Abstract
σB is an alternative σ factor that controls the general stress response in Bacillus subtilis. In the absence of stress, σB is negatively regulated by anti-σ factor RsbW. RsbW is also a protein kinase which can phosphorylate RsbV. When cells are stressed, RsbW binds to unphosphorylated RsbV, produced from the phosphorylated form of RsbV by two phosphatases (RsbU and RsbP) which are activated by stress. We now report the values of the Km for ATP and the Ki for ADP of RsbW (0.9 and 0.19 mM, respectively), which reinforce the idea that the kinase activity of RsbW is directly regulated in vivo by the ratio of these nucleotides. RsbW, purified as a dimer, forms complexes with RsbV and σB with different stoichiometries, i.e., RsbW2-RsbV2 and RsbW2-σB1. As determined by surface plasmon resonance, the dissociation constants of the RsbW-RsbV and RsbW-σB interactions were found to be similar (63 and 92 nM, respectively). Nonetheless, an analysis of the complexes by nondenaturing polyacrylamide gel electrophoresis in competition assays suggested that the affinity of RsbW2 for RsbV is much higher than that for σB. The intracellular concentrations of RsbV, RsbW (as a monomer), and σB measured before stress were similar (1.5, 2.6, and 0.9 μM, respectively). After ethanol stress they all increased. The increase was greatest for RsbV, whose concentration reached 13 μM, while those of RsbW (as a monomer) and σB reached 11.8 and 4.9 μM, respectively. We conclude that the higher affinity of RsbW for RsbV than for σB, rather than a difference in the concentrations of RsbV and σB, is the driving force that is responsible for the switch of RsbW to unphosphorylated RsbV.
In Bacillus subtilis, the general stress response is induced by adverse conditions of growth and provides the cell with multiple resistances (4, 10, 29). This response is controlled by σB, an alternative σ factor, which directs RNA polymerase to the promoters of more than 100 genes that encode the general stress proteins (13, 14, 22, 23). The genes of the σB regulon are induced either by starvation for glucose, phosphate, or oxygen, which leads to a decrease in the intracellular concentration of ATP (energy stress), or by factors such as salt, ethanol, acid, and heat, which produce a physical insult but which leave unchanged the intracellular concentration of ATP (environmental stress) (5, 12, 26).
Two pathways for the activation of σB that correspond to these two classes of stress have been identified. For energy stress, the regulation of σB involves only RsbV, RsbW, and RsbP (8, 25, 27). An additional set of proteins, RsbU, RsbT, RsbS, RsbR, and RsbX, is required to regulate σB in environmental stress (1, 16, 30, 31). All the genes for these proteins, except rsbP, belong to the sigB operon, which is driven by a σA-dependent promoter and by an internal σB-dependent promoter that lies upstream of rsbV, rsbW, sigB, and rsbX (30).
Biochemical and genetic studies have revealed the protein-protein interactions among the Rsb proteins and the functions of these proteins (8, 17, 28). The activation of σB is based on a “partner-switching” mechanism involving RsbW, the anti-σ factor, and RsbV, the anti-anti-σ factor. σB is inhibited by the formation of a noncovalent complex with RsbW (3). RsbW is also a specific protein kinase whose substrate is RsbV, and it “switches” from σB, and thus releases it, when unphosphorylated RsbV is available. The phosphorylation state of RsbV, which is controlled by the balance between the kinase activity of RsbW and the phosphatase activities of RsbU and RsbP, is therefore the key factor for RsbW switching and the release of σB. The two phosphatases are activated by different signals: RsbU is activated by environmental stress, and RsbP is activated by energy stress. The gene for rsbP encodes an N-terminal PAS domain, a family of proteins that sense redox potentials, oxygen concentration, and light (24), and it is believed that RsbP is activated by energy stress through its PAS domain (5). Moreover, it has been suggested that, when the ATP concentration falls in the cell, the kinase activity of RsbW diminishes, the concentration of unphosphorylated RsbV increases, and RsbW then binds to RsbV to release σB (2).
Although there is good evidence that σB is regulated by such partner-switching behavior (see references already cited), the mechanism is not understood in molecular detail. To obtain more biochemical data on this regulatory system, we have begun a study of the biochemical properties that regulate protein-protein interactions between RsbV, RsbW, and σB. In this paper, we report the Km for ATP and the Ki for ADP of RsbW, the values of which suggest that the activity of RsbW can indeed be regulated by these two nucleotides. We establish the stoichiometry of the RsbW-σB and RsbW-RsbV complexes, the affinity of RsbW for its two alternative partners, and the concentrations of these three proteins in the cell. Taken together, these data are supportive of a mechanism where the kinetic and equilibrium constants of RsbW regulate the stress response of B. subtilis.
MATERIALS AND METHODS
Cloning and purification.
Wild-type sigB was amplified by PCR from the genomic DNA of B. subtilis SG38 and cloned into an expression vector with a self-cleavable intein tag (New England BioLabs; IMPACT T7). After overexpression in Escherichia coli (BL21), cells were disrupted by sonication in an extraction buffer containing 50 mM Tris-Cl, pH 7.5, 500 mM NaCl, 1 mM dithiothreitol (DTT), 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The intein-σB fusion was retained on a chitin column, and the native σB was eluted after cleavage of the intein by 60 mM DTT. Contaminants were further removed by S75 (Pharmacia) gel filtration; no contaminants can be seen by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Wild-type rsbW and rsbV were also amplified by PCR and cloned into a pET11a expression vector. After overexpression in E. coli (BL21), cells were sonicated in buffer A containing 50 mM Tris-Cl, pH 8, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, and 1 mM PMSF and centrifuged at 27,200 × g for 45 min at 4°C. The supernatant was loaded on a DEAE column, washed with 30 ml of buffer A, and eluted with a 100-ml linear gradient of buffer A plus 1 M NaCl. The RsbV-containing fractions, which eluted at about 250 mM NaCl, were precipitated by 50% (wt/vol) ammonium sulfate overnight. The contaminants were then removed by S75 gel filtration. The same procedure was used to purify RsbW, which eluted from the DEAE column at about 480 mM NaCl, but the precipitation step with ammonium sulfate was omitted. For both proteins, after SDS-PAGE and Coomassie staining, the purity was estimated to be higher than 98%.
Equilibrium constants of the protein-protein interactions.
Surface plasmon resonance (SPR) with a BIAcore 2000 (Pharmacia) was used to determine the kinetic and equilibrium constants of the protein-protein interactions, as described by Magnin et al. (20). Purified RsbV and σB were dialyzed overnight against phosphate-saline buffer containing 1 mM DTT. For immobilization on the sensor chip (CM5) samples of σB and RsbV at concentrations of 1.5 and 4.0 mg/ml were diluted 10-fold in 10 mM sodium acetate buffer (pH 5.6 and 5.2, respectively). A sample of RsbW solution was dialyzed overnight against running buffer (50 mM Tris-Cl [pH 7.5], 50 mM KCl, 5 mM MgCl2, 1 mM DTT). Several concentrations of RsbW, ranging from 0.5 to 4 μM, were prepared for use as the analyte by dilution with the running buffer. After injection, attempts to remove undissociated RsbW from the σB flow cell by using a running buffer containing 500 mM NaCl failed, while this buffer appeared to be active in regenerating the RsbV flow cell. To fully remove the undissociated RsbW from immobilized σB, 70 μl of 2 μM RsbV was injected, leading to a rapid return to the baseline. The BIAevaluation software (Pharmacia) was used to determine the binding constants.
Determination of the Km for ATP and the Ki for ADP.
Assays for phosphorylation were essentially performed as described in Magnin et al. (20) with slight modifications. Before the start of the reaction, 15 μM RsbV was preincubated for 10 min at 30°C in phosphorylation buffer (50 mM Tris-Cl [pH 7.5], 50 mM KCl, 1 mM DTT, 2 mM MgCl2) containing 100 μCi of [γ-32P]ATP and a range of concentrations of nonradiolabeled ATP from 0.1 to 2 mM. The reactions were initiated by the addition of RsbW (1 μM), which had been preincubated in the phosphorylation buffer for 10 min at 30°C. At intervals, 20-μl samples were precipitated in 20% (wt/vol) trichloroacetic acid (TCA) containing 1 mg of bovine serum albumin ml−1. The precipitated proteins were collected on cellulose nitrate filters and washed with 30 ml of 10% TCA containing 1 or 5 mM ATP. The filters were transferred into 2 ml of scintillation liquid (Ecoscint; National Diagnostics) and counted in a Beckman LS 5000TD scintillation counter.
Intracellular concentrations of RsbV, RsbW, and σB.
B. subtilis cells (SG38) were grown in Luria-Bertani medium at 20°C to an A600 of 0.7. The culture was divided into several flasks, and to one-half of them ethanol was added to a final concentration of 4%. After 30 min cells were harvested by centrifugation in tubes containing ice chips to avoid any additional stress (27). Cell extracts were obtained after disruption of the cells by sonication in a 1× SDS-PAGE buffer and centrifugation for 30 min at 10,000 × g at 4°C. The proteins were separated by SDS-PAGE alongside known quantities of purified RsbV, RsbW, or σB. After transfer to nitrocellulose membranes and immunoblotting with purified antibodies for each of these proteins, the autoradiography films were scanned and the intensities of the bands were analyzed by using GIMP, version 1.2 (http://www.gimp.org). Only those samples whose intensities fell within the range of standards were further considered, and the intracellular concentrations quoted in Results are the averages from three experiments.
RESULTS
Quaternary structure of RsbV, RsbW, and σB.
The apparent molecular masses of purified RsbV and σB, as determined with a calibrated Superdex-75 filtration column, suggested that they were present as monomers (data not shown). Under the same conditions, purified RsbW had an apparent molecular mass of 38 kDa, suggesting that RsbW is a dimer (data not shown). The existence of a dimer of RsbW and monomers of RsbV and σB had previously been predicted by analysis of the interactions among the Rsb proteins by a two-hybrid system (28).
Kinetic properties of RsbW.
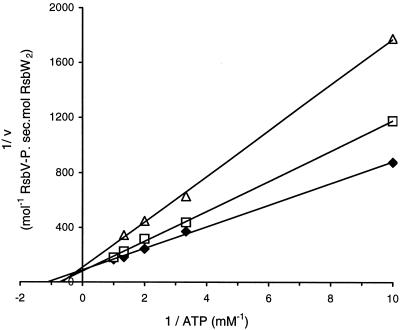
The current model for the regulation of σB during energy stress assumes that the kinase activity of RsbW is diminished by a decrease in the concentration of ATP in the cell (2). This suggestion implies that the Km for ATP of RsbW should be in the physiological range of the intracellular concentration of ATP. To verify this idea, we studied the kinetic properties of RsbW. Studies of the phosphorylation of RsbV catalyzed by RsbW revealed that the reaction is linear with time, which is not the case for the phosphorylation of the anti-anti-σF, SpoIIAA (RsbV paralogue), by SpoIIAB (RsbW paralogue) (20). In our reaction conditions, the maximum turnover was 9.5 mmol of RsbV phosphorylated per mol of RsbW dimer per s and the Km for ATP was 0.9 mM (average of four independent experiments producing values ranging from 0.86 to 1.05 mM) (Fig. 1). The Ki for ADP was about 0.2 mM, and ADP acted as a competitive inhibitor of the reaction (Fig. 1). These values for Km and Ki are much higher than those reported for SpoIIAB, which were found to be in the micromolar range (21). It has been estimated that the intracellular concentration of ATP is between 0.8 and 3 mM (11, 15, 18) and that the ratio of ATP to ADP is 10:1. Therefore, the intracellular ADP concentration, which is unchanged by glucose limitation or ethanol stress (26), is in the range 0.1 to 0.3 mM. Hence, both the Km for ATP and the Ki for ADP are in the physiological range of their intracellular concentrations, reinforcing the proposal (2) that the kinase activity of RsbW is directly regulated by changes in the ATP/ADP ratio. RsbW can therefore be considered a sensor of energy in the cell. RsbP, the phosphatase for RsbV of the energy stress signaling pathway, which is known to be essential for conveying the energy stress signal, contains a PAS domain, suggesting that it too has a role in sensing the energy level (25). Moreover, it has recently been demonstrated that RsbQ, an α/β hydrolase/acyltransferase, activates RsbP (5), presumably by direct modification of RsbP. The catalytic activity of RsbQ is required for stress-dependent activation of RsbP. Therefore, during energy stress, the phosphatase activity of RsbP is stimulated, and this, combined with the decrease in the kinase activity of RsbW, rapidly increases the concentration of unphosphorylated RsbV to liberate σB from the inactive RsbW-σB complex. Since it is known that the GTP/GDP ratio is susceptible to change during energy stress, we assayed the effects of GTP and GDP on the phosphotransfer reaction catalyzed by RsbW. No phosphorylation of RsbV by RsbW was observed when 2 mM GTP was used instead of 2 mM ATP as the phosphate donor in this reaction. Furthermore, no inhibition of the phosphorylation reaction was observed in the presence of 0.2 mM GDP in a reaction buffer containing 0.5 mM ATP. Therefore, it is the ATP/ADP ratio, and not the GTP/GDP ratio, which directly affects the regulation of σB by RsbW and RsbV.
FIG. 1.
Lineweaver-Burk plot used to determine the apparent Km for ATP in the absence of ADP (diamonds) or in the presence of 0.1 mM ADP (squares) or 0.2 mM ADP (triangles). The Ki for ADP was deduced from these values.
Stoichiometry of the complexes between RsbW and RsbV or σB.
The stoichiometry of the RsbW-σB complex has been reported to be 2:2, based on the size of the complex in gel filtration experiments (8, 9) and based on electrophoretic mobility in SDS-PAGE after the cross-linking of the two proteins (2). Experiments to determine the stoichiometry of the RsbW-RsbV complex have given conflicting results. Heterotetramer RsbW2-RsbV2 was proposed based on the results of gel filtration experiments (8), while cross-linking experiments argued for the existence of RsbW1-RsbV1, RsbW2-RsbV1, and RsbW1-RsbV2 complexes. We have tried to distinguish between these possibilities by means of SPR, gel filtration, and mass spectrometry techniques.
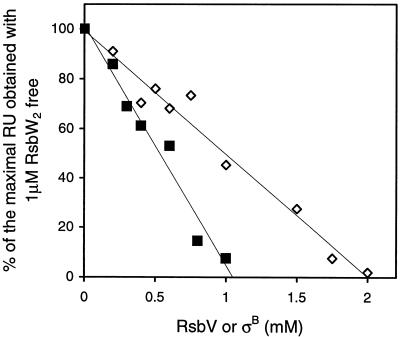
In preliminary SPR experiments, we immobilized σB to the chip and discovered that there was a linear relationship between the concentration of RsbW passed over σB and the height of the sensorgram. We used this property to set up the following experiment. RsbW (2 μM) was mixed with increasing concentrations of σB or RsbV, and the mixture was then injected over an SPR chip on which σB had been immobilized to act as a reporter of the concentration of free RsbW. The response disappeared (that is, the concentration of free RsbW became zero) when 2 μM RsbW was mixed with 1 μM σB or with 2 μM RsbV (Fig. 2). These results suggest that the stoichiometry of the RsbW-σB complex is 2:1. This value differs from that obtained by chemical cross-linking (2), where the authors concluded that the stoichiometry of the complex of RsbW and σB was 2:2. However, it was clear from those experiments that a range of molecular masses was observed, from 60 to 100 kDa. The smallest of these complexes probably corresponds to the RsbW2-σB1 complex described here.
FIG. 2.
Analysis of the stoichiometries of RsbW-RsbV and RsbW-σB complexes. σB was immobilized on the chip, and 1 μM RsbW dimer was injected. The height of this sensorgram was taken as 100%. The same concentration of RsbW dimer was then preincubated for a few minutes at room temperature with increasing concentrations of RsbV or σB and then injected. The heights of the sensorgrams were recorded and plotted against the concentrations of RsbV (diamonds) and σB (squares). RU, response units.
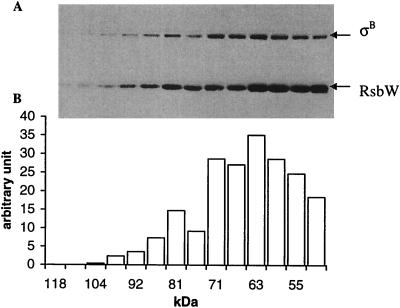
To validate the ratio derived from SPR, we used gel filtration, which revealed that an equimolar mixture of the RsbW dimer and σB was eluted as a complex centered on 63 kDa, suggesting that the complex formed between RsbW and σB has the ratio of 2:1, i.e., RsbW2-σB1 (Fig. 3). Similarly, it was reported previously that, by gel filtration, the stoichiometry of the RsbW-σB complex is 2:2 (8). Our results do not support this conclusion, perhaps because Superdex-75, used in this study, is the most appropriate column to determine molecular masses of around 60 kDa. In this context, it is perhaps relevant that the stoichiometry of the SpoIIAB-σF complex has also been reported to be 2:1 (6, 7).
FIG. 3.
Analysis of the apparent size of the RsbW-σB complex by gel filtration. The RsbW dimer and σB were mixed in an equimolar ratio and loaded onto a Superose-12 gel filtration column (Pharmacia). (A) Samples of the fractions were separated by SDS-12% PAGE and blotted with anti-RsbW and anti-σB polyclonal antibodies. (B) The intensities of the σB bands were measured by using Adobe Photoshop software after digital scanning of the autoradiographic film.
Given that RsbW is a dimer, what is the nature of its complex with RsbV? From the cross-linking experiments (2), it is not possible to determine the relative ratios and the absolute number of discrete RsbW-RsbV complexes. The results from our SPR experiments argue that the ratio of this complex is 1:1, but we cannot determine whether the complex is RsbW2-RsbV2 or RsbW1-RsbV1. Gel filtration experiments ruled out the existence of an RsbW1-RsbV1 complex, since no protein expected to correspond to a molecular mass of 30 kDa was found in those fractions (data not shown).
However, these results do not rule out the possibility of the simultaneous existence of two complexes (RsbW2-RsbV1 and RsbW2-RsbV2) if RsbW is in excess in comparison to RsbV, and indeed the resolution of the gel filtration column was insufficient to distinguish between these potential RsbW2-RsbV1 and RsbW2-RsbV2 complexes. Careful analysis of the SPR data in combination with mathematical probabilities of the existence of these complexes could provide the answer to this question. Theoretically, both RsbW2-RsbV1 and RsbW2-RsbV2 complexes can exist since, when an excess of RsbW is mixed with RsbV, there must be either one or two molecules of RsbV bound to a dimer of RsbW. Assuming that the RsbW dimer is symmetric and that RsbV can bind randomly to either protomer of the RsbW dimer, we can predict the relative proportions of these RsbW-RsbV complexes when RsbW is titrated with RsbV from the general equation P(r) = (n!/r! (n − r)!) Pr (1 − P)n − r, where P is the maximum probability, r is the number of RsbV molecules bound to RsbW, and n is the maximum number of binding sites, i.e., two. This equation predicts that, if both RsbW2-RsbV1 and RsbW2-RsbV2 complexes can exist, the curve for the titration of RsbW by RsbV is parabolic. However, the experimental data (Fig. 2) clearly showed not a parabolic but a linear decrease in the response units as RsbW was titrated by RsbV. This result strongly suggests that only the RsbW2-RsbV2 complex exists. This conclusion implies in turn that, in the interaction between RsbV and RsbW, the binding of one molecule of RsbV favors the binding of a second molecule. Presumably, once the first molecule of RsbV binds, a conformational change in RsbW occurs, leading to an increase in the affinity for a second molecule of RsbV. Alternatively, or in addition, the presence of one molecule of RsbV bound to RsbW might allow some stabilizing interactions between the two molecules of RsbV in addition to the existing interactions with RsbW.
Nanospray ionization mass spectrometry analysis of RsbW and its complexes.
Purified RsbW-RsbV and RsbW-σB complexes were analyzed by nanospray ionization mass spectrometry. Most samples gave a series of well-resolved charge states. For RsbW alone m/z values were around 3,000, and for both RsbW-RsbV and RsbW-σB m/z values were about 4,000. The ions observed for RsbW alone suggest an apparent molecular mass of 36,220 Da. The predicted mass of RsbW from the sequence of its gene is 17,993 Da, and therefore these values confirm our finding by gel filtration that RsbW is a dimer. The difference in mass of ∼200 Da is probably a consequence of the tight binding of water molecules or buffer ions to RsbW.
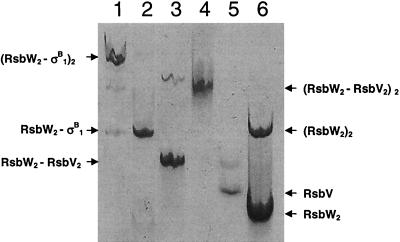
During chromatographic purification of the RsbW-RsbV and RsbW-σB complexes, two complexes, which had different electrophoretic mobilities by nondenaturing PAGE, were observed for each (Fig. 4). In each case, the faster-migrating species was the predominant form, but the precise ratio of the predominant to the minor species varied from preparation to preparation. These predominant species coelectrophoresed with the complexes shown in Fig. 5 and 6. The charge state ensemble of the mass spectrum for the predominant RsbW-RsbV complex was centered at an m/z of 4,320, consistent with a molecular mass of 60,440 Da. Given that the expected mass of RsbV is 11,939 Da, these results imply that the predominant form of the RsbW-RsbV complex has a stoichiometry of 2:2, i.e., one dimer of RsbW is bound by two monomers of RsbV, which is entirely consistent with our SPR data. Mass spectrum analysis of the minor RsbW-RsbV complex revealed a spectrum that was dominated by a series of charge states of low m/z values (<1,000), indicative of an unfolded sample, suggesting that this species is unlikely to have a significant function.
FIG. 4.
Nondenaturing electrophoresis of RsbW-RsbV and RsbW-σB purified complexes. The different complexes were identified by nanospray mass spectrometry. Lane 1, RsbW-σB slow-migrating complex identified as (RsbW2-σB1)2; lane 2, RsbW2-σB1; lane 3, RsbW2-RsbV2, the main, faster-migrating band, and (RsbW2-RsbV2)2, which is visible above; lane 4, σB; lane 5, RsbV; lane 6, RsbW2, the faster-migrating species, and (RsbW2)2, the slower-migrating species.
FIG. 5.
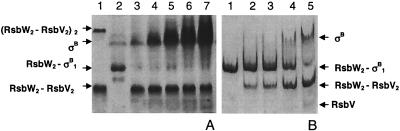
Competition between RsbV and σB for binding to RsbW as analyzed by using a nondenaturing 10% PAGE gel stained with Coomassie blue. (A) A mixture of RsbW and RsbV was incubated for a few minutes at room temperature to allow the formation of an RsbW2-RsbV2 complex before being challenged by increasing concentrations of σB. Lane 1, 5 μM RsbW2-RsbV2 complex (the higher band corresponds to the [RsbW2-RsbV2]2 complex); lane 2, RsbW2-σB complex; lanes 3 to 7, 5 μM RsbW2-RsbV2 complex challenged with 5, 10, 20, 30, and 50 μM σB, respectively. (B) Reciprocal experiment. A purified RsbW2-σB1 complex was challenged with increasing concentrations of RsbV. Lane 1, RsbW2-σB1; lanes 2 to 5, 13.7 μM purified RsbW2-σB1 complex challenged with 4, 8, 16, and 32 μM RsbV, respectively.
FIG. 6.
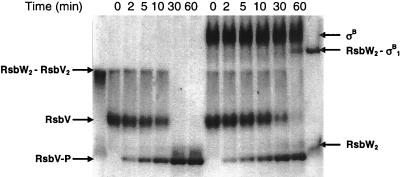
Effect of the presence of ATP on the partition of RsbW between RsbV and σB. During the course of the phosphorylation of 15 μM RsbV by 1 μM RsbW dimer in the presence of 1 mM ATP and 3 mM MgCl2 and in the absence (left) or in the presence (right) of σB (15 μM), samples were taken at time intervals as indicated above the gel and subjected to nondenaturing 12% PAGE. Mixtures with an equimolar ratio of the RsbW monomer to RsbV and an equimolar ratio of the RsbW dimer to σB (first and last lanes, respectively) were used as markers of the positions of the two complexes.
Similarly, we analyzed both RsbW-σB complexes by nanospray ionization mass spectrometry. Here, the charge state of the predominant RsbW-σB fraction was centered at around an m/z of 4,440, corresponding to a species with a molecular mass of 66,460 Da. This mass value is consistent only with a complex in which one RsbW dimer binds one σB monomer (predicted mass of 29,770 Da). The mass spectrum of the minor RsbW-σB fraction exhibited many of the same ions as the major species but in addition contained charge states with m/z values of <1,000 and >6,000. We conclude that these species represent unfolded proteins and that the species with a molecular mass of 133,140 Da most likely represents a (RsbW2-σB)2 complex, the physiological significance of which is unclear. Our observation that one dimer of RsbW binds a single monomer of σB is similar to that made by Campbell et al. in their studies of the SpoIIAB-σF complex both in solution and in the crystal (6, 7). However, there is a crucial difference between these two systems: the interactions between SpoIIAB and σF and between SpoIIAB and SpoIIAA are nucleotide dependent while the interactions between RsbW and σB and RsbW and RsbV are nucleotide independent. The molecular basis for this difference is unknown (2).
Relative affinities of RsbW for RsbV and for σB.
In unstressed cells, RsbV is mainly phosphorylated, but the imposition of energy or environmental stress activates RsbP or RsbU. Phosphorylated RsbV is then dephosphorylated, inducing the switch of RsbW from σB to RsbV. This switch can be explained if the affinity of RsbW for RsbV is higher than that for σB and/or if the concentration of RsbV is higher than the σB concentration. We first measured the affinity of RsbW for its partners by SPR. All attempts to immobilize RsbW on the chip were unsuccessful. We therefore immobilized RsbV and σB and passed RsbW over these two proteins. The calculated Kds for the RsbW-RsbV and the RsbW-σB interactions were found to be similar (Table 1). However, our determination of the stoichiometries of the complexes revealed that a dimer of RsbW binds to two monomers of RsbV, which suggests that the results of the SPR experiments might not represent the affinity of RsbW for its two partners in solution since RsbW is physically unlikely to bind simultaneously to two RsbV monomers immobilized on the chip. We therefore studied the ability of increasing quantities of σB to displace RsbV from an RsbW-RsbV complex. The results of this competition were analyzed by nondenaturing PAGE (Fig. 5). We found that even a fivefold excess of σB over RsbV could not induce RsbW to switch from RsbV to σB (Fig. 5A). Similar results were obtained when σB was present in an eightfold excess over RsbV (results not shown). In a reciprocal experiment, a complex between RsbW and σB was formed and challenged with increasing concentrations of RsbV. The results showed that RsbW switched easily from σB to RsbV when RsbV was available (Fig. 5B).
TABLE 1.
Direct measurement of the dissociation constants by using RsbW2 as the analyte and immobilized RsbV and σB as the ligandsa
| Ligand | kon (105 M−1 s−1) | koff (10−3 s−1) | Kd (nM) |
|---|---|---|---|
| σB | 0.61 | 5.7 | 93 |
| RsbV | 0.93 | 5.8 | 62 |
kon and koff, rate constants for association and dissociation, respectively.
We conclude that, in solution, the affinity of RsbW for RsbV is higher than that for σB by at least a factor of 8. The discrepancy between the results of these experiments and the SPR results is probably due to the fact that, in SPR experiments, RsbW can bind to only one molecule of immobilized RsbV. The measured affinity of RsbW2 for RsbV as measured by SPR would then reflect the affinity of RsbW2 for binding the first molecule of RsbV. Since we have not observed a stable RsbW2-RsbV1 complex, it appears that the binding of a second molecule of RsbV is a thermodynamically favorable process. Therefore, the real dissociation constant for the RsbW-RsbV interaction is significantly lower than that calculated from SPR measurements.
Effect of the presence of ATP on the partition of RsbW between RsbV and σB.
The experiments for determining the relative affinities of RsbW for RsbV and σB were all performed in the absence of a nucleotide. In an attempt to visualize the partition of RsbW between its two partners in the presence of ATP, we studied the phosphorylation of RsbV in the absence or presence of σB. The reaction was started by the addition of RsbW. Samples were taken at intervals, mixed with 50 mM EDTA to stop the reaction, and subsequently subjected to nondenaturing PAGE (Fig. 6). In the absence of σB, and as long as RsbV was not fully phosphorylated, a complex with the electrophoretic mobility of the RsbW2-RsbV2 complex (Fig. 5) could be seen. In the presence of σB, this RsbW2-RsbV2 complex was the only one visible for the first 30 min of the reaction. After 30 min, both RsbW2-RsbV2 and RsbW2-σB1 complexes could be seen, and after 60 min the RsbW2-σB1 complex was clearly visible whereas RsbV was completely phosphorylated. This result shows that, in the presence of ATP, RsbW remains associated with RsbV, rather than with σB, until RsbV is nearly completely phosphorylated. It is also apparent from this gel that the presence of σB slightly affects the rate of phosphorylation of RsbV by RsbW. This effect has been repeatedly observed, but its extent has been variable. It is not clear how or why σB affects this reaction, but this variability has precluded any further investigation. We therefore conclude that, as no nucleotide is required for the formation of the two different complexes (2), the binding of ATP to RsbW does not significantly alter its affinities for its partners.
Intracellular concentrations of RsbW, RsbV, and σB.
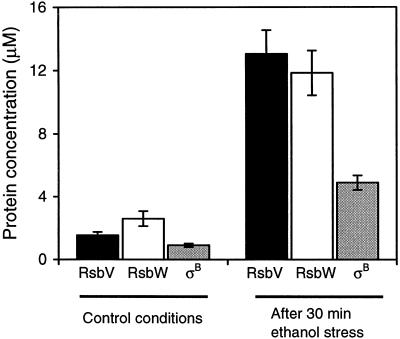
We have shown above that the affinity of RsbW for RsbV is higher than that for σB and that RsbW can switch from σB to unphosphorylated RsbV as long as the concentration of σB is not at least eightfold higher than that of RsbV. We next measured the intracellular concentrations of these three proteins by the use of quantitative Western blotting for B. subtilis cells before and after stress. We found that the induction of the σB operon when B. subtilis cells were challenged with 1 mM MnCl2, a mimic of energy stress, was irreproducible. Since it is known that energy stress induces the synthesis of the proteins of the σB operon to the same extent as ethanol stress (26), we used the latter, more reproducible treatment to determine the concentrations of RsbV, RsbW, and σB. We found that, in the absence of stress, the concentrations of RsbV, RsbW (considered as a monomer), and σB were roughly similar and constant (1.5, 2.6, and 0.9 μM, respectively) with a slight excess of RsbW (Fig. 5).
After 30 min of ethanol stress, the concentration of σB reached a maximum, 4.9 μM, by which point the concentration of RsbW had increased to 11.8 μM. The concentration of RsbV seemed to increase slightly more than those of the other two proteins, reaching 13 μM (Fig. 7), so that the concentration of RsbV was more than twice the concentration of σB. Given the preference of RsbW for RsbV over σB, this concentration of RsbV is expected to be sufficient to allow the release of most of the σB from the complexes with RsbW.
FIG. 7.
Changes in the intracellular concentrations of RsbV, RsbW, and σB after 30 min of 4% ethanol stress. The concentrations of these three proteins were determined as described in Materials and Methods. The total intracellular volume was estimated after cell counts by assuming that the volume of each Bacillus cell is 1.8 × 10−15 liter.
DISCUSSION
The ratio of the concentrations of ATP and ADP is an important feature of the status of the cell. It is physiologically relevant that the kinase activity of the anti-σB factor, RsbW, can be directly affected by a change in this ratio. When the ATP concentration decreases, the phosphorylation of RsbV by RsbW is impaired, RsbW binds to RsbV without phosphorylating it, and σB is set free to interact with core RNA polymerase. Compared to the kinetics for the RsbW paralogue, SpoIIAB (20), the kinetics for RsbW reported here strongly suggest that they have evolved to fit the intracellular concentrations of ATP and ADP. In the regulation of σB, there are therefore two enzyme systems that monitor the energy level in the cell, i.e., the couple RsbQ-RsbP and RsbW.
What is the molecular mechanism by which a decrease in ATP concentration induces the σB regulon? Our results suggest that the switch of RsbW from σB to RsbV relies not on a difference in the cellular concentrations of these two proteins but rather on a higher affinity of RsbW for RsbV than for σB. We propose that a decrease in ATP concentration sets in train the following events. As the rate of phosphorylation of RsbV by RsbW diminishes, the balance between phosphorylation by RsbW and dephosphorylation by RsbP tilts in favor of the latter. Voelker et al. (27) have shown that, after stress but before induction of the σB operon, only one-half (or less) of the preexisting RsbV is dephosphorylated. At this point (i.e., when the concentration of nonphosphorylated RsbV is 0.8 μM), 0.4 μM σB is released. This concentration of σB is apparently sufficient to generate σB-containing RNA polymerase and induce the σB operon, since we have found that the cellular concentrations of the proteins encoded by this operon (RsbV, RsbW, and σB) increase by severalfold during the 30 min following the imposition of an ethanol stress (Fig. 7). As the cellular concentration of σA is about 8 μM and fairly constant (19) and as the affinity of the core RNA polymerase for σB is about 60-fold less than that for σA (C. Rollenhagen and M. D. Yudkin, unpublished data), we cannot rule out the possibility that an anti-σA factor is required to facilitate the induction of the σB operon. When RsbV, RsbW, and σB are present at the high concentrations characteristic of stressed cells (Fig. 7), the balance between phosphorylation and dephosphorylation of RsbV continues to favor, we assume, the accumulation of nonphosphorylated RsbV and thus ensures that RsbW is present in the RsbW2-RsbV2 complex and that σB is free. This high concentration of σB is presumably now sufficient to ensure expression of all the genes of the σB regulon in order to ensure the survival of the cell.
Acknowledgments
We are grateful to D. A. Harris for his valuable advice on using probability theory in the mathematical analysis of the SPR data. We thank E. A. Campbell and S. A. Darst for communicating results prior to publication, David J. Scott for help in nanospray ionization mass spectrometry, and Helen Prescott for technical assistance.
R.J.L. is the recipient of a Wellcome Trust Research Career Development Fellowship. This work was supported by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Akbar, S., C. M. Kang, T. A. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567-578. [DOI] [PubMed] [Google Scholar]
- 2.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 3.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an α/β hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, E. A., and S. A. Darst. 2000. The anti-sigma factor SpoIIAB forms a 2:1 complex with sigma(F), contacting multiple conserved regions of the sigma factor. J. Mol. Biol. 300:17-28. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, E. A., J. L. Sun, O. Muzzin, S. Masuda, C. A. Olson, and S. A. Darst. 2002. Crystal structure of the Bacillus stearothermophilus anti-σ factor SpoIIAB with the sporulation σ factor σF. Cell 109:795-807. [DOI] [PubMed] [Google Scholar]
- 8.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour, A., U. Voelker, A. Voelker, and W. G. Haldenwang. 1996. Relative levels and fractionation properties of Bacillus subtilis σB and its regulators during balanced growth and stress. J. Bacteriol. 178:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaidenko, T., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guffanti, A. A., S. Clejan, L. H. Falk, D. B. Hicks, and T. A. Krulwich. 1987. Isolation and characterization of uncoupler-resistant mutants of Bacillus subtilis. J. Bacteriol. 169:4469-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecker, M., W. Schumann, and U. Völker. 1996. Heat-shock and general stress in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 13.Hecker, M., and U. Volker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 14.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 15.Jolliffe, L. K., R. J. Doyle, and U. N. Streips. 1981. The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25:753-763. [DOI] [PubMed] [Google Scholar]
- 16.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, C. M., K. Vijay, and C. W. Price. 1998. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol. Microbiol. 30:189-196. [DOI] [PubMed] [Google Scholar]
- 18.Karl, D. W. 1980. Cellular nucleotide measurements and application in microbial ecology. Microbiol. Rev. 44:739-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lord, M., D. Barilla, and M. D. Yudkin. 1999. Replacement of vegetative σA by sporulation-specific σF as a component of the RNA polymerase holoenzyme in sporulating Bacillus subtilis. J. Bacteriol. 181:2346-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnin, T., M. Lord, and M. D. Yudkin. 1997. Contribution of partner switching and SpoIIAA cycling to regulation of σF activity in sporulating Bacillus subtilis. J. Bacteriol. 179:3922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najafi, S. M., D. A. Harris, and M. D. Yudkin. 1997. Properties of the phosphorylation reaction catalyzed by SpoIIAB that help to regulate sporulation of Bacillus subtilis. J. Bacteriol. 179:5628-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 26.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-σB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. The yeast two-hybrid system detects interactions between Bacillus subtilis σB regulators. J. Bacteriol. 178:7020-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wise, A. A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]