted proteins. At the time of harvesting, the optical density at 600 nm (OD600) of the culture was determined. Bacterial cells were removed by centrifugation in a microcentrifuge at 8,000 × g for 5 min. The upper two-thirds of the supernatant was removed and centrifuged again. The upper two-thirds of the supernatant was then removed and passed through a 0.22-μm-pore-size filter. Proteins were concentrated by precipitation with 10% (wt/vol) ice-cold trichloroacetic acid and washed with ice-cold acetone. All samples were resuspended in sample buffer containing 2-mercaptoethanol and heated to 95°C for 5 min. Resuspension volumes were adjusted according to the OD600 of the cultures so that an equivalent amount of each sample was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were visualized by staining them with silver (3) or were transferred to nitrocellulose membranes for Western blot analysis. The membranes were blocked with 2% skim milk in phosphate-buffered saline (PBS) for 1 h. Rabbit polyclonal antibody in 0.2% skim milk and 0.05% Tween 20 in PBS were then added at the following dilutions: α-YopN, 1:20,000; α-YopP, 1:2,000; and α-YopE, 1:20,000. After being extensively washed, the membranes were incubated with goat α-rabbit immunoglobulin G-horseradish peroxidase (Sigma, St. Louis, Mo.) at a dilution of 1:20,000. Western blots were visualized by chemiluminescence (ECL kit; Amersham).
TNF-α biological assays.
The use of a biological assay to monitor the effects of Yersinia on the production of tumor necrosis factor alpha (TNF-α) by host cells was established previously (4, 27). The methods used in this study were the same as those previously described with slight modifications. Murine J774A.1 macrophage-like cells (ATCC TIB-67) were plated at 5 × 104/ml in Dulbecco's modified Eagle's medium (DMEM)-10% fetal bovine serum (FBS) the day before assays were to be performed. One hour before infection, the medium was removed, the cells were washed in serum-free DMEM, and the medium was replaced with DMEM-5% FBS. Prior to the infection of macrophages, bacteria were grown overnight in L broth and subcultured to an OD600 of 0.2 in appropriate medium. To induce the Ysa TTSS, bacteria were cultured in L medium with 0.29 M NaCl for 1 h at 26°C. To induce the Ysc TTSS, bacteria were cultivated in calcium-limited L medium at 37°C for 1 h. Just prior to infection, the OD600 of the cultures was again determined to establish the appropriate volume of culture needed to infect macrophages at a multiplicity of infection of 80. Infection was initiated by adding bacteria to cell monolayers and synchronized by centrifugation of the culture plates for 5 min at 250 × g. Incubation was continued at 26°C in 5% CO2 for induction of the Ysa TTSS and at 37°C in 5% CO2 for induction of the Ysc TTSS, after which aliquots of the supernatants were harvested at appropriate times and tested for the presence of TNF-α. Detection of TNF-α was completed using a biological assay with WEHI-13VAR cells (ATCC CRL-2148). The day before assays were to be performed, 100-μl aliquots of WEHI-13VAR cells were plated in 96-well plates at 3 × 105/ml in RPMI containing 10% FBS. Immediately before the assays, the medium was removed from the WEHI-13VAR cells and replaced with 50 μl of RPMI containing 5% FBS, 1 μg of actinomycin D (Sigma)/ml, and 200 μg of gentamicin/ml. Supernatants from infected macrophages (50 μl) were added to the WEHI-13VAR cells, and the plates were incubated for 20 to 24 h at 37°C in 5% CO2. The medium was removed, the cells were washed once with PBS, and 50 μl of 0.5% crystal violet in 20% methanol was added. After 5 min at room temperature, the wells were rinsed with distilled H2O and 100 μl of 1% SDS was added to solubilize the crystal violet. The plates were incubated for 30 min at 37°C, after which the OD562 of each sample was measured in a microplate reader. The responsiveness of WEHI-13VAR cells to TNF-α was calculated as described previously (4). Accordingly, the amount of TNF-α production is reported as the percent maximal release and is the average ± standard deviation of four independent experiments performed in triplicate. As a control to be sure that culture media did not induce toxic effects, the growth rates of the WEHI-13VAR cells in 50% DMEM-50% RPMI and in 100% RPMI were compared. There was no difference in cell counts after 48 h at 37°C in 5% CO2 (data not shown).
RESULTS
Secretion of YspG, YspH, and YspJ requires the Ysa TTSS and pYVe8081 but does not require a functional Ysc TTSS.
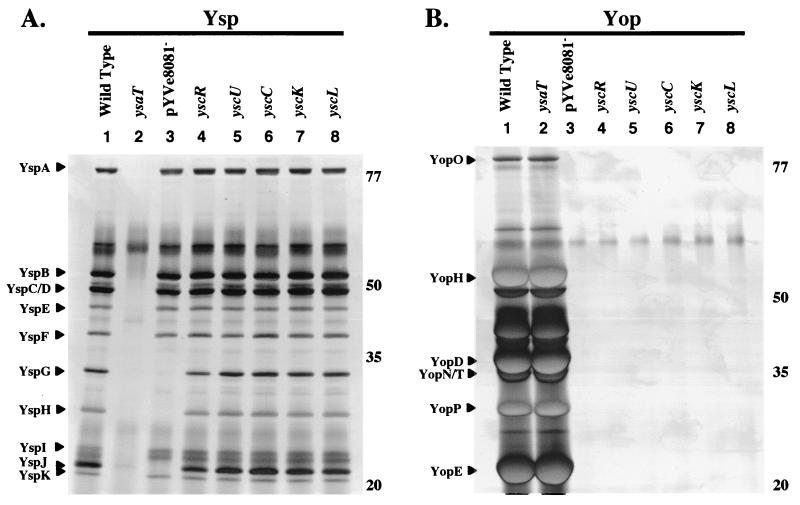
In a previous study, we observed that the loss of pYVe8081 from Y. enterocolitica prevented the secretion of two Ysp proteins of about 32 and 30 kDa (46). Secretion of an additional Ysp that migrated with an apparent molecular mass of 20 kDa was also affected by loss of pYVe8081 (Fig. 1A). This result was overlooked in a previous analysis because this protein tends to comigrate with other Ysps (46). We referred to these proteins as YspG, YspH, and YspJ, respectively, and hypothesized that they are encoded by genes located on pYVe8081. To further test this hypothesis, several strains of Y. enterocolitica that maintained pYVe8081 but carried mutations which inactivated the Ysc TTSS were examined. The purpose of these experiments was to establish that the Ysc TTSS was not contributing to export of YspG, YspH, and YspJ. The strains that were examined had insertion mutations in yscC, yscK, yscL, yscR, and yscU (Table 1). Each of these genes maps to the virB or virC operon, both of which encode components of the Ysc protein export apparatus (42). Each strain was grown under conditions to induce secretion of either Ysps or Yops, and the profile of secreted proteins was examined. Wild-type, Ysa TTSS-defective, and pYVe8081− strains of Y. enterocolitica were included in the analysis as controls (Fig. 1, lanes 1 to 3). The results showed that the Ysp secretion of these ysc mutants was not affected (Fig. 1A, lanes 4 to 8), but secretion of Yops was blocked (Fig. 1B, lanes 4 to 8). This indicated that the secretion of YspG, YspH, and YspJ by the Ysa TTSS requires the presence of pYVe8081 but is not affected by the loss of Ysc TTSS function. The requirement for pYVe8081 for the secretion of YspG, YspH, and YspJ suggested that these proteins might be identical to Yops.
FIG. 1.
Secretion of YspG, YspH, and YspJ is affected by the loss of pYVe8081 but does not require the Ysc TTSS. Secreted proteins were isolated, concentrated from culture supernatants, and separated by SDS-PAGE. The proteins were then visualized by staining them with silver. (A) Cultures were grown at 26°C in L medium supplemented with 290 mM NaCl to induce Ysp production. Lanes: 1, JB580v (wild type); 2, GY4499 (ysaT); 3, GY4478 (pYVe8081−); 4, YVM356 (yscR); 5, YVM351 (yscU); 6, YVM373 (yscC); 7, GY4555 (yscK); 8, YVM377 (yscL). Each lane contains the equivalent of 1 ml of culture supernatant at an OD600 of 1.0. Ysps are assigned according to size on the left, and the approximate locations of molecular mass standards (in kilodaltons) are indicated on the right. (B) Cultures were grown at 37°C in L medium depleted of calcium by the addition of 20 mM sodium oxalate and 20 mM MgCl2 to induce Yop production. The lanes are as listed for panel A. Each lane contains the equivalent of 0.5 ml of culture supernatant at an OD600 of 1.0. Yops are assigned according to size on the left, and the approximate locations of molecular mass standards (in kilodaltons) are indicated on the right.
YspG, YspH, and YspJ correspond to YopN, YopP, and YopE.
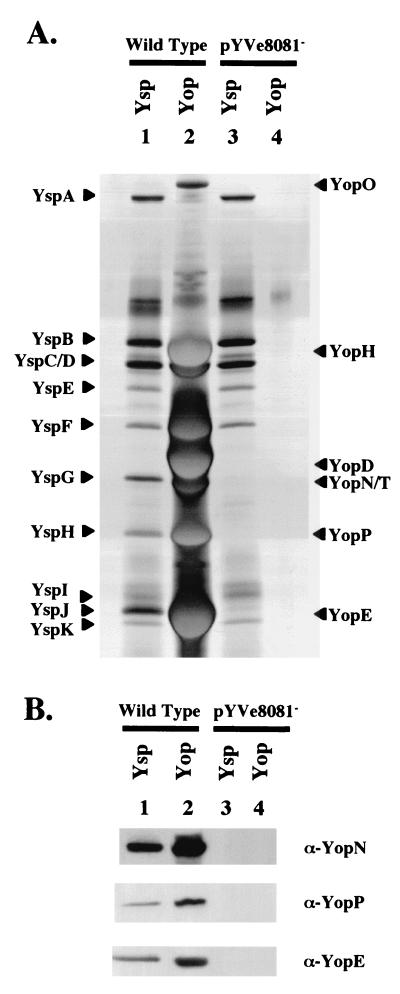
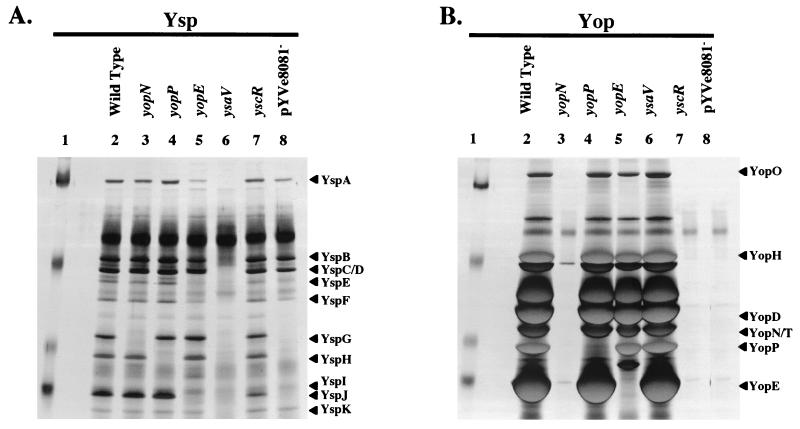
To begin to identify which Yop each of the Ysp proteins might represent, the profiles of Ysps and Yops were compared for wild-type and pYVe8081− strains of Y. enterocolitica (Fig. 2A ). This examination revealed that YspG, YspH, and YspJ migrated with apparent masses similar to those of YopN, YopP, and YopE, respectively. Additional examination by immunoblot analysis showed that antibodies directed against YopN, YopP, and YopE correspondingly recognized YspG, YspH, and YspJ (Fig. 2B). To further investigate the possibility that YspG, YspH, and YspJ correspond to YopN, YopP, and YopE, individual mutants of Y. enterocolitica that had an insertion mutation in yopN, yopP, or yopE were constructed (Fig. 3). Each mutant was examined for the secretion of Ysps and Yops. The results of this analysis showed that the yopN mutant was defective for YspG secretion, the yopP mutant was defective for YspH secretion, and the yopE mutant was defective for YspJ secretion (Fig. 3A, lanes 3, 4, and 5). With regard to Yop secretion, the yopN mutant was defective for the secretion of essentially all Yops (Fig. 3B, lane 3). This result was expected, since yopN is the first gene in the yopN tyeA sycN yscXY lcrD operon and several of these genes are required for Ysc TTSS function. In contrast, yopP and yopE are not included with other genes required for Ysc TTSS function. Accordingly, the yopP mutant did not secrete YopP and the yopE mutant did not secrete YopE, but both mutants retained the ability to secrete other Yops (Fig. 3B, lanes 4 and 5).
FIG. 2.
Comparison of proteins secreted by selected strains of Y. enterocolitica grown under Ysa TTSS- or Ysc TTSS-inducing conditions. (A) Analysis of secreted proteins by SDS-PAGE. Lanes: 1 and 2, Ysp and Yop preparations from cultures of JB580v (Wild Type); 3 and 4, Ysp and Yop preparations from cultures of GY4478 (pYVe8081−). The secreted proteins were prepared and separated as described in the legend to Fig. 1. The cultures for lanes 1 and 3 were grown at 26°C in L medium supplemented with 290 mM NaCl to induce Ysp production, and the lanes contain the equivalent of 1 ml of culture supernatant at an OD600 of 1.0. The cultures for lanes 2 and 4 were grown at 37°C in L medium depleted of calcium by the addition of 20 mMsodium oxalate and 20 mM MgCl2 to induce Yop production, and the lanes contain the equivalent of 0.5 ml of culture supernatant at an OD600 of 1.0. The proteins were visualized by staining them with silver. The locations of selected Ysps and Yops are assigned according to size on the left and right, respectively. (B) Immunoblot analysis of secreted proteins with polyclonal antibodies directed against YopN, YopP, and YopE. The lanes are as described for panel A. Proteins were prepared and separated as indicated in the legend to panel A and then transferred to nitrocellulose membranes and probed with either α-YopN, α-YopP, or α-YopE antibody as indicated on the right side of each panel. The lanes contain the equivalent of 2 and 0.25 ml of culture supernatant at an OD600 of 1.0 for cultures induced for Ysp production and Yop production, respectively.
FIG. 3.
Secretion of YspG, YspH, and YspJ is blocked by mutations in yopN, yopP, and yopE, respectively. (A) Cultures were grown at 26°C in L medium supplemented with 290 mM NaCl to induce Ysp production. (B) Cultures were grown at 37°C in L medium depleted of calcium by the addition of 20 mM sodium oxalate and 20 mM MgCl2 to induce Yop production. Lanes: 1, molecular mass markers; 2, JB580v (Wild Type); 3, GY4764 (yopN); 4, GY4618 (yopP); 5, GY4765 (yopE); 6, GY4428 (ysaV); 7, YVM356 (yscR); 8, GY4488 (pYVe8081−). The locations of selected Ysps and Yops are assigned according to size on the left of each panel. The secreted proteins were isolated and analyzed as described in the legend to Fig. 1. The molecular mass standards are approximately 77, 50, 35, and 20 kDa.
Complementation analysis confirms that yopP encodes YspH.
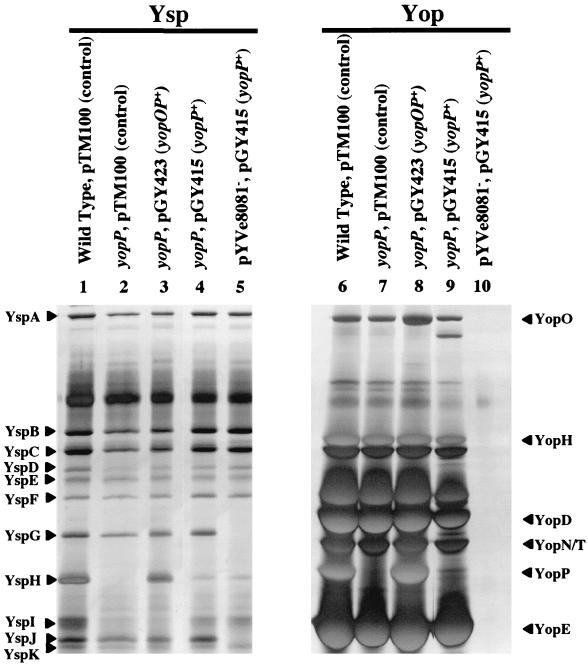
At this stage of the study, we decided to focus our efforts on YspH. Both genetic and biochemical evidence supported the hypothesis that YspH and YopP represent the same protein. Previous studies have indicated that yopP is located downstream of yopO, and based on studies of a similar locus in Y. pseudotuberculosis, these two genes may form an operon (15). Also, the DNA sequence of pYVe8081 revealed that yopP is followed by yscM2, which has a regulatory function for the Ysc TTSS (42). These two genes are separated by ca. 600 bp and would not be expected to be cotranscribed. However, to eliminate the possibility that an insertion mutation in yopP might have a polar effect on yscM2, complementation analysis was used to establish that the loss of YspH secretion could be restored in trans by a plasmid-borne copy of yopOP or yopP. Plasmids pGY423 (yopOP+) and pGY415 (yopP+) were constructed by cloning a 4.4-kb region containing yopOP and a 1.3-kb region containing yopP into pTM100. The cloning vector and each plasmid containing yopP was delivered into the yopP mutant, and the resulting strains were examined for the secretion of Ysps and Yops (Fig. 4). The results showed that genetic complementation of the yopP mutation by either plasmid-encoded yopOP or yopP restored both YspH and YopP secretion (Fig. 4). Therefore, the phenotype of the yopP mutant was not due to polar effects on yscM2. This again indicates that YspH and YopP represent the same protein. The data also indicate that yopO and yopP do not necessarily need to be cotranscribed in order for YopP (YspH) to be secreted by the Ysa TTSS. However, more YopP (YspH) was detected in culture supernatants from strains with pGY423 (yopOP) than from strains with pGY415 (yopP). For clarity, we will refer to the protein encoded by yopP as YopP and use the term YspH only when necessary.
FIG. 4.
Restoration of YspH and YopP secretion by complementation of the yopP mutation. Cultures were grown at 26°C in L medium supplemented with 290 mM NaCl to induce Ysp production for the samples in lanes 1 to 5. Cultures were grown at 37°C in L medium depleted of calcium by the addition of 20 mM sodium oxalate and 20 mM MgCl2 to induce Yop production for the samples in lanes 6 to 10. Lanes: 1 and 6, JB580/pTM100 (Wild Type); 2 and 7, GY4618/pTM100 (yopP, control); 3 and 8, GY4618/pGY423 (yopP, yopOP+); 4 and 9, GY4618/pGY415 (yopP and yopP+); 5 and 10, GY4478/pGY415 (pYVe8081− and yopP+). The locations of selected Ysps and Yops are assigned according to size on the left and right, respectively. The secreted proteins were isolated and analyzed as described in the legend to Fig. 1.
A role for the Ysa TTSS in suppressing TNF-α production by macrophages.
It is well documented that translocation of YopP (YopJ in Y. pseudotuberculosis and Y. pestis) into macrophages leads to suppression of TNF-α production (4, 27). It has also been shown that YopP is required for stimulating macrophage apoptosis (22, 23, 37). Because evidence presented here indicates that YopP is secreted by the chromosomally encoded Ysa TTSS, we hypothesized that secretion of YopP by this system would lead to YopP-dependent phenotypes in macrophages. To address this possibility, we examined the effect of YopP on the release of TNF-α by infected macrophages (Fig. 5). First, it was demonstrated that bacteria grown in calcium-limited media at 37°C exhibited a requirement for a functional Ysc TTSS and YopP in order to suppress TNF-α production by macrophages (Fig. 5A). This result is consistent with those of previous studies (4, 27). Under these conditions, the abilities of mutants defective for the Ysa TTSS to suppress TNF-α production did not appear to be affected. The experiment was then modified to favor expression of the Ysa TTSS by growing the bacteria used to infect macrophages in a medium containing 290 mM NaCl at 26°C and infecting macrophages at 26°C (Fig. 5A). Interestingly, Y. enterocolitica retained the ability to suppress secretion of TNF-α by macrophages under these conditions. Suppression of TNF-α production required both the Ysa TTSS and YopP but no longer required the Ysc TTSS (Fig. 5A). This suggested that YopP was targeted to macrophages by the Ysa TTSS. Consistent with these results, complementation of the yopP mutation with a plasmid-encoded copy of the yopP locus restored the ability of this mutant to suppress TNF-α production under both Ysa and Ysc TTSS-inducing conditions (Fig. 5B). Taken together, these results confirm that the absence of YopP leads to increased TNF-α production by Y. enterocolitica-infected macrophages. It also indicates that YopP is exported and targeted by both the Ysa and Ysc TTSSs.
FIG. 5.
Mutations in yopP affect Ysa and Ysc TTSS-dependent suppression of TNF-α release by Y. enterocolitica-infected macrophages. (A) Macrophages were infected for 2 h with selected strains of Y. enterocolitica as indicated from left to right: control (mock infection), JB580v (wild type [WT]), GY4618 (yopP), GY4499 (ysaT), GY4428 (ysaV), GY4478 (pYVe8081−), YVM356 (yscR), and GY1087 (yscR, ysaV). Macrophage infections were conducted under Ysa TTSS-inducing conditions or Ysc TTSS-inducing conditions as described in the text. The detection of TNF-α production by macrophages was completed using a biological assay as described in Materials and Methods. The results are displayed as the average ± standard deviation of four independent experiments performed in triplicate. The data were statistically analyzed using Student's t test to compare values obtained for infection with the WT strain to infections with each mutant. Values that have differences with a P value of <0.01 (*) and <0.001 (**) are indicated. (B) Complementation of a yopP mutation restores Ysa TTSS- and Ysc TTSS-dependent suppression of TNF-α release by infected macrophages. Macrophages were infected for 2 h with selected strains of Y. enterocolitica as indicated from left to right: control (mock infection), JB580v (WT), GY4618/pTM100 (yopP), GY4618/pGY423 (yopP, yopOP+), and GY4618/pGY415 (yopP and yopP+). Macrophage infections were conducted under Ysa TTSS-inducing conditions or Ysc TTSS-inducing conditions as described in the text. Detection of TNF-α production by macrophages was completed and the data were analyzed as described for panel A.
DISCUSSION
Export of polypeptides to host environments by TTSSs of Y. enterocolitica is important for virulence. Similarities among different TTSSs suggest that some proteins may be exported by more than one pathway. In agreement with this hypothesis, it was determined that YspG, YspH, and YspJ, which are secreted by the chromosomally encoded Ysa TTSS, correspond to the previously characterized YopN, YopP, and YopE proteins that have been shown to be exported by the plasmid-encoded Ysc TTSS. Thus, under the laboratory conditions used in this study, each of these proteins appears to be a substrate of both the Ysa and Ysc TTSSs. The reason that export of these Yops by the Ysa TTSS was not recognized previously may be that induction of the Ysa TTSS occurs under specific conditions. Previous examinations of YopN, YopP, and YopE secretion have established that these proteins are Ysc TTSS substrates (15). However, conditions that induce the Ysc TTSS do not lead to Ysa TTSS induction (references 17 and 46 and this study). It is also possible that other studies used strains of Yersinia that do not possess a TTSS that is equivalent to the Ysa system. For example, a chromosomal locus predicted to encode a second TTSS is present in Y. pestis, but it shows only distant homology to the ysa region of Y. enterocolitica (28).
The observation of Ysa TTSS-dependent secretion of YopP provided the opportunity to examine how this system might affect the activities of host cells. It is already established that the activity of YopP as an effector requires it to be translocated into host cells. The potential for YopP targeting by the Ysa TTSS was tested by examining the effects of Ysa and Ysc TTSS functions on YopP-dependent suppression of TNF-α release by infected macrophages. This analysis confirmed that previously described laboratory conditions known to induce Ysc TTSS function lead to YopP-dependent suppression of TNF-α production by macrophages. However, Ysa TTSS function was required for YopP-dependent suppression of TNF-α production by infected macrophages when experimental conditions were adjusted to induce the Ysa TTSS. These inducing conditions require a temperature that is lower than the expected temperature of a mammalian host. This apparent discrepancy may simply indicate that other environmental conditions in the host contribute to induction of the Ysa TTSS. Nonetheless, the results presented here illustrate the potential for different TTSSs of Y. enterocolitica to export an overlapping group of substrates. They also demonstrate that the Ysa TTSS can deliver effector proteins into host cells.
The first functional evidence that indicated sharing of substrates by different TTSSs of Y. enterocolitica was possible came from a study of the secreted phospholipase YplA (46). Under laboratory conditions, this protein is exported by the flagellar TTSS, but it can also be exported by the Ysa and Ysc TTSSs. Because regulation of YplA production is coupled with the expression of flagellar genes, it is difficult to establish if this protein is exported by alternate pathways in vivo (40). Additional studies will be required to determine if proteins other than YopN, YopP, and YopE are exported by the Ysa and Ysc TTSSs under conditions that are different from those used in this study and those in vivo. Studies of Salmonella enterica serovar Typhimurium have suggested that this bacterium also utilizes two different TTSSs to target some proteins to the host environment (20).
Yersinia infection of mammalian cells has been shown to lead to a rapid induction of multiple MAP kinase pathways. This rapid induction is followed by an inhibition that requires the activity of YopP in Y. enterocolitica and its equivalent, designated YopJ, in Y. pestis and Y. pseudotuberculosis (4, 27, 34-36, 38). One effect of YopP/J activity is that blocking MAP kinase cascades limits production of inflammatory mediators like TNF-α (4, 27). In addition, YopP/J stimulates apoptosis in macrophages by modifying the caspase cascade (12, 22, 23, 37). The mechanism by which YopP/J may influence these pathways appears to be quite complicated (for a review, see reference 24). While it is not fully understood how YopP/J causes these effects, YopP/J binds to members of the MKK family and IKKβ (25). YopP/J is also thought to act as a ubiquitin-like protease, resulting in decreased cellular levels of free SUMO-1 and increased levels of SUMO-1-conjugated proteins, which would affect the stability of putative host regulatory proteins (26). The net effect of YopP/J activity, whether direct or indirect, leads to multiple effects on host cells, including down-regulation of the NF-κB signaling pathway, MAP kinase pathways, and cytokine induction (27, 36). YopE also affects the activities of host cells by acting as a GTPase-activating protein for the Rho family of small GTP-binding proteins, including RhoA, Rac1, and Cdc42 (1, 45). This family of proteins is involved in modulating host cell actin polymerization. The net effect of YopE function is the loss of host cell actin filaments and impairment of phagocytic activity. YopN has been shown to be involved in coordinating the secretion of Yop proteins by the Ysc TTSS (5, 13). It is possible that YopN also has a role in coordinating protein secretion by the Ysa TTSS. Given the multiple effects of YopN, YopP/J, and YopE on host cellular responses, the secretion of effectors by both the Ysa and Ysc TTSSs of Y. enterocolitica may be an attempt by this species to influence the host immune response by targeting different types of cells or by acting at different stages of an infection.
Other evidence also indicates that the Ysa and Ysc TTSSs may contribute to different stages of infection. The Ysa TTSS appears to influence pathogenesis in mice when they are orally infected with Y. enterocolitica but not when they are infected by an intraperitoneal route (17). In contrast, the Ysc TTSS is essential for pathogenesis in mice regardless of the route of infection (9). Perhaps the Ysa TTSS is primed to affect Y. enterocolitica interactions with the host at an early stage of infection in order to limit initial innate immune responses. Subsequently, induction of the Ysc TTSS may be necessary to extend the influence of YopP and YopE on the immune response. The consequence of targeting effectors by two different TTSSs may be that Y. enterocolitica can influence both the innate immune system and the subsequent adaptive immunity of the host.
Acknowledgments
We thank Krista Venecia, Shane Petersen, Virginia Miller, and Andrew Darwin for providing strains and plasmids. We also thank Olaf Schneewind for providing α-YopN, α-YopP, and α-YopE polyclonal antibodies.
This work was supported by University of California start-up funds and an Academic Senate Faculty Research Award.
REFERENCES
- 1.Black, D. S., and J. B. Bliska. 2000. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol. Microbiol. 37:515-527. [DOI] [PubMed] [Google Scholar]
- 2.Bleves, S., and G. R. Cornelis. 2000. How to survive in the host: the Yersinia lesson. Microbes Infect. 2:1451-1460. [DOI] [PubMed] [Google Scholar]
- 3.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 4.Boland, A., and G. R. Cornelis. 1998. Role of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection. Infect. Immun. 66:1878-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland, A., M.-P. Sory, M. Iriatre, C. Kerbourch, P. Wattiau, and G. R. Cornelis. 1996. Status of YopM and YopN in Yersinia yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 15:5191-5201. [PMC free article] [PubMed] [Google Scholar]
- 6.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, P. B. 1975. Oral Yersinia enterocolitica infection of mice. Am. J. Pathol. 81:703-705. [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, P. B., and F. M. Collins. 1974. Experimental Yersinia enterocolitica infection in mice: kinetics of growth. Infect. Immun. 9:851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., T. Biot, C. Lambert de Rouvolt, T. Michiels, B. Mulder, C. Sluters, M.-P. Sory, M. Van Bouchaute, and J.-C. Vanootegham. 1989. The Yersinia yop regulon. Mol. Microbiol. 3:1455-1459. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriate, C. Neyt, M.-P. Sory, and I. Stainer. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 12.Dennecker, G., W. G. Declercq, C. A. A. Boland, R. Banabdilah, M. van Gurp, M. P. Sory, P. Vandenbeele, and G. R. Cornelis. 2001. Yersinia enterocolitica YopP-induced apoptosis of macrophages involves the apoptotic signaling cascade upstream of Bid. J. Biol. Chem. 276:19706-19714. [DOI] [PubMed] [Google Scholar]
- 13.Forsberg, A., A.-M. Viitanen, M. Skurnik, and H. Wolf-Watz. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5:977-986. [DOI] [PubMed] [Google Scholar]
- 14.Galyov, E. E., S. Hakansson, A. Forsberg, and H. Wolf-Watz. 1993. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature 361:730-732. [DOI] [PubMed] [Google Scholar]
- 15.Galyov, E. E., S. Håkansson, and H. Wolf-Watz. 1994. Characterization of the operon encoding the YpkA ser/thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J. Bacteriol. 176:4543-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, K. L., and J. E. Dixon. 1991. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J. Biol. Chem. 266:17026-17030. [PubMed] [Google Scholar]
- 17.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 18.Iriarte, M., and G. R. Cornelis. 1999. The 70-kilobase virulence plasmid of Yersiniae, p. 91-126. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile genetic elements. American Society for Microbiology, Washington, D.C.
- 19.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 20.Miao, E., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted by the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 21.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills, S. D., A. Boland, M.-P. Sory, P. Van Der Smissen, C. Kerbourch, B. B. Finley, and G. R. Cornelis. 1997. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc. Natl. Acad. Sci. USA 94:12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monack, D. M., J. Mecsas, N. Ghori, and S. Falkow. 1997. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc. Natl. Acad. Sci. USA 94:10385-10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orth, K. 2002. Function of Yersinia effector YopJ. Curr. Opin. Microbiol. 5:38-43. [DOI] [PubMed] [Google Scholar]
- 25.Orth, K., L. E. Palmer, Z. Q. Bao, S. Stewart, A. E. Rudolph, J. B. Bliska, and J. E. Dixon. 1999. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science 285:1920-1923. [DOI] [PubMed] [Google Scholar]
- 26.Orth, K., Z. Xu, M. B. Mudgett, Z. Q. Bao, L. E. Palmer, J. B. Bliska, W. F. Mangel, B. Staskawicz, and J. E. Dixon. 2000. Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594-1597. [DOI] [PubMed] [Google Scholar]
- 27.Palmer, L. E., S. Hobbie, J. E. Galan, and J. B. Bliska. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-α production and down regulation of the MAP kinases p38 and JNK. Mol. Microbiol. 27:953-965. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 29.Petersen, S., and G. M. Young. 2002. An essential role for cAMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70:3665-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Portnoy, D. A., and S. Falkow. 1982. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J. Bacteriol. 148:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portnoy, D. A., S. L. Moseley, and S. Falkow. 1981. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect. Immun. 31:775-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portnoy, D. A., H. Wolf-Watz, I. Bolin, A. B. Beeder, and S. Falkow. 1984. Characterization of common virulence plasmids in Yersinia species and their role in the expression of outer membrane proteins. Infect. Immun. 43:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosquist, R., K.-E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruckdeschel, K., S. Harb, A. Roggenkamp, M. Hornef, R. Zumbihl, S. Kohler, J. Heesemann, and B. Rouot. 1998. Yersinia enterocolitica impairs activation of transcription factor NF-κB: involvement in the induction of programmed cell death and in suppression of macrophage tumor necrosis factor production. J. Exp. Med. 187:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruckdeschel, K., J. Machold, A. Roggenkamp, S. Schubert, J. Pierre, R. Zumbihl, J. P. Liautard, J. Heesemann, and B. Rouot. 1997. Yersinia enterocolitica promoted deactivation of macrophage mitogen-activated protein kinases, extracellular signal-regulated kinase-1/2, p38, and cJun NH2-terminal kinase. Correlation with its inhibitory effect on tumor necrosis factor-alpha production. J. Biol. Chem. 272:15920-15927. [DOI] [PubMed] [Google Scholar]
- 36.Ruckdeschel, K., O. Mannel, K. Richter, C. A. Jacobi, K. Trulzsch, B. Rouot, and J. Heesemann. 2001. Yersinia outer protein P of Yersinia enterocolitica simultaneously blocks the nuclear factor-kappa B pathway and exploits lipopolysaccharide signaling to trigger apoptosis in macrophages. J. Immunol. 166:1823-1831. [DOI] [PubMed] [Google Scholar]
- 37.Ruckdeschel, K., A. Roggenkamp, V. Lafont, P. Mangeat, J. Heesemann, and B. Rouot. 1997. Interaction of Yersinia enterocolitica with macrophages leads to macrophage cell death through apoptosis. Infect. Immun. 65:4813-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schesser, K., A. K. Spiik, J. M. Dukuzumuremyi, M. F. Neurath, S. Pettersson, and H. Wolf-Watz. 1998. The yopJ locus is required for Yersinia-mediated inhibition of NF-κB activation and cytokine expression: YopJ contains a eukaryotic SH2-like domain that is essential for its repressive activity. Mol. Microbiol. 28:1067-1079. [DOI] [PubMed] [Google Scholar]
- 39.Schmiel, D. H., E. Wagar, L. Karamanou, D. Weeks, and V. L. Miller. 1998. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect. Immun. 66:3941-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmiel, D. S., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 42.Snellings, N. J., M. Popek, and L. E. Linder. 2001. Complete DNA sequence of Yersinia enterocolitica serotype O:8 low-calcium response plasmid reveals a new virulence plasmid-associated replicon. Infect. Immun. 69:4627-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sory, M.-P., and G. R. Cornelis. 1990. Translocation of hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 44.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 2002. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 92:11928-11931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Von Pawel-Rammingen, U., M. V. Telepnev, G. Schmidt, K. Aktories, H. Wolf-Watz, and R. Rosqvist. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737-748. [DOI] [PubMed] [Google Scholar]
- 46.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]