Abstract
CzcR is the Rickettsia prowazekii homolog of the Caulobacter crescentus global response regulator CtrA. CzcR expression partially compensates for developmental defects in ctrA mutant C. crescentus cells, and CzcR binds to all five CtrA binding sites in the C. crescentus replication origin. Conversely, CtrA binds to five similar sites in the putative R. prowazekii replication origin (oriRp). Also, Escherichia coli IHF protein binds over a central CtrA binding site in oriRp. Therefore, CtrA and IHF regulatory proteins have similar binding patterns in both replication origins, and we propose that CzcR is a global cell cycle regulator in R. prowazekii.
Chromosomal replication control is poorly understood in most gram-negative α-proteobacteria. These bacteria have diverse lifestyles despite their close affiliations based on 16S rRNA signature sequences (for reviews see references 16 and 22). Notable members of this group include the nonpathogenic organisms Rhodobacter capsulatus, Caulobacter crescentus, and Sinorhizobium meliloti, as well as the pathogenic organisms Agrobacterium tumefaciens, Brucella abortus, and Rickettsia prowazekii. Recently, the 4-Mb C. crescentus (27), 3.6-Mb S. meliloti (9), 2.8- and 2.0-Mb A. tumefaciens (15), and 1-Mb R. prowazekii (2) genomes have been completely sequenced. Despite the ∼3-Mb difference in genome size between C. crescentus and R. prowazekii, the hemE and RP001 genes flanking the C. crescentus replication origin were also found to border the putative R. prowazekii replication origin (7). Thus, it is apparent that essential replication sequences survived an extensive genomic reduction. The structure and genomic organization of the C. crescentus replication origin (Cori) are significantly different from the structure and genomic organization of the Escherichia coli replication origin (4, 5). Therefore, it has been hypothesized that Cori represents a new class of α-proteobacterial replication origins (7). Determinations of the hemE and RP001 genes flanking the putative replication origins in S. meliloti and A. tumefaciens further supported this hypothesis (9, 15).
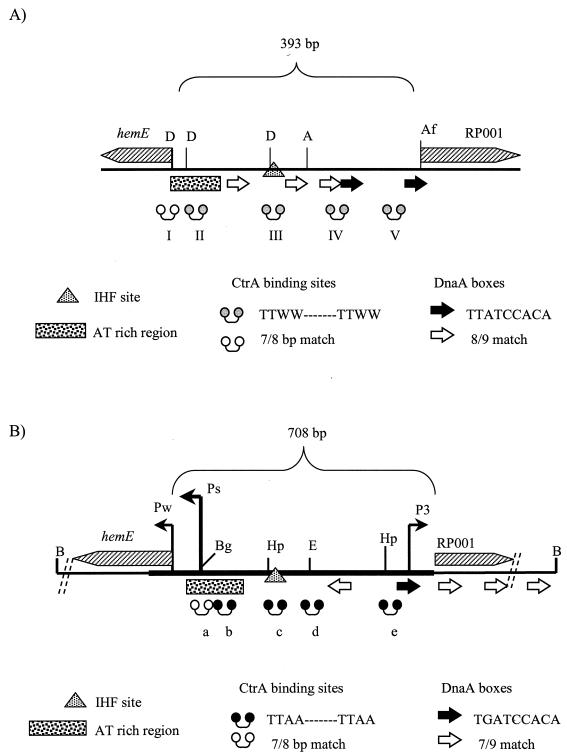
C. crescentus is an excellent model organism for the study of chromosomal replication in α-proteobacteria as extensive studies have been conducted to elucidate the mechanism of regulation of its complex developmental life cycle. This freshwater bacterium has a doubling time of 90 min and asymmetrically divides into two types of morphologically different progeny, the replication-incompetent swarmer cells and the replication-competent stalk cells (8, 26). In order for chromosomal replication to occur, a swarmer cell must differentiate into a stalk cell. Completion of chromosomal replication in the stalk cell signals the transition to the predivisional cell, and then asymmetric division occurs. Cori, identified by autonomous plasmid replication (ARS) assays and in vivo 32P DNA labeling, initiates bidirectional replication in the stalk cell (6, 23). Sequence analysis of Cori revealed features shared with E. coli oriC, such as an AT-rich region and DnaA boxes (23) (Fig. 1B). In addition, Cori possesses unique features, which are described below (Fig. 1B). The hemE operon (23) and the RP001 homolog (7) flank and overlap Cori. A hemE weak promoter (Pw) accounts for most of HemE protein synthesis. A strong promoter (Ps) produces nontranslated transcripts that have been proposed to regulate chromosome replication (24). Five TTAA-n7-TTAA motifs (sites a to e) are binding sites for the response regulator, CtrA (30, 33) (Fig. 1B and 2B). CtrA sites a and b overlap Ps, site c overlaps an IHF site, and site e overlaps an essential DnaA box (24, 33; R. Siam and G. T. Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. H91, p. 368, 2000). Thus, CtrA is essential for regulation of chromosomal replication and other key events in C. crescentus, and therefore it is a global response regulator.
FIG. 1.
Schematic illustrations of oriRp and Cori, including overlapping flanking hemE and RP001 genes. (A) Schematic map of 393-bp intergenic region of putative oriRp. The dumbbells indicate CzcR binding sites of consensus sequence TTWW-N7-TTWW, where W is A or T (see Fig. 3C); the shaded dumbbells indicate a perfect match with the consensus sequence, and the open dumbbells indicate a match at 7 of 8 bp. The arrows indicate potential DnaA boxes identified based upon the E. coli consensus sequence TTATCCACA; the solid arrows indicate a perfect match, and the open arrows indicate a match at 8 of 9 bp. An AT-rich region overlaps sites I and II, and an IHF binding site overlaps site III. The locations of the hemE and RP001 promoters are not known. D, DraI; A, AseI; Af, AflIII. (B) Schematic map of the 708-bp intergenic region of Cori. The thick line indicates the minimal DNA segment that can support ARS activity. The location of the strong (Ps), weak (Pw), and third promoters are indicated. The dumbbells indicate CtrA binding sites a to e; the solid dumbbells indicate a perfect match with consensus sequence GTTAA-N7-TTAA, and the open dumbbells indicate a match at 7 of 8 bp with the consensus sequence. The arrows indicate potential DnaA boxes based upon the proposed C. crescentus consensus sequence TGATCCACA; the solid arrows indicate a perfect match with the consensus sequence, and the open arrows indicate a match at 7 of 9 bp with the consensus sequence. An AT-rich region overlaps CtrA binding sites a and b. An IHF site overlaps CtrA binding site c. B, BamHI; Bg, BglII; Hp, HpaI; E, EcoRI. Note the similarities of oriRp and Cori as discussed in the text.
FIG. 2.
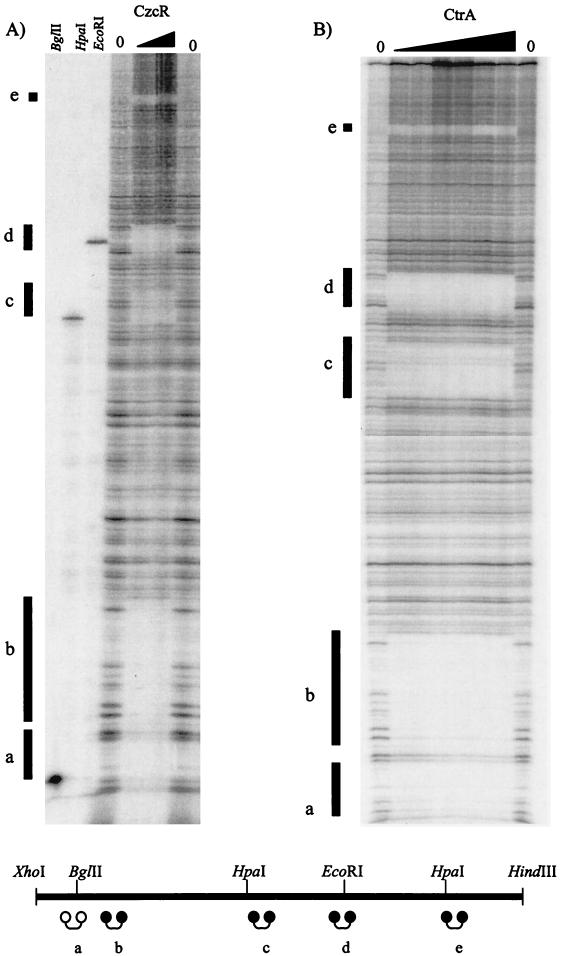
Binding of CzcR and CtrA to CtrA binding sites in Cori. DNase I footprinting assays were performed with different amounts of protein His-CzcR (1 and 2 μg) (A) or thrombin-cleaved CtrA (0.5, 1, 2, 3, 4, 4.5, and 5 μg) (B) by using 32P end label at the HindIII site in Cori (pGM1877). The vertical lines indicate CtrA binding sites a to e. In panel A, to create reference markers in the adjacent lanes, the 32P-end-labeled Cori DNA was also cut with BglII, HpaI, or EcoRI (standard restriction digestion reactions), whose sites are present at CtrA binding sites a, c, and d (see Fig. 1B). A schematic diagram of Cori with the locations of the HindIII and XhoI sites is shown at the bottom, and dumbbells indicate CtrA binding sites a to e; the solid dumbbells indicate a perfect match with consensus sequence GTTAA-N7-TTAA, and the open dumbbell indicates a match with the consensus sequence at 7 of 8 bp. Note the identical binding patterns of CzcR and CtrA at all five CtrA binding sites.
The C. crescentus CtrA protein belongs to the OmpR family of winged helix-turn-helix response regulators (17, 29). Members of this family have an N-terminal domain that is phosphorylated by a sensor histidine kinase, and they have a C-terminal DNA binding domain. The protein structure of OmpR is very similar to that of CtrA. The CtrA-like proteins form their own subgroup within the OmpR family, and this subgroup is unique to α-proteobacteria (3).
Throughout the C. crescentus cell cycle, CtrA is present in the swarmer cell and the swarmer cell component of the predivisional cell, but CtrA is degraded before the onset of the S phase in the stalk cell by the protease ClpXP (11, 19). Transcription of ctrA is autoregulated by a negative-positive feedback mechanism involving two promoters, P1 and P2 (10). As part of a two-component signal transduction system, CtrA has a histidine kinase counterpart, CckA, which phosphorylates CtrA (CtrA∼P) (18). CtrA∼P binds to its five sites and apparently represses chromosomal replication in swarmer cells (30, 31). CtrA phosphorylation stimulates cooperative binding at sites a and b, but increased binding to sites c, d, and e is independent (33).
Although related to C. crescentus, R. prowazekii, the cause of louse-borne typhus, is an obligately intracellular parasite that reproduces only within the eukaryotic host cell cytoplasm (35). Research on R. prowazekii is difficult, as R. prowazekii can be cultivated only in eukaryotic cells and has a doubling time of 10 h (36). The bioenergetic mechanisms of this bacterium are well established; however, the method or control of chromosomal replication remains to be elucidated (1). The R. prowazekii replication origin was putatively identified by G-C skew analysis (2). Upon sequence analysis, an AT-rich region and several DnaA boxes based upon the E. coli DnaA box consensus sequence were identified (2). However, unique lysine residues within a highly conserved portion of the DNA binding region in the DnaA protein suggest that DnaA may recognize an alternative DnaA box sequence (34). In addition, a CtrA homolog with 59% identity and 70% homology, CzcR, has been found in R. prowazekii, as well as in Rickettsia conorii (3, 28). The homology is especially significant in the DNA binding carboxy-terminal region. Also, like CtrA, CzcR has a conserved aspartate at position 51 within the putative receiver domain that has the potential for phosphorylation from a histidine kinase.
Here we describe evidence that the R. prowazekii CtrA homolog, CzcR, is functionally similar to CtrA and that the genomic structure of the R. prowazekii putative replication origin (oriRp) is also similar to that of Cori. As determined by DNase I footprinting assays, CtrA binds to five sites matching the CtrA consensus sequence identified in oriRp. Conversely, CzcR binds to the five CtrA binding sites (sites a to e) in Cori. In addition, a 40-bp IHF binding site was found to overlap a central CtrA binding site in oriRp. In vivo complementation assays showed that CzcR can compensate for defective CtrA in C. crescentus cells. Although the ARS assays were negative for the putative oriRp in C. crescentus host cells, our results and previously reported results (2, 7) strongly suggest that oriRp is the actual replication origin. This study provides the first evidence of a conserved functional role for a CtrA homolog at the replication origin in an α-proteobacterium other than C. crescentus. In addition, the implicated role of the CtrA homolog CzcR at the R. prowazekii replication origin further supports the previously reported hypothesis that there is a new class of replication origins in α-proteobacteria.
Bacterial strains and ARS assays.
Strains and plasmids used in this study are listed in Table 1. C. crescentus Cori supports autonomous plasmid replication in C. crescentus but not in E. coli (23). To determine whether oriRp can replicate in C. crescentus, oriRp was cloned into three different plasmid vectors (Bluescript pSKII, pAGMT, and pAYC177) that do not by themselves replicate in C. crescentus. The oriRp plasmid constructs were electroporated into electrocompetent SC1107 CB15NΔbla C. crescentus cells (23). For positive and negative controls, Cori plasmids and vector plasmids were individually electroporated into C. crescentus CB15NΔbla host cells. A broad-host-range replicating plasmid (pGM976) was included in all electroporation samples as an internal positive control to confirm that electroporation was successful. Electrocompetent cells were prepared by growing cells in 50 ml of PYE medium to an optical density at 660 nm (OD660) of ∼0.5. These cells were washed three times in sterile 10% glycerol and resuspended in 1 ml (final volume) of sterile 10% glycerol. For electroporation, 0.100 ml of cells was mixed with ∼1 μg of plasmid DNA in a 1-mm electrocuvette and electroporated by using the EC1 program of a Bio-Rad MicroPulser electroporator. The following plasmid constructs were used: pMW1070, pMW1095, pGM2703, pGM2667, pGM2668 with pSK, pAGMT∗, pGM2656, and pGM2657 (vectors for negative controls) and pGM1500, pAGMT∗1E, pGM2671, and pGM2672 (positive controls). Electroporated cells were recovered in 1 ml of PYE broth, incubated at 30°C for 90 min, and then spread on PYE medium plates with the appropriate antibiotic(s) for selection (20 μg of ampicillin per ml and 5 μg of gentamicin per ml or 15 μg of tetracycline per ml). Observation of the electroporated oriRp plasmid plates, normalized for background colony growth, revealed negative results for ARS activity (Table 2). To discount the possibility that oriRp may not entirely include the potential ARS element, a larger 1,753-bp (pMW1095) fragment containing oriRp was subjected to the same assays as pMW1070 (pGM2704, pGM2669, pGM2670). Negative ARS assay results were obtained for pMW1095, which supported the initial conclusion that oriRp does not support autonomous plasmid replication in C. crescentus. We initially surmised that the ARS assays failed due to the small size of the fragment (492 bp) containing oriRp that may not have included the entire ARS element. Thus, a much larger fragment (1,753 bp) was tested; however, positive ARS assay results were not obtained. The reasons for the inability of oriRp plasmids to autonomously replicate in C. crescentus are unknown at this time. However, it should noted that R. prowazekii is an obligately intracellular organism; thus, the failure to obtain autonomous plasmid replication may have been due to an inadequate supply of factors, perhaps from the eukaryotic host cell, that may be required for the replication process.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F′ endA1 hsdR17 (rk− mk+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR (φ80dlacΔ(lacZ)M15), host cells for all plasmid constructs | Michael S. DuBow |
| M15 | F′ lacIq Δ(lacZ)M15 supE44 lacY1 lacZ ara-14 galK2 xyl-5 mtl-1 leuB6 proA2 Δ(mcrC-mrr) recA+rpsL20 thi-1 λ− | Michael S. Dubow |
| BL21(DE3) | B F−ompT hsdS(rB− mB−) dcm+ Tetrgal 1 (DE3) endA Hte | Stratagene |
| CodonPlus RIL | [argU ileY leuW Camr] | |
| GM2589 | pGex 2T::CtrA in E. coli M15 | This study |
| GM2773 | pTrcHisA::CzcR in E. coli BL21(DE3) CodonPlus RIL | This study |
| K5746 | pPLhip.himA-5 in E. coli N5271 (IHF overexpressing strain) | 25 |
| C. crescentus strains | ||
| SC1107 | CB15N Δbla::Tn5, AmpS | 12 |
| LS1094 | ctrA401 (Ts) | 29 |
| GM2785 | pUJ142 in LS1094 | This study |
| GM2786 | pGM2456 in LS1094 | This study |
| GM2787 | pGM2767 in LS1094 | This study |
| Plasmids | ||
| pGM976 | pRKlac290 Cori BamHI to Ps::lacZ | 23 |
| pGM2656 | pACYC177 BglII PL+, Ampr | Lucille Shapiro |
| pGM2657 | pACYC177 BglII PL− Ampr | Lucille Shapiro |
| pGM2667 | pGM2656 EcoRI/SalI oriRp from pDW1070 | This study |
| pGM2668 | pGM2657 SalI/EcoRI oriRp from pDW1070 | This study |
| pGM2669 | pGM2656 EcoRI/SalI large fragment of oriRp from pDW1095 | This study |
| pGM2670 | pGM2657 SalI/EcoRI large fragment of oriRp from pDW1095 | This study |
| pGM2671 | pGM2656 BamHI/BamHI Cori from pGM1500 | This study |
| pGM2672 | pGM2657 BamHI/BamHI Cori from pGM1500 | This study |
| pAGMT* | pACYC184 derivative with RK2 oriT, Gmr Tcs | 23 |
| pGM2703 | SalI/BamHI oriRp from pDW1070 in pAGMT* | This study |
| pGM2704 | SalI/BamHI large fragment of oriRp from pDW1095 in pAGMT* | This study |
| pAGMT*E | pAGMT* BamHI fragment E from cosmid I | 23 |
| pUJ142 | Xylose-inducible promoter cloning vector, Cmr | Urs Jenal |
| pGM2767 | pUJ142::czcR | This study |
| pGM2456 | pUJ142::ctrA | This study |
| pBluescript(II) | SK(+) cloning vector, Ampr | Stratagene |
| pGM1500 | Cori BamHI/BamHI in pSK(+) | 23 |
| pGM1877 | Cori HindIII at position 214 to XhoI in pSK(+) | 33 |
| pMW1070 | PCR 492-bp oriRp blunt ligated into EcoRV site in pSK(+) | This study |
| pMW1089 | PCR 758-bp czcR blunt ligated into SmaI with engineered NdeI and BamHI sites of pSK(+) | This study |
| pMW1095 | PCR 1,753-bp large fragment containing oriRp blunt ligated into EcoRv in pSK(+) | This study |
TABLE 2.
ARS assaysa
| Electroporated plasmid | Growth on:
|
||
|---|---|---|---|
| PYE- tetracycline | PYE- ampicillin | PYE- gentamicin | |
| pGM2656 (vector) | ++ | − | NA |
| pGM2657 (vector) | +++ | − | NA |
| pGM2667 (oriRp small fragment) | +++ | − | NA |
| pGM2668 (oriRp small fragment) | +++ | − | NA |
| pGM2669 (oriRp large fragment) | +++ | − | NA |
| pGM2670 (oriRp large fragment) | ++ | − | NA |
| pGM2671 (Cori) | +++ | +++ | NA |
| pGM2672 (Cori) | +++ | +++ | NA |
| pAGMT* | ++ | NA | − |
| pGM2703 (oriRp small fragment) | ++ | NA | − |
| pGM2704 (oriRp large fragment) | ++ | NA | − |
| pAGMT*E (Cori) | ++ | NA | +++ |
Colony growth from plasmids electroporated into C. crescentus host strain SC1107. Each plasmid was mixed with pGM976 (Tetr), which served as an internal positive control. +, ++, and +++, different degrees of growth; −, no growth; NA, not applicable.
Binding of CtrA to Cori and oriRp.
Sequence analysis of oriRp suggested that there are five CtrA binding sites similar to the consensus sequence TTAA-N7-TTAA; one of these sites is a perfect match, three sites have 1-bp mismatches, and one site has a 2-bp mismatch (Fig. 3C). To determine the ability of CtrA to recognize and bind to these five sites, the fusion protein glutathione S-transferase (GST)-CtrA was overexpressed and purified from E. coli host strain M15 (GM2589). GM2589 was grown in 500 ml of Luria-Bertani medium containing 0.2% glucose and 100 μg of ampicillin per ml to an OD660 of ∼0.500 and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 30 min, pelleted by centrifugation at 4,000 rpm for 20 min at 4°C, resuspended in 20 ml of cold 1× phosphate-buffered saline-10% glycerol, and sonicated on ice. For extraction, purification, and thrombin cleavage of GST-CtrA we used the protocol provided with a GST fusion protein expression kit (Amersham Pharmacia Biotech). Phenylmethylsulfonyl fluoride (1 mM) and dithiothreitol (1 mM) were added to all reagents. Phosphorylation of CtrA by using purified maltose-binding protein (MBP)-EnvZ and DNase I footprinting assays with poly(dI-dC) (Pharmacia Biotech) employed as nonspecific DNA were performed as described previously (17, 33). The oriRp DNA fragment was prepared from pMW1070 that was cleaved and end labeled at the HincII site with [γ-32P]ATP (Amersham Pharmacia) and then cleaved with BamHI (both sites located in the pSK polylinker) or vice versa. The Cori DNA fragment was prepared from pGM1877 that was cleaved and end labeled at the HindIII site with [γ-32P]ATP and then cleaved with XhoI (both sites oppositely located in peripheral ends of Cori) or vice versa. For restriction site markers, end-labeled DNA fragments were digested with appropriate enzymes. Sequencing of pMW1070 and pGM1877 with universal pSK or pKS primers was performed with a Sequitherm kit (Epicentre Technologies Inc.).
FIG. 3.
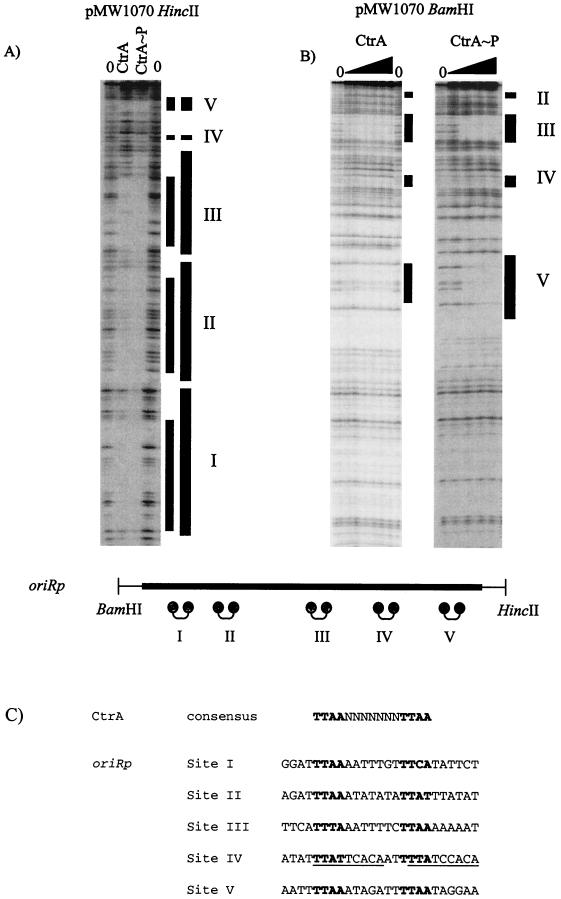
CtrA binding sites on oriRp. A DNase I footprinting assay of oriRp was performed with thrombin-cleaved CtrA (4 μg) and CtrA∼P (phosphorylated CtrA) (4 μg) (A) by using a HincII 32P-end-labeled fragment (A) and with different amounts of thrombin-cleaved CtrA and CtrA∼P (0.05, 0.5, 1, and 2 μg) by using BamHI 32P-end-labeled pMW1070 (B). The lines labeled with roman numerals indicate binding sites. The thick lines indicate enhanced binding by CtrA∼P. Five CtrA binding sites (sites I to V) on the HincII end-labeled oriRp fragment (A) and three of the five CtrA binding sites (sites III, IV, and V) on the BamHI end-labeled oriRp fragment (B) were detected. The schematic diagram of oriRp shows the positions of BamHI and HincII restriction sites flanking oriRp; the dumbbells indicate CtrA binding sites I to V. (C) Comparison of the sequences of all five oriRp CtrA binding sites to the Cori CtrA binding consensus sequence. The potential CzcR binding consensus sequence is TTWW-N7-TTWW, where W is A or T. Note that in site IV there are two potential DnaA boxes (underlined) overlapping the halves of the binding site.
Thrombin-cleaved CtrA was utilized in DNase I footprinting assays with a 492-bp oriRp (Fig. 3) and was resolved next to a sequencing ladder and restriction site markers (data not shown). It was found that CtrA bound to five sites, designated sites I to V. As shown in Fig. 3A, pMW1070 (oriRp) was 32P end labeled at the HincII site, and all five CtrA binding sites are shown in Fig. 3A. As shown in Fig. 3B, pMW1070 (oriRp) was 32P end labeled at the BamHI site, and CtrA binding sites III, IV, and V are shown in Fig. 3B. The general binding sequence for CtrA in oriRp appears to be TTWW-N7-TTWW, where W is either A or T (Fig. 3C). At all five sites (sites I to V), CtrA binding is enhanced upon phosphorylation with E. coli EnvZ (Fig. 3A and B). Enhanced binding of CtrA upon phosphorylation has been observed previously in Cori (33).
Binding of CzcR to CtrA binding sites in Cori.
To determine if CzcR possesses DNA binding specificity similar to that of CtrA, DNase I footprinting assays were performed with His-CzcR and 32P-end-labeled pGM1877 (Cori) (Fig. 2A). The czcR gene was directionally PCR cloned (5′ AAAAAAGGATCCATGAGAGTTTTATTAATT 3′ and 5′ AAAAAAGAATTCTTATTAT GCTCCTTGTGC 3′) with engineered 5′ BamHI and 3′ EcoRI sites from pMW1089 and ligated into pTrcHisA (Invitrogen). However, overexpression of histidine-tagged CzcR was not successful in E. coli BL21 and M15 host cells due to differential tRNA codon usage. Histidine-tagged CzcR was overexpressed in E. coli BL21(DE3) CodonPlus RIL (Stratagene) by using 5 mM (final concentration) IPTG induction at an OD660 of ∼0.500 in 1 liter of Luria-Bertani medium with 50 μg of ampicillin per ml for 90 min. Extraction and purification were performed as described previously for histidine-tagged CtrA (29). The E. coli BL21(DE3) strain hosts a plasmid that encodes genes for arginine, isoleucine, and leucine tRNA codons. CzcR was phosphorylated by using purified MBP-EnvZ as described previously (33). The percentage of CzcR phosphorylated was measured as described previously (33). Five CzcR binding sites were detected in Cori; by using restriction site markers and sequencing ladders, these sites were found to be the same as the previously established five CtrA binding sites (sites a to e) in Cori (see Fig. 2B for comparison with CtrA control footprints). Note that the footprint patterns for His-CzcR binding to Cori and CtrA binding to Cori are identical. These results indicate that R. prowazekii CzcR binds to a DNA sequence similar to that to which C. crescentus CtrA binds. In addition, sequence analysis of the upstream region of czcR revealed a putative binding site that matches the CzcR binding consensus sequence, TTWW-N7-TTWW (data not shown). This evidence suggests that CzcR, like CtrA, may also have an autoregulation mechanism.
In vivo CzcR complementation of the C. crescentus ctrA401(Ts) mutation.
To examine CzcR activity in vivo, we tested the complementation of a C. crescentus mutant ctrA401 strain (LS1094). This strain produces a defective CtrA protein at 28°C that becomes nonfunctional, and therefore lethal, at 37°C (29). Compared to wild-type C. crescentus cells, the LS1094 cells at 28°C are prominently elongated and curled, and their motility is reduced due to the stunted growth or the absence of flagella. Stalk synthesis is initiated; however, stalk biogenesis does not occur (29). As the binding properties of CzcR are similar to those of CtrA, can CzcR compensate for a lack of functional CtrA? To investigate this question, a xylose-inducible plasmid was employed. For xylose-conditional expression of CzcR, czcR was directionally PCR cloned (5′ AAAAGAATTCATGAGAGTTTTATTAATTGA 3′ and 5′ AAAAAAGCTTTTATTATGCT CCTTGTGCTA 3′) at HindIII and EcoRI sites into pUJ142, which has a xylose-inducible promoter (pGM2767). For xylose-conditional expression of CtrA, pUJ142::ctrA (LS2890) was constructed as described by Domian et al. (11). For positive and negative complementation controls, plasmids pUJ142+CtrA (pLS2890) and pUJ142 were electroporated into electrocompetent LS1094 C. crescentus cells and plated on PYE antibiotic selection plates. All three electroporated plasmid strains (GM2785, GM2786, GM2787) were grown in M2G (0.2% glucose) medium to saturation and subcultured into M2G and M2GX (0.3% xylose) media. Every 2 h for 10 h, OD600 were determined for each strain in M2G and M2GX media at an OD660 of ∼0.1. Comparison of the growth curves established by using time course OD660 values of each of the three strains grown in M2G or M2GX medium did not reveal any significant differences in the growth rates (data not shown). However, light microscopic observation at 6 h revealed morphological differences between cells grown in M2G and M2GX media (Fig. 4). For electron microscopy, samples were centrifuged at low speed and resuspended to an OD660 of ∼1.00, and then a drop of each sample was placed on a formvar-carbon-coated copper 300 grid and left undisturbed for 2 min. The excess liquid was wicked off, and the grid was stained by floating it in unbuffered saturated ammonium molybdate for 10 s; the excess liquid was wicked off, and the grid was air dried. The grids were viewed with a Phillips EM300 microscope with an accelerating voltage of 60 kV.
FIG. 4.
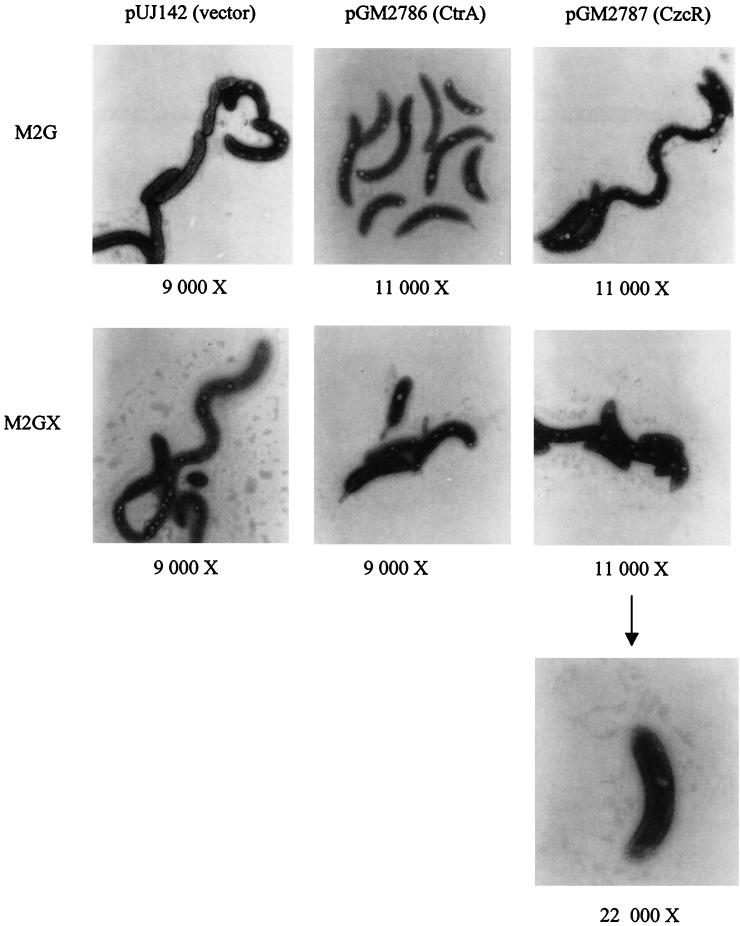
In vivo CzcR and CtrA complementation assays: electron micrographs of C. crescentus LS1094 [ctrA401(Ts)] harboring xylose-inducible plasmids containing czcR and ctrA and the vector control grown in M2G or M2GX (xylose) medium for 6 h at 28°C. The magnification for each electron micrograph is indicated. Note the electron micrograph having a magnification of ×22,000 depicting a swarmer cell with double flagella at the pole. See text for details.
The cells of all three strains grown in M2G medium exhibited similar characteristics; the cells were elongated and curled and had reduced motility (Fig. 4). Strains grown in M2GX medium, however, were found to have significant differences in cell morphology (Fig. 4). Xylose induction of CzcR from plasmid pGM2676 partially complemented the multiple phenotypes seen in the ctrA401 strain. With pUJ142 (vector control), the cells remained elongated and curly without stalks and with very little motility in M2GX medium, as well as in M2G medium. With the positive control pGM2456 (CtrA), the cells showed wild-type characteristics, such as short motile swarmer cells, asymmetric predivisional cells, and stalk biogenesis. Analysis of the morphology of the test pGM2767 (CzcR) strain placed it between the morphologies exhibited by pUJ142 (vector) and pGM2456 (CtrA). The pGM2767 (CzcR) cells were clearly more motile than pUJ142 (vector) cells but not as motile as pGM2456 (CtrA) cells. The majority of the pGM2767 (CzcR) cells were similar to pGM2456 (CtrA) cells, and stalk cells were observed. However, cells with double flagella at the pole were also observed (Fig. 4). This cell morphology may have been due to the effect of partial complementation of CzcR for defective CtrA in which there is difficulty in ejecting the flagellum (hence, the double flagella at the pole). A minority of the pGM2767 (CzcR) cell population resembled the pUJ142 (vector) cells. These microscopic observations indicate that CzcR partially compensates for defective CtrA in C. crescentus LS1094 cells.
The partial restoration of the wild-type phenotype by CzcR further supports the hypothesis that there is functional similarity between CtrA and CzcR, as initially suggested by protein homology and in vitro CzcR binding to CtrA binding motifs in Cori. Xylose induction of CzcR had an in vivo effect similar to that of wild-type CtrA on C. crescentus ctrA401 cells, resulting in an almost normal progression of the cell cycle. However, it should be noted that complete reversion to the wild-type phenotype by expression of CzcR was not achieved, and this may have been due to poor expression of CzcR in C. crescentus LS1094 cells. Like E. coli, C. crescentus has significantly different tRNA codon usage for isoleucine and leucine than R. prowazekii has (www.kazusa.or.jp/codon). It is quite possible that not all expressed CzcR proteins are full length; thus, the compensatory effect is intermediate.
IHF binding site overlaps a CtrA binding site in oriRp.
IHF is a small heterodimeric histone-like protein that is composed of α and β subunits and is found in many gram-negative eubacteria (14), including C. crescentus. R. prowazekii presumably has an IHF protein since its genome contains genes for both α and β subunits and the protein is identified as DNA binding protein HU in The Institute for Genome Research database (2). The E. coli IHF α subunit exhibits 29% identity and 52% homology with the R. prowazekii homolog, and the E. coli IHF β subunit exhibits 32% identity and 53% homology with the R. prowazekii homolog. The carboxy-terminal DNA binding domains of the E. coli α and β subunits exhibit significant homology with the R. prowazekii homologs (data not shown). The E. coli IHF protein binds in the center of the chromosomal replication origin (oriC) and bends DNA more than 180° (32). This IHF binding assists DnaA in unwinding the AT-rich region of E. coli oriC during the initiation of replication (4, 5). Until recently, IHF binding to other chromosomal replication origins has not been reported.
Interestingly, C. cresentus Cori has one IHF binding site that overlaps CtrA binding site c and matches the consensus sequence proposed by Goodman and colleagues (14), and this site was implicated in the control of chromosomal replication (14; Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). CtrA∼P binding in Cori at site c is altered in the presence of the histone-like protein IHF as IHF binding to its site overlapping site c inhibits binding of CtrA∼P, thereby possibly altering the state of replication repression (Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). In fact, mutations at the IHF binding site resulted in elimination of autonomous plasmid replication (Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). Thus, since IHF is more abundant in the stalked cells (13), it is hypothesized that in the absence of CtrA prior to the onset of the S phase, IHF binds to Cori and bends the DNA, allowing the DnaA-bound boxes in close proximity to the AT-rich region to set the scene for replication initiation (Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). Thus, analogous to the E. coli oriC replication initiation mechanism, it is suggested that the IHF protein plays an important role in priming Cori for replication initiation in C. crescentus (Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). It is also hypothesized that as chromosomal replication is completed and the stalk cell moves into the predivisional cell stage, the level of CtrA protein increases and CtrA may displace IHF (Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). Thus, the roles of IHF and CtrA at Cori provide a model for replication control.
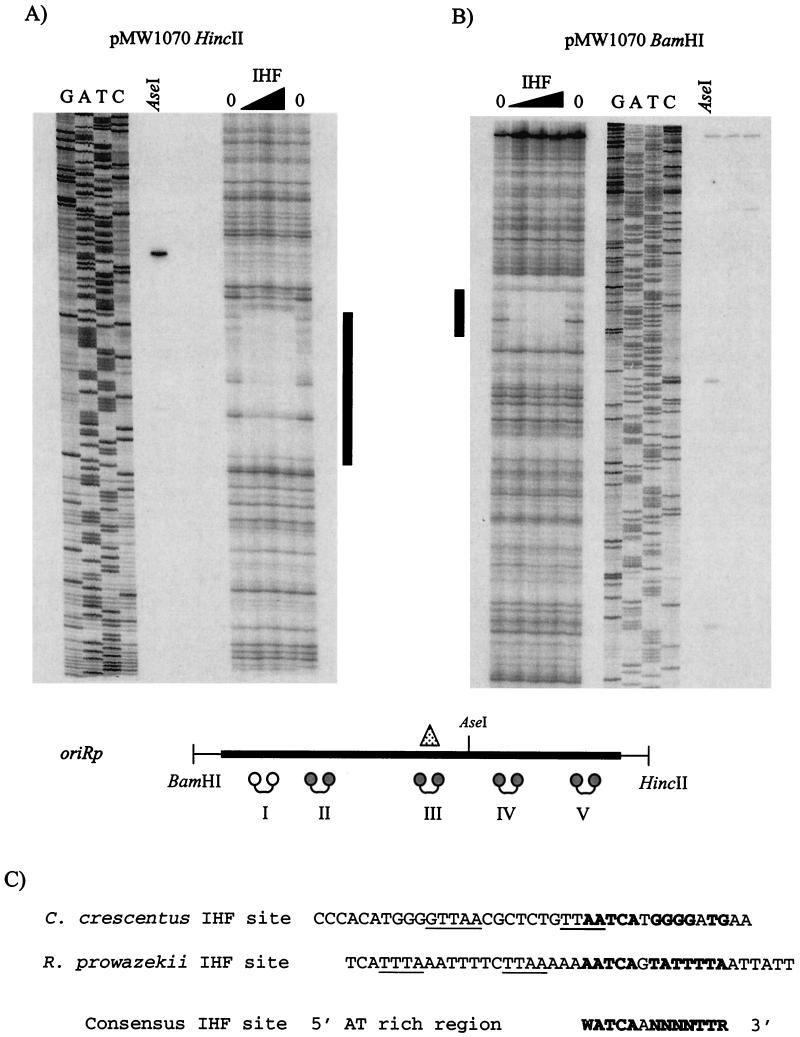
Accordingly, we tested for an IHF binding site in oriRp. E. coli IHF was overexpressed and purified as described previously (25). DNase I footprinting assays were performed with E. coli IHF by using pMW1070 32P end labeled at HincII and BamHI sites (Fig. 5A and B). Only one IHF binding site was detected. The footprint contains a sequence that is similar to the proposed consensus sequence in which the 5′ region is AT rich, and the 3′ region matches the consensus sequence (WATCAANNNNTTR) (14) (Fig. 5C). The sequence of the Cori IHF binding site, which overlaps CtrA binding site c, also matches the proposed consensus sequence (Fig. 5C) (Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). As observed inside Cori, the IHF binding site in oriRp also overlaps a central CtrA binding site (site III) (Fig. 5A and B).
FIG. 5.
IHF binding site in oriRp. A DNase I footprinting assay was performed with different amounts of protein (1, 5, and 10 μg) of E. coli IHF with AseI as a restriction site marker and sequencing ladder by using HincII 32P-end-labeled oriRp (pMW1070) (A) and BamHI 32P-end-labeled oriRp (pMW1070) (B). One 40-bp IHF binding site was detected (indicated by a vertical line). The schematic diagram of oriRp shows the positions of BamHI and HincII restriction sites flanking oriRp; the dumbbells indicate CtrA binding sites, and the triangle indicates the E. coli IHF binding site that overlaps site III. (C) Comparison of proposed C. crescentus and R. prowazekii IHF binding site sequences with the E. coli IHF proposed consensus binding site sequence proposed by Goodman and colleagues (14). Cori CtrA binding site c and oriRp CtrA binding site III in the IHF binding sites are underlined.
Likewise, the putative role of IHF in chromosomal replication initiation in C. crescentus is also implicated in R. prowazekii. Although homologs of IHF have been identified in other α-proteobacteria, it has not been determined whether IHF binding sites are present within the corresponding chromosomal replication origins. Nevertheless, based upon the molecular data pertaining to the replication origins in C. crescentus and R. prowazekii, it is hypothesized that the role of IHF is conserved in chromosomal replication initiation in α-proteobacteria.
Homologs of the C. crescentus CtrA protein have been identified in the α-proteobacteria S. meliloti, R. capsulatus, A. tumefaciens, B. abortus, and R. prowazekii (3); however, relatively little is known about the biological functions of these proteins. Their N-terminal receiver domains (including the position 51 aspartate residue), as well as their C-terminal DNA binding domains, are highly conserved. The S. meliloti CtrA, like the C. crescentus CtrA, is essential, and transcription of S. meliloti ctrA may also be autoregulated (3). The R. capsulatus CtrA homolog, on the other hand, is not essential for viability, and its cellular functions are different from those of C. crescentus CtrA (21). R. capsulatus CtrA regulates the synthesis of a phage-like agent that transfers genes between R. capsulatus cells. CtrA homologs in A. tumefaciens and B. abortus have not been subjected to similar analyses, and it is not known if they are essential for viability. However, potential CtrA binding sites have been found in the transcription promoter of the methyltransferase gene, ccrM, in A. tumefaciens (20). The C. crescentus ccrM gene is essential and is regulated by CtrA (30).
In this study, the R. prowazekii CtrA homolog, CzcR, was purified and analyzed. DNase I footprinting with R. prowazekii CzcR detected five distinct binding sites in Cori (Fig. 2A) that exactly matched the sites (sites a to e) bound by C. crescentus CtrA in Cori (Fig. 2B). Thus, recognition of the consensus binding sequence TTAA-N7-TTAA is conserved in CzcR. The actual binding specificities of other CtrA homologs have not been investigated. However, since CzcR is less homologous to C. crescentus CtrA than the other homologs are, it is very likely that the S. meliloti, R. capsulatus, A. tumefaciens, and B. abortus CtrA proteins also recognize the TTAA-N7-TTAA sequence.
Based upon our results, a schematic diagram of the placement of the hemE and RP001 genes, the AT-rich region, potential DnaA boxes, the CtrA binding sites, and the IHF binding site in oriRp is shown in Fig. 1A. A similar schematic diagram of Cori is shown for comparison in Fig. 1B. The intergenic regions of oriRp and Cori are different sizes (393 and 708 bp, respectively) (Fig.1). Both origins have five CtrA/CzcR binding sites with comparable spacing patterns. The only exception is CtrA binding site I, which overlaps the coding region of hemE. The similarities in the genetic organizations of Cori and oriRp strongly support the previous proposal that essential replication origin elements are retained during genome reduction in R. prowazekii and the proposal that the flanking hemE and RP001 genes serve as proximal markers for the chromosomal replication origin in α-proteobacteria (7). Very recently, it has been determined that the chromosomal replication origins in S. meliloti (9) and A. tumefaciens (15) also possess flanking hemE and RP001 genes.
Putative DnaA boxes were identified in Cori and oriRp on the basis of comparison to the E. coli consensus sequence. There are identical numbers of DnaA boxes in both origins, although the positions and the DnaA box sequences differ slightly for the two origins. In Cori, a DnaA box overlaps the right half of the CtrA binding site (site e), whereas in oriRp, a DnaA box overlaps both halves of CtrA binding site IV. In oriRp, an IHF binding site overlaps the central response regulator binding site, suggesting that the mechanism of replication initiation is similar to that proposed for E. coli (4, 5) and C. crescentus (Siam and Marczynski, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.), bending the DNA to bring the DnaA boxes in close proximity to the AT-rich region. The locations of the hemE and RP001 promoters and potential strong promoters in oriRp are not known at this time.
Acknowledgments
We thank W. Spencer, B. Gorbatyuk, C. Khursigara, and M. Arbing for helpful discussions of this work. We thank Steven Goodman for his kind gift of the overexpressing IHF strain. We also thank C. Ng for his invaluable assistance with PCR, E. C. S. Chan for kind help with dark-field microscopy, and T. Owens for assistance with PhosphorImager scans. We are grateful for the assistance of Paul S. Hoffman and Rafael Garduno and their laboratories and the Electron Microscope Imaging Centre of the Faculty of Medicine of Dalhousie University.
This work was supported by a Fonds pour la Formation de Chercheurs et l'Aide á la Recherche (FCAR) Ph.D. fellowship and a Department of Microbiology & Immunology F.C. Harrison Fellowship to A.K.C.B. and R.S. and by Medical Research Council of Canada (MRC) grant MT-13453 and MRC Scholarship Award 273-46 to G.T.M.
REFERENCES
- 1.Andersson, S. G. E. 1998. Bioenergetics of the obligate intracellular parasite Rickettsia prowazekii. Biochim. Biophys. Acta 1365:105-111. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. M. Alsmark, R. M. Podowski, K. Näslund, A.-S. Eriksson, H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, M. J., D. Y. Hung, A. Reisenauer, L. Shapiro, and S. R. Long. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J. Bacteriol. 183:3204-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boye, E., A. Løber-Olesen, and K. Skarstad. 2000. Limiting DNA replication to once and only once. EMBO Rep. 1:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramhill, D., and A. Kornberg. 1988. A model for initiations of origins of DNA replication. Cell 54:915-918. [DOI] [PubMed] [Google Scholar]
- 6.Brassinga, A. K. C., and Marczynski, G. T. 2001. Replication intermediate analysis confirms that chromosomal replication origin initiates from an unusual intergenic region in Caulobacter crescentus. Nucleic Acids Res. 29:4441-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brassinga, A. K. C., R. Siam, and G. T. Marczynski. 2001. Conserved gene cluster at replication origins of the α-proteobacteria Caulobacter crescentus and Rickettsia prowazekii. J. Bacteriol. 183:1824-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brun, Y., G. T. Marczynski, and L. Shapiro. 1994. The expression of asymmetry during Caulobacter cell differentiation. Annu. Rev. Biochem. 63:419-450. [DOI] [PubMed] [Google Scholar]
- 9.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domian, I. J., A. Reisenauer, and L. Shapiro. 1999. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. USA 96:6648-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcription regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 12.Ely, B. 1987. Genetic map of Caulobacter crescentus, p. 242-244. In S. J. O'Brien (ed.), Genetic maps 1987: a compilation of linkage and recombination maps of genetically studied organisms, vol. 4. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Gober, J. W., M. R. Alley, and L. Shapiro. 1991. Positional information during Caulobacter cell differentiation. Curr. Opin. Genet. Dev. 1:324-329. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, S. D., N. J. Velten, Q. Gao, S. Robinson, and A. M. Segall. 1999. In vitro selection of integration host factor binding sites. J. Bacteriol. 181:3246-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Lartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, R. S. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Rev. 62:1435-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, K., and M. I. Igo. 1996. Identification of the bases in the ompF regulatory region, which interacts with the transcription factor OmpR. J. Mol. Biol. 262:615-623. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, C., I. J. Domian, J. R. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111-120. [DOI] [PubMed] [Google Scholar]
- 19.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 19:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahng, L. S., and L. Shapiro. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 183:3065-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang, A. S., and T. J. Beatty. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Sci. Acad. USA 97:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang, B. F., M. W. Gray, and G. Burger. 1999. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33:351-397. [DOI] [PubMed] [Google Scholar]
- 23.Marczynski, G. T., and L. Shaprio. 1992. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J. Mol. Biol. 226:959-977. [DOI] [PubMed] [Google Scholar]
- 24.Marczynski, G. T., K. Lentine, and L. Shapiro. 1995. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 9:1543-1557. [DOI] [PubMed] [Google Scholar]
- 25.Nash, H. A., C. A. Robertson, E. Flamm, R. A. Weisberg, and H. I. Miller. 1987. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J. Bacteriol. 169:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newton, A., and N. Ohta. 1990. Regulation of the cell division cycle and differentiation in bacteria. Annu. Rev. Microbiol. 44:689-719. [DOI] [PubMed] [Google Scholar]
- 27.Nierman, W. C., T. V. Feldblyum, I. T. Laub, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and Rickettsia prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 29.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 30.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reisenauer, A., K. Quon, and L. Shapiro. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol. 181:2430-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, P. A., S. Yang, K. Mizuuchi, and H. A. Nash. 1996. Crystal structure of an IHF-DNA complex: a protein induced DNA U turn. Cell 87:1295-1306. [DOI] [PubMed] [Google Scholar]
- 33.Siam, R., and G. T. Marczynski. 2000. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 19:1138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waite, T. T., E. I. Shaw, H. H. Winkler, and D. O. Wood. 1998. Isolation and characterization of the dnaA gene of Rickettsia prowazekii. Acta Virol. 42:95-101. [PubMed] [Google Scholar]
- 35.Winkler, H. H. 1990. Rickettsia species (as organisms). Annu. Rev. Microbiol. 44:131-153. [DOI] [PubMed] [Google Scholar]
- 36.Winkler, H. H. 1995. Rickettsia prowazekii, ribosomes and slow growth. Trends Microbiol. 3:196-198. [DOI] [PubMed] [Google Scholar]