Abstract
The DegP protein, a multifunctional chaperone and protease, is essential for clearance of denatured or aggregated proteins from the inner-membrane and periplasmic space in Escherichia coli. To date, four natural targets for DegP have been described: colicin A lysis protein, pilin subunits and MalS from E. coli, and high-molecular-weight adherence proteins from Haemophilus influenzae. In vitro, DegP has shown weak protease activity with casein and several other nonnative substrates. We report here the identification of the major pilin subunit of the Pap pilus, PapA, as a natural DegP substrate and demonstrate binding and proteolysis of this substrate in vitro. Using overlapping peptide arrays, we identified three regions in PapA that are preferentially cleaved by DegP. A 7-mer peptide was found to be a suitable substrate for cleavage by DegP in vitro. In vitro proteolysis of model peptide substrates revealed that cleavage is dependent upon the presence of paired hydrophobic amino acids; moreover, cleavage was found to occur between the hydrophobic residues. Finally, we demonstrate that the conserved carboxyl-terminal sequence in pilin subunits, although not a cleavage substrate for DegP, activates the protease and we propose that the activating peptide is recognized by DegP's PDZ domains.
The DegP (HtrA, protease Do) protease is a multifunctional protein essential for the removal of misfolded and aggregated proteins in the periplasm of Escherichia coli. DegP is one of several dozen proteases in E. coli and is known to have homologues in virtually all gram-negative bacteria, cyanobacteria, and mycobacteria as well as in higher-order organisms such as yeasts and humans (33). DegP homologues have recently been described for a number of gram-positive bacteria (18, 32, 36, 45). There are also two homologues of DegP, referred to as DegQ and DegS, in E. coli (22, 54). DegP has been rediscovered several times, as is revealed by the nomenclature (30, 33, 42). The DegP (for “degradation”) nomenclature refers to the initial mapping of a mutation in E. coli that allowed the accumulation of unstable fusion proteins in the periplasm (50, 51). The HtrA (for “heat shock regulated”) designation indicates that a transposon insertion in the same gene resulted in a temperature-sensitive growth phenotype (28). Lastly, DegP was also designated protease Do, again as a mutation that conferred a temperature-sensitive growth phenotype in E. coli (42). The DegP designation is used throughout this report.
Purified DegP exhibited functional protease activity in in vitro assays using casein as a substrate, although its activity on this substrate was weak (29). Lipinska et al. (29) demonstrated that the activity with casein was inhibitable by diisopropyl fluorophosphate and not by any other known protease inhibitors, suggesting that DegP contains an active site serine residue. Interestingly, DegP is not inhibited by the classic serine protease inhibitor, phenylmethyl-sulfonyl fluoride, suggesting differences in the mode of action of DegP (21, 29). Site-directed mutagenesis at serine 210 and histidine 105, two components of the serine protease catalytic triad, compromised DegP function in vitro and in vivo; i.e., strains carrying serine 210 or histidine 105 mutant derivatives were sensitive for growth at elevated temperatures (43). A recent study has shown that DegP functions as both a chaperone and a protease: at low temperature (28°C), the chaperone function predominates; following a shift to high temperature (45°C), the protease function is activated (48). Synthesis of DegP is controlled by σE, the so-called “stress” sigma factor that controls genes essential for survival in the face of extracellular stress (37, 39). This response is regulated, in part, by the CpxA/CpxR two-component regulatory system (6, 7). Recently, Jones et al. (19) demonstrated that expression of pilin subunits in the absence of the cognate periplasmic chaperone (such conditions result in pilin subunit aggregation) resulted in activation of the degP promoter.
The first identified in vivo target for DegP was colicin A lysis protein (Cal) (4). DegP was found to clip the acylated precursor form of Cal into two fragments. Mature Cal also accumulated to higher levels in degP mutant strains (4). A second family of DegP targets was identified as bacterial pilins. The K88 and K99 pilin subunits were found to accumulate to higher levels in degP mutant strains (1). A more detailed study of this phenomenon demonstrated that P pilins, specifically PapA and PapG, are substrates for the DegP protease (19). The Haemophilus influenzae nonpilus adhesin proteins HMW1 and HMW2 were also found to be in vivo substrates for DegP (49). In addition, Spiess et al. (48) recently demonstrated that the MalS protein of E. coli was fully degraded in vitro by DegP.
The crystal structure of E. coli DegP was recently solved (23), revealing that DegP is a hexamer composed of two trimeric rings. The proteolytic sites are located in the central cavity, and the PDZ domains from all six monomers associate to form the walls of the protease machine and are well positioned to mediate substrate binding and access to the central catalytic cavity (23). Further, the structural constraints of the central cavity are such that DegP substrates must be partially unfolded to gain access to the catalytic sites (23).
In the last several years, a significant body of data has accumulated which has demonstrated that DegP is a virulence factor for several pathogenic organisms. In Salmonella enterica serovar Typhimurium, Salmonella enterica serovar Typhi, Brucella abortus, Brucella melitensis, and Yersinia enterocolitica, htrA nulls were found to reduce or abolish virulence (9, 10, 17, 27, 35, 52). The htrA null mutants were found to be more sensitive to oxidative stress and killing by immune cells. Moreover, an htrA lesion was found to be useful in attenuating both Salmonella enterica serovar Typhi (52, 53) and Salmonella enterica serovar Typhimurium (38) for implementation as vaccine strains. Boucher et al. (2) demonstrated that Pseudomonas aeruginosa conversion to mucoidy, the so-called cystic fibrosis phenotype, involves two htrA homologues. In a recent report, Pedersen et al. (34) demonstrated that an htrA null mutant in Legionella pneumophila was attenuated for virulence. Recently, degP was insertionally inactivated in Streptococcus pyogenes, resulting in temperature and oxidative sensitivity as well as in compromising of virulence in a mouse model (18).
Early in vivo data suggested that pilin subunits of the so-called chaperone-usher assembly pathway were DegP substrates (1). Expression of pilin subunit proteins in the absence of the cognate chaperone (PapD) resulted in failure to accumulate subunit in the periplasm of wild-type (DegP+) bacteria, and degP mutant strains were found to accumulate higher levels of pilin subunit in the periplasm (1, 14, 15, 19). Moreover, subunit expression in the degP mutant was highly toxic (19). Both toxicity and accumulation were suppressed by complementation with papD, which encodes the periplasmic chaperone, or degP (19). In the present study, we developed a scheme for purification of the PapA subunit of the P pilus and demonstrated that this protein is a natural substrate that is efficiently cleaved by the DegP protease. We also identified three cleavage sites within the PapA sequence and characterized the determinants essential for proteolytic cleavage of one of the peptide substrates. Lastly, we demonstrated that the conserved carboxyl-terminal sequence in pilin subunits is a potent activator of the protease.
MATERIALS AND METHODS
Strains and genetic constructs.
DH5α (13) and INVαF' (Invitrogen, Carlsbad, Calif.) were used in cloning steps. KS272 (MC1000) [F− Δ(ara-leu)7697 galE galK ΔlacX74 rpsL (Strr)] was used for expression of DegP (51). KS474 (KS272 degP::kan) was used for production of PapA-6H4A. pKS17 (51), a gift of T. Silhavy, was used for overexpression and purification of DegP. PCR cloning of papA was performed as previously described (31), using primers designed to insert six histidine codons and two alanine codons immediately downstream of the PapA leader-processing site. The mutated papA gene was amplified to include BamHI and EcoRI restriction sites to facilitate cloning into pMMB66 (11), creating pCL101. The PapD expression plasmid, pHJ9203, was described previously (19).
PapA-6H4A expression and purification.
PapA-6H4A was purified from whole periplasm by denaturing metal-affinity chromatography (MAC). In brief, periplasm, prepared as previously described, was applied to Talon (Clontech, Palo Alto, Calif.) metal affinity beads in batch and washed in buffer (20 mM Tris [pH 8], 100 mM NaCl) containing 6.0 M guanidine-HCl to remove the chaperone (PapD). For efficient cleavage by DegP, PapA-6H4A was subjected to reduction and carboxymethylation. Guanidine-HCl-denatured PapA-6H4A was reduced in a buffer consisting of 400 mM Tris (pH 8.5), 5.0 mM EDTA, and 0.11 M 2-mercaptoethanol and incubated at 4°C for 2 h. The reduced protein was carboxymethylated by incubation with 0.21 M iodoacetic acid for 1 h at 4°C. The protein was then dialyzed into 20 mM Tris (pH 8.5) for use in cleavage assays.
Purification of DegP protease.
DegP was purified from whole periplasm, which was prepared as previously described (19). Approximately 30 g (wet weight) of cells was used to prepare periplasm. Following dialysis against 33 mM MES-33 mM HEPES-33 mM acetate (pH 5.9), the periplasm was applied to a HiTrap S cation exchange column (Amersham Pharmacia Biotech, Uppsala, Sweden). DegP eluted at approximately 100 mM NaCl in a linear salt gradient. Peak protein fractions were adjusted to 0.5 M ammonium sulfate and applied to an HIC butyl column (Amersham Pharmacia Biotech). DegP eluted from the hydrophobic column at approximately 0.3 M ammonium sulfate. All five bands appearing in the peak fractions eluting from the butyl column were identified as DegP species by amino-terminal sequence analysis.
Whole protein cleavage assay.
Proteolysis assays were performed in a buffer consisting of 20 mM Tris (pH 8) at 45°C for 30 min to 15 h. MgCl2 or CaCl2 (5 mM) was added to later assays, and to verify dependence on divalent cation, EDTA was added at 10 mM. Where indicated, the assay was supplemented with the noncleavable peptide with the sequence KSMCMKLSFS, SPCJ-1 (200 μM), which is derived from the carboxyl terminus of the PapG adhesin.
Peptide scanning (ProteaseSpot).
Overlapping peptide scanning libraries of PapA sequences in 7-mer and 12-mer formats, which were shifted by 3 amino acid residues between neighboring peptides, were prepared by the SPOT synthesis technique (U. Reineke, D. Kurzhals, A. Kohler, C. Blex, J. McCarthy, P. Li, L. Germeroth, and J. Schneider-Mergener, abstract from the Proceedings of the 26th European Peptide Symposium, abstr. P327, 2000), conjugated to aminobenzoic acid at the amino terminus, linked (carboxyl terminus) to continuous cellulose membranes (ProteaseSpots), and assayed by Jerini AG (Berlin, Germany). Purified DegP was prepared as described above and assayed (0.28 mg/ml) in a 96-well microplate format, using 20 mM phosphate buffer (pH 7)-5 mM MgCl2. At 1, 4, and 28 h, aliquots were removed from the assay plate and quantified for liberated fluorescence (excitation, 325 nm; emission, 420 nm).
Peptide cleavage assay.
The following three peptides were used in the fluorescence-proximity peptide cleavage assay: SPCJ-12 [Abz-HYTAVVKKSSAV-DAP(Dnp)-NH2], SPCJ-13 [Abz-HYTAVVK-DAP(Dnp)-NH2], and SPCJ-14 [Abz-HYTASSK-DAP(Dnp)-NH2], where Abz, the fluorescent group, is aminobenzoic acid and DAP(Dnp)-NH2, the quench group, is diaminopropionamide 2,4-dinitrophenyl. The peptides were used in the assay at concentrations ranging from 2 μM to 10 μM. DegP was added into the assay at concentrations ranging from 0.5 to 5 μg. The assay buffer consisted of 25 mM HEPES (pH 7.5)-5 mM CaCl2. The cleavage reactions were incubated at 45°C and read in a Wallac Victor2V 1420 multilabel HTS counter (Wallac Oy, Turku, Finland) at 30-min intervals. Fluorescence detection utilized excitation at 340 nm, and emission was monitored at 420 nm.
Cleavage site mapping.
Purification of peptides and products of the cleavage reaction was performed on a C18 5μ ST 4.6/250 Sephasil column (Amersham Pharmacia Biotech), with a gradient to 100% acetonitrile-0.05% trifluoroacetic acid. Lyophilized cleavage reactions (5 ml) were resuspended in 2% acetonitrile-0.065% trifluoroacetic acid for application to the C18 column. Fractions containing peptide and cleavage products were lyophilized to dryness and mass was determined by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) (MALDI-TOF MS). MALDI analysis was conducted using a 5- mg/ml solution of the matrix α-cyano-4-hydroxycinnamic acid (HCCA) in 0.1% trifluoroacetic acid-33% acetonitrile. Analyte was mixed with the matrix at a ratio of 1:3. Spectra were produced using a custom-built MALDI-TOF mass spectrometer.
Other methods.
Proteins were prepared for sequencing by transfer to polyvinylidene difluoride membrane as previously described (8, 44) and delivered to Midwest Analytical Inc. (St. Louis, Mo.) for amino-terminal sequence determination. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis were performed as previously described (8, 44). Toxicity assays were performed as previously described (19). The EnzChek assay kit (Molecular Probes, Eugene, Oreg.) was used according to the manufacturer's instructions and read using a Tecan SpectraFluor Plus (Research Triangle Park, N.C.) apparatus with the appropriate filter sets.
RESULTS
Construction and purification of PapA-6H4A.
A significant obstacle to the study of chaperone-mediated pilus biogenesis is the inability to purify pilin subunits in the absence of the periplasmic chaperone. This handicap can be overcome, in part, by inactivation of the DegP protease (1). Using the method of Morrison and Desrosiers (31), a sequence encoding a polyhistidine affinity tag (6-His tag) was inserted into the papA gene. By taking advantage of MAC under denaturing conditions, the pilin subunit was stably expressed and purified (data not shown). As with wild-type PapA, the PapA-6H4A protein was toxic when induced in the absence of the PapD chaperone (19). The toxicity of PapA-6H4A was suppressed by coexpression of papD or degP in trans (data not shown).
In order to purify sufficient PapA for the planned experiments, PapA-6H4A was synthesized in the presence of the PapD chaperone in KS474 (degP::kan) (19). The PapD-PapA complex was purified from whole periplasm on MAC in batch, followed by a denaturing step to remove PapD (see Materials and Methods).
Purification of DegP protease.
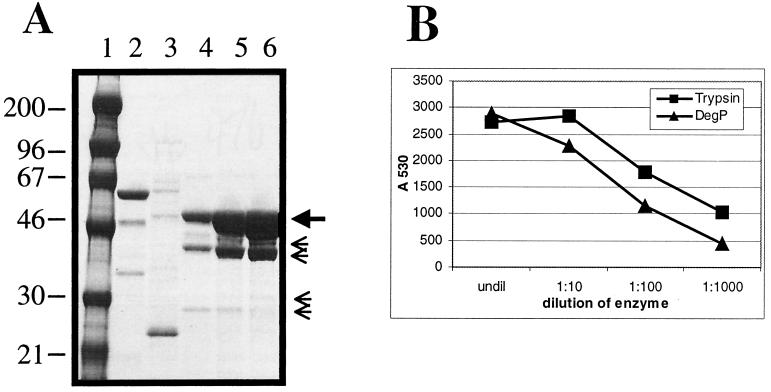
DegP was purified from KS272/pKS17 (51) following overnight (15 to 20 h) growth to saturation, which was sufficient to induce high-level expression of DegP. Whole periplasm prepared from 6 liters of culture was fractionated by cation exchange chromatography on SP Sepharose (HiTrap SP; Amersham Pharmacia Biotech) followed by hydrophobic interaction chromatography on butyl Sepharose (HiTrap butyl; Amersham Pharmacia Biotech) (Fig. 1A). This two-step purification process resulted in approximately 98%-pure DegP protease. The contaminating bands seen on SDS-PAGE analysis, as shown in Fig. 1A (small arrows), were identified as DegP truncates by amino-terminal sequence analysis and presumably result from autocatalytic cleavage (data not shown).
FIG. 1.
Purification of DegP. (A) Periplasm prepared from 30 g of cells was applied to a 5-ml HiTrap SP column (Amersham Pharmacia Biotech) and eluted with a linear salt gradient. Peak fractions from the cation exchange fractionation were pooled, brought to 0.5 M ammonium sulfate, and applied to a HiTrap HIC butyl column (Amersham Pharmacia Biotech). The flow-through fraction is shown in lane 2, and a portion of the gradient elution is shown in lanes 3 to 6. DegP eluted in approximately 0.3 M NaCl, as shown in lanes 4 to 6. The small arrows indicate truncated forms of DegP, all of which were identified by amino-terminal sequencing (unpublished data). Lane 1 contains high-molecular-weight markers. (B) In vitro protease assay. The pooled peak HIC butyl fractions were checked for general protease activity on a commercial casein substrate. The EnzChek assay kit (Molecular Probes) uses a sensitive fluorescein-labeled casein substrate that is internally quenched until cleaved. The 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene signal is read following excitation at 485 nm and monitoring emission at 530 nm. DegP (1.7 mg/ml) was incubated with the substrate according to the manufacturer's instructions and read after 1 h in a plate fluorometer (SpectraFluor Plus; Tecan). Trypsin (1 mg/ml) was used as a control.
Assay of protease activity.
A commercially available protease assay, EnzChek (Molecular Probes), was used to test cleavage activity of the DegP preparations on a casein substrate. DegP activity with casein was previously described by Lipinska et al. (29). Peak cleavage activity on the EnzChek substrate followed the elution profile of DegP through both chromatography steps (data not shown). Figure 1B shows a titration of purified DegP protease and trypsin on the casein substrate.
Soluble PapA cleavage assay.
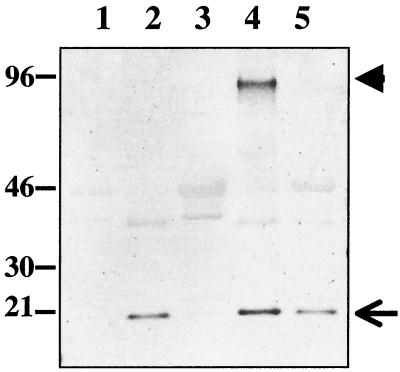
Previous studies of DegP cleavage activity indicate that the preferred substrates were denatured, aggregated, or unfolded proteins (20, 22, 33). To provide DegP with a suitable substrate for cleavage in a soluble format, PapA-6H4A was denatured with 6 M guanidine-HCl, reduced and carboxymethylated (PapA-6H4A-rcm), and dialyzed into 20 mM Tris (pH 8.5). PapA-6H4A-rcm was mixed with DegP and incubated at 45°C overnight, and the resulting reaction products were resolved on SDS-PAGE and further visualized by Western blotting using antisera prepared against whole Pap pili (Fig. 2). As seen in Fig. 2, lane 4, PapA-6H4A-rcm forms polymers (or aggregates) that are stable in SDS in addition to the 21-kDa monomer. Both forms of PapA-6H4A-rcm are sensitive to degradation by DegP protease (Fig. 2, lanes 3 and 5).
FIG. 2.
In vitro DegP cleavage assay. Reduced and carboxymethylated PapA-6H4A (PapA-6H4A-rcm) was mixed with DegP in 20 mM Tris (pH 8) and incubated overnight at 45°C. The reaction mixtures were resolved on SDS-PAGE, transferred to polyvinylidene difluoride membrane, and developed with a polyclonal antibody raised against whole P pili. PapA-6H4A-rcm (lanes 2 and 3, 0.25 μg; lanes 4 and 5, 0.5 μg) was incubated in the presence (lanes 3 and 5) and absence (lanes 2 and 4) of approximately 50- and 100-fold molar excess of DegP (lanes 3 and 5, respectively). The large arrowhead indicates a PapA aggregate in lane 4. Densitometric quantification of the cleaved PapA-6H4A band indicated that DegP cleaved 84% of the input protein (compare lanes 2 and 3) and nearly 100% of the aggregate band and 49% of the monomer in lane 5. Lane 1 contains DegP alone as a control. Some cross-reactivity between the PapA antisera and DegP was seen in the blot.
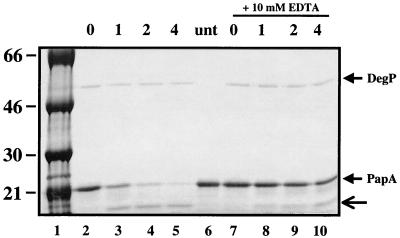
Previous reports have described several different buffer systems suitable for monitoring DegP proteolytic activity (20, 22, 29, 48). Divalent cation (Mg2+) was reported to be required in some systems (20). Addition of 5 mM MgCl2, MnCl2, or CaCl2 (Fig. 3 and data not shown) stimulated DegP cleavage of PapA-6H4A-rcm. Moreover, addition of EDTA to the cleavage reaction inhibited cleavage activity (Fig. 3; compare lanes 3 to 5 and 7 to 10).
FIG. 3.
DegP cleavage of PapA-6H4A substrate is inhibited by chelating agent and stimulated by addition of a noncleavable peptide, SPCJ-1. DegP (5 μg) was incubated with carboxymethylated PapA-6H4A (40 μg) in 20 mM Tris (pH 8)-5 mM MgCl2 in the presence of 200 μM SPCJ-1. EDTA (10 mM) was added to the reaction mixtures loaded in lanes 7 to 10. Aliquots were taken at the indicated time points (0, 1, 2, and 4 h), and the reaction was stopped by the addition of SDS loading buffer and incubation on ice. The reaction mixtures were resolved on SDS-PAGE and stained with Coomassie brilliant blue. An untreated sample (unt) was run in lane 6 as a reference. The ∼12-kDa cleavage product (lanes 3 to 5 and 10) is indicated by the arrow. In excess of 50% of input PapA-6H4A was degraded in 60 min, and 90% was degraded at the 4-h time point.
Activation of DegP by a carboxyl-terminal pilin subunit peptide.
The DegP protease has two so-called PDZ domains (postsynaptic density 95, discs large, ZO-1) located downstream of the catalytic serine residue (33). PDZ domains have been implicated in both substrate recognition and protein multimerization in a number of proteins (26, 40, 46). The carboxyl-terminal motif of pilin subunits has some homology to the motif reportedly recognized by PDZ domains (26, 46). We hypothesized that titration of the carboxyl-terminal peptide from the PapG adherence protein (SPCJ-1; KSMCMKLSFS) into the DegP cleavage reaction would inhibit cleavage of the PapA-6H4A-rcm protein through competition for the substrate-binding site in the PDZ domain. Much to our surprise, addition of SPCJ-1 to the cleavage reaction enhanced degradation of PapA-6H4A-rcm 10- to 20-fold (Fig. 3 and data not shown).
Identification of DegP recognition sites within PapA by peptide scanning.
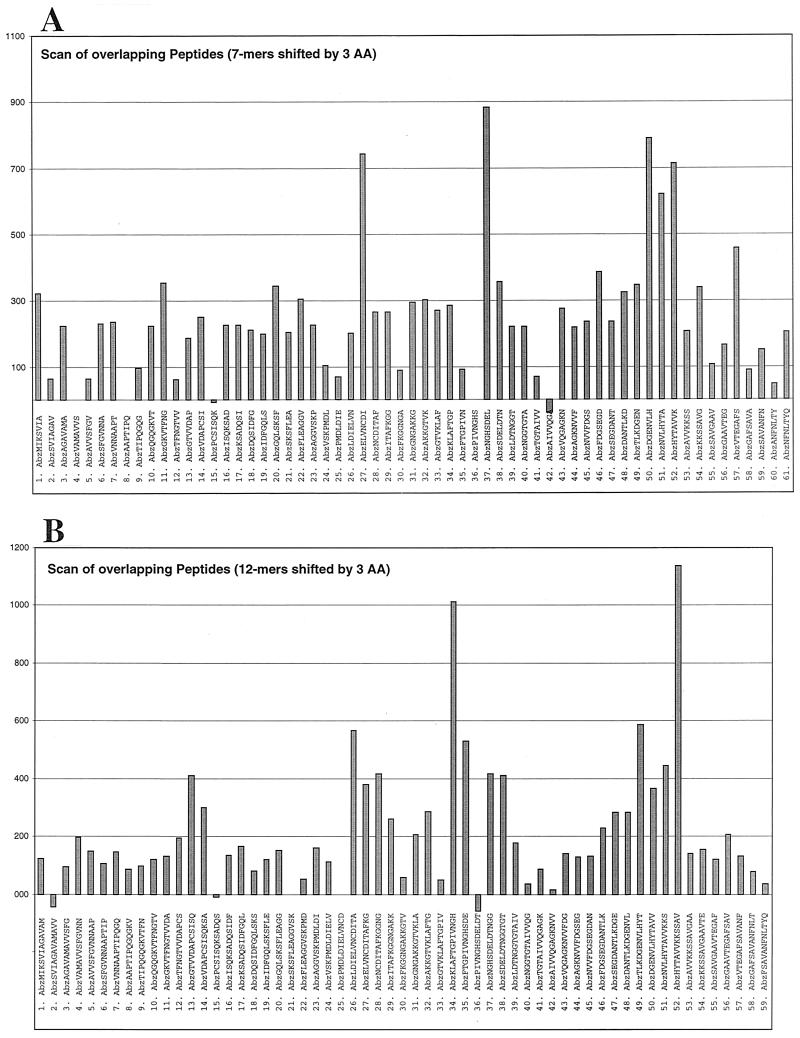
Utilizing SPOT synthesis technology (Reineke et al., 26th European Peptide Symposium, 2000) (ProteaseSpot; Jerini AG), two overlapping peptide scanning libraries of the complete PapA amino acid sequence were constructed and assayed to identify 7-mer and 12-mer peptides that were suitable substrates for DegP proteolysis (Fig. 4). The peptides carried an aminobenzoic acid (Abz) fluorescent tag at the amino terminus and were synthesized linked to a cellulose membrane. In both the 7-mer and the 12-mer ProteaseSpot scans, three regions of PapA were identified as having sequences cleavable by DegP, as represented by enhanced release of fluorescent counts from the cellulose-bound peptides.
FIG.4.
Identification of the DegP protease cleavage site in PapA. SPOT synthesis technology was used to construct two overlapping peptide libraries (7-mer and 12-mer formats) of the PapA sequence. The peptides were synthesized linked to a continuous cellulose membrane (ProteaseSpots) and had a fluorescent tag, aminobenzoic acid (Abz), conjugated to the amino terminus. The ProteaseSpots were assayed in a 96-well format with 0.28 mg of DegP/ml in 20 mM PO4 (pH 7.5)-5 mM MgCl2. The data presented are those of the liberated fluorescence (excitation, 325 nm; emission, 420 nm), following 28 h of incubation. The data show relative levels of cleavage for each of the peptides in the 7-mer (A) and 12-mer (B) libraries. AA, amino acid.
Peptide cleavage assay.
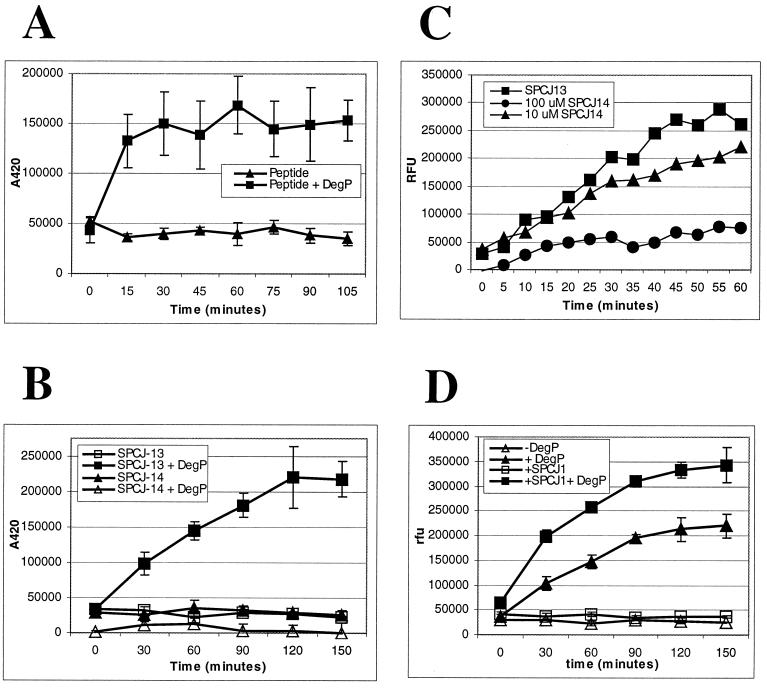
In order to establish a soluble peptide cleavage assay to monitor DegP protease activity, the most efficiently cleaved region, represented by the sequence HYTAVVKKSSAV, was used as a model substrate and a peptide, SPCJ-12, was prepared for use in a fluorescent-quench detection assay. SPCJ-12 was prepared with an aminobenzoic acid (Abz) group, the fluorescent moiety, on the amino terminus and a diaminopropionamide 2,4-dinitrophenyl [DAP(Dnp)-NH2] group, the quench moiety, on the carboxyl terminus. As shown in Fig. 5A, DegP cleaved SPCJ-12 in a time-dependent fashion. To further narrow the recognition sequence and define the determinants of cleavage, two additional peptide reagents, SPCJ-13 and SPCJ-14, were prepared. SPCJ-13 has the sequence HYTAVVK (the first 7 residues of SPCJ-12), whereas SPCJ-14 has the sequence HYTASSK. The double serine replacement was utilized to test the essentiality of the paired hydrophobic residues in SPCJ-13. As shown in Fig. 5B, DegP cleaved SPCJ-13 in a time-dependent fashion; however, SPCJ-14 was not cleaved. In order to gain insight into the failure of DegP to cleave the double serine replacement peptide, we set up an inhibition assay to see if SPCJ-14 could block cleavage of SPCJ-13. Such a result would suggest that DegP still recognized SPCJ-14 but could not cleave the peptide. As shown in Fig. 5C, when added in 10-fold molar excess, SPCJ-14 blocked cleavage of SPCJ-13. Lastly, addition of the noncleavable peptide, SPCJ-1, to the peptide cleavage assay enhanced cleavage of SPCJ-13 (Fig. 5D). This finding supports that seen with the whole protein cleavage assay (Fig. 3).
FIG. 5.
In vitro peptide cleavage assay. (A) SPCJ-12 (8 μM) was incubated with and without 5 μg of DegP in reaction buffer (25 mM HEPES [pH 8], 5 mM CaCl2) for the indicated times.(B) SPCJ-13 and SPCJ-14 (10 μM) were incubated with and without 5 μg of DegP in reaction buffer for the indicated times. (C) Inhibitory activity of SPCJ-14 was demonstrated by preincubating DegP (5 μg) with 10 or 100 μM SPCJ-14 for 30 min and then adding 10 μM SPCJ-13 and continuing the incubation for 60 min. (D) SPCJ-13 was incubated with and without 5 μg of DegP in reaction buffer. To test the stimulatory effect of SPCJ-1, 200 μM peptide was added at the onset of the incubation. Assays were monitored (excitation, 340 nm; emission, 420 nm) at the indicated times in a Victor2 V (Wallac Oy) plate reader.
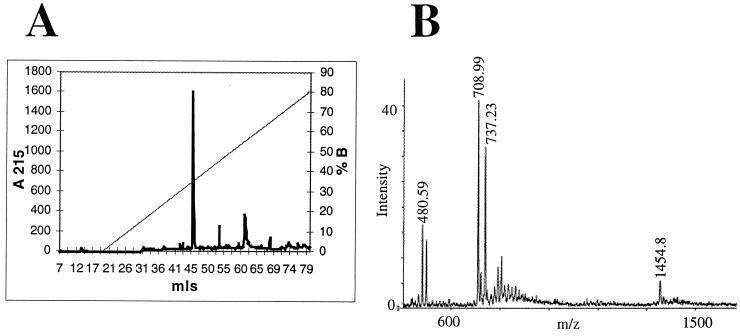
In order to map the precise cleavage sites in SPCJ-12 and SPCJ-13, the reaction products of the cleavage reactions were resolved on reverse-phase high-pressure liquid chromatography (RP-HPLC) using a C18 Sephasil column (Fig. 6A). Untreated SPCJ-13 had a retention volume of 53.06 ml (data not shown), whereas after cleavage, a single new peak was seen with a retention volume of 45.72 ml and the 53.06-ml peak was greatly diminished (Fig. 6A). The mass of the material in the new peak was determined by MALDI-TOF MS analysis. The new peak contained two species having masses [VK-DAP (Dnp)-NH2 = 480.59 Da; Abz-HYTAV = 708.99 Da] consistent with cleavage between the paired valine residues in the peptide (Fig. 6B). As a confirmation, the cleavage site in SPCJ-12 was also mapped by MALDI-TOF MS, and again it was demonstrated that DegP cleaved between the paired valine residues (data not shown).
FIG. 6.
Mapping DegP cleavage site in substrate peptides. (A) RP-HPLC was employed to resolve products of a scaled-up (5 ml) cleavage reaction. The peak at 45.72 ml (actually two peaks that showed absorbance at 215 nm, 340 nm, and 420 nm) (data not shown) was lyophilized to dryness, and the substituents were identified by MALDI-TOF MS. (B) The single peak contained two species resulting from cleavage of the peptide. The first species had a m/z of 480.59, corresponding to VK-DAP(Dnp)-NH2, while the other species had a m/z of 708.99, corresponding to Abz-HYTAV. This analysis was repeated using the SPCJ-12 peptide (12-mer) and mapped the same cleavage site (data not shown).
DISCUSSION
The degradation and clearance of misfolded and/or misassembled proteins in the cytoplasm and periplasm are essential for vigorous growth of bacteria (12, 25, 33). In the periplasm of E. coli and most gram-negative organisms, this task falls to the DegP protease (33). When a functional copy of degP is absent, bacteria fail to grow at high temperature (29, 33). Moreover, recent data show that in addition to compromising growth at high temperature, degP null mutants are more sensitive to oxidative stress and are also avirulent (2, 9, 10, 27, 33-35, 39, 52, 53). The current model suggests that the oxidative response of the host to an invading pathogen results in oxidative denaturation of proteins in the periplasm. In several recently described animal models of infection (2, 9, 10, 27, 33-35, 38, 52, 53), the inability of a degP mutant strain to clear or refold denatured protein placed a sufficient burden on the pathogen to result in a reduction in virulence.
In keeping with this model, the preferred substrate for DegP appears to be proteins that are globally or transiently denatured, supporting the hypothesis that its role in vivo is to remove misfolded or denatured proteins from the periplasm (22). Kolmar et al. (22) demonstrated that DegP would cleave slow-folding mutants of λ repressor and Arc repressor and that the cleavage site P1 residue was a hydrophobic residue, most often a valine. These data support the hypothesis, since the hydrophobic cleavage site would be unavailable in properly folded proteins. Therefore, the recognition site for DegP substrates would be exposed when the proteins went off pathway or were denatured due to an environmental insult. Although an attractive model, this study was hampered by the fact that neither protein used is a periplasmic protein. In support of this finding, however, Laskowska et al. (25) demonstrated in vitro that purified DegP protein would degrade thermally aggregated proteins fractionated from E. coli extracts. Moreover, they showed that the DnaJ chaperone would antagonize DegP degradation; i.e., the chaperone would aid in refolding the proteins such that they were no longer targets for degradation by DegP.
We have clearly demonstrated that the PapA pilin from the pyelonephritis-associated pilus (Pap) is an in vitro target for DegP. Using an amino-terminal polyhistidine affinity tag coupled with denaturing MAC, sufficient PapA-6H4A could be purified away from the periplasmic chaperone, PapD, for implementation in a soluble cleavage assay. DegP protease was purified by standard chromatographic means (Fig. 1) and showed proteolytic activity with both a casein substrate and denatured, reduced, and carboxymethylated PapA-6H4A (Fig. 1 and 3). DegP cleavage activity with PapA-6H4A that had not been reduced and carboxymethylated was minimal by comparison to its activity with PapA-6H4A-rcm (data not shown). However, defining the precise “folded state” of PapA lacking complex formation with PapD or assembly into the pilus is a difficult proposition. Recent cocrystallization studies have revealed that pilin subunits have an incomplete immunoglobulin fold due to the absence of a seventh carboxyl-terminal strand, which is needed to complete a beta sheet in the native fold (5, 41). During pilus biogenesis, this missing strand is provided by either the periplasmic chaperone or the neighboring pilin subunit through donor strand complementation or donor strand exchange, respectively (5, 41). How the folded state of PapA-6H4A, when denatured, reduced, and carboxymethylated, relates to the unfolded or final folded state is unknown.
An amino-terminal truncate of the cleaved PapA was identified using the denatured substrate on the affinity resin as well as in the soluble cleavage assay, suggesting that one site of cleavage was near the middle of the protein (Fig. 3 and data not shown). To further define the specificity of DegP cleavage and to map the cleavage sites within PapA, an overlapping peptide scan was utilized (ProteaseSpot; Jerini AG) (Reineke et al., 26th European Peptide Symposium, 2000). Treatment of both scanning libraries (7-mer and 12-mer) (Fig. 4) with DegP pointed to three regions of PapA that had enhanced cleavage by DegP in the solid-phase assay. Interestingly, the cleavable regions identified by this analysis all lie within or proximal to beta strands and are in highly conserved regions of pilin subunit proteins (see Fig. 4 in reference 47). The scanning results were verified for the carboxyl-terminal-most peptide sequence, SPCJ-12 (HYTAVVKKSSAV), as well as the shorter version, SPCJ-13 (HYTAVVK), in a soluble cleavage assay utilizing a fluorescence quench assay system (Fig. 5). The specificity of cleavage of this sequence was further defined using the control peptide, SPCJ-14 (HYTASSK), which challenged the protease to cleave the peptide lacking the paired valine residues. The lack of cleavage of SPCJ-14 is in agreement with previously reported studies from the Sauer laboratory that showed that the P1 residue of model DegP substrates was most often a valine residue (22). Moreover, SPCJ-14 effectively competed for cleavage of SPCJ-13 in an inhibition assay, suggesting that DegP still recognizes the mutant substrate although the substituted polar residues block cleavage. Of the other sequences identified by the peptide scanning, LDIELVNCDITA, ELVNCDI, KLAFTGPIVNGH, FTGPIVNGHSDE, TLKDGENVLHYT, DGENVLH, and NVLHYTA also have paired hydrophobic residues (italics); however, the NGHSDEL sequence does not follow this pattern. These peptides have yet to be tested in the soluble cleavage assay system or confirmed with control sequences. Clearly, peptide structure, in addition to sequence, has a role to play in recognition by DegP, as can be seen in several instances in which neighboring peptides, which shared partial sequence, were not equivalently cleaved in the solid-phase ProteaseSpot assay (Fig. 4). An additional factor to be considered is the solid-phase presentation of the peptides in the ProteaseSpot system, which may sterically block cleavage of some of the peptides.
Precise cleavage site mapping was carried out with one of the mapped cleavage sites by RP-HPLC and MALDI-TOF MS (Fig. 6). Mapping of the cleavage site in both the 12-mer peptide (SPCJ-12) and the 7-mer peptide (SPCJ-13) revealed that cleavage occurred between the paired valine residues in the sequence. This result is supported by those of earlier studies of Kolmar et al. (22), in which slow-folding variants of Arc repressor and the N-terminal domain of the λ repressor were used as in vitro DegP substrates (22). These investigators mapped several cleavage sites in each protein and demonstrated that the P1 residue was preferentially a hydrophobic residue with a β-branched side chain (e.g., valine or isoleucine). A preference for residues at the P1′ position was not defined by Kolmar et al. (22). We have not tested substitutions exclusively at either the P1 or the P1′ position. Clarification of the sequence requirements for DegP cleavage will require further substitution analysis of the SPCJ-13 peptide as well as cleavage site mapping within the other mapped cleavage sites in PapA.
The PapA cleavage fragment (∼12 kDa) discovered in the solution assay (Fig. 3) was identified as an amino-terminal fragment of PapA by amino-terminal sequencing. This cleavage product most likely results from cleavage at one of the upstream sites (LDIELVNCDITA or FTGPIVNGHSDE), although this has yet to be confirmed, and cleavage of a peptide bearing either sequence has not been evaluated in the solution assay. The cognate carboxyl-terminal cleavage product of PapA was not identified on gels, as additional cleavage at downstream sites may have resulted in products below the resolution limit of our SDS-PAGE system.
In terms of substrate recognition, Songyang et al. (46) recently defined a carboxyl-terminal motif in target proteins recognized by PDZ domains. As implied by this finding, PDZ domains in bacterial proteases may recognize a sequence that signals a misfolded state of the protein that differs from that of the protease cleavage site. The most striking feature of the motif identified by Songyang et al. (46) is a conserved residue (threonine, serine, or tyrosine) at the −3 position (>60% of sequences identified) and a highly conserved (>80%) hydrophobic carboxyl-terminal residue (valine, isoleucine, or leucine). Interestingly, an alignment of the carboxyl terminus of 44 pilin subunits revealed that the −3 position was often (∼50%) a serine, threonine, or tyrosine (47). Bacterial pili, such as Pap pili, assembled by the chaperone-usher assembly pathway were recently divided into two families, based on structural and sequence features: namely, the so-called FGS and FGL subfamilies (16). In the FGL subfamily, the conservation at the −3 position (serine, threonine, or tyrosine) is greater than 80%. Our initial results with SPCJ-1 (KSMCMKLSFS), which conforms to the carboxyl-terminal 10 residues of PapG, suggests that the PDZ domains in DegP play a role in substrate recognition through an interaction with the carboxyl-terminal sequence. Addition of SPCJ-1 and not of an irrelevant peptide (data not shown) enhanced the cleavage activity of the protease (Fig. 3 and 5). Whether the noncleavable peptide (SPCJ-1) stimulates multimerization of DegP, the multimer is the proposed active state of the enzyme (22, 33, 40), or it represents a signal that unfolded substrate is present and activates DegP through another mechanism is unclear. As described in detail in two recent papers (5, 41) and as supported by earlier genetic data (3, 14, 24, 44, 47, 55), the carboxyl terminus of pilin subunits is an integral component of the recognition site for chaperone-subunit complex formation. As discussed briefly above, the lack of the carboxyl-terminal seventh strand of the immunoglobulin fold results in a deep groove on the surface of the pilin that exposes the hydrophobic core. One edge of this groove is lined by the carboxyl-terminal beta strand of the pilin (the F strand) (see references 5 and 41 for a complete discussion). During donor strand complementation, the chaperone donates its G strand to complete the immunoglobulin fold of the pilin. The donated G strand completes the hydrophobic core of the pilin through interactions with both sides of the groove. Both genetic data and molecular modeling support the model that the carboxyl terminus of pilin subunits, the F strand, is a part of the pilin-pilin interface that drives pilus assembly (3, 5, 14, 41). Therefore, this motif would rarely be exposed to the solvent. The exposure of this motif, in the event of misfolding or misassembly, would signal the need for either a chaperone to catalyze refolding or a protease to degrade the protein.
These studies illustrate the mechanism by which the major periplasmic protease, DegP, cleaves misfolded pilin subunits and may relate to the general proteolytic pathway for DegP substrates. DegP's role in the pathogenesis of both gram-negative and gram-positive pathogens provides the impetus to develop assays suitable for high-throughput screening and the identification of small molecule inhibitors of this important virulence target. The described studies pave the way for the development of a novel class of anti-infectives developed against the DegP protease.
Acknowledgments
This work was supported by SBIR grant 1R43AI46828 (C.H.J.) from the NIAID.
We thank Kevin Jones and Tove' Bolken for critical reading of the manuscript and Jennifer Yoder and Susan Yeh for technical assistance. The authors also thank Elizabeth Barovsky at the Environmental Health Sciences Center, Oregon State University, for mass spectrometry analysis.
REFERENCES
- 1.Bakker, D., C. E. Vader, B. Roosendaal, F. R. Mooi, B. Oudega, and F. K. de Graaf. 1991. Structure and function of periplasmic chaperone-like proteins involved in the biosynthesis of K88 and K99 fimbriae in enterotoxigenic Escherichia coli. Mol. Microbiol. 5:875-886. [DOI] [PubMed] [Google Scholar]
- 2.Boucher, J. C., J. Martinez-Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullitt, E., C. H. Jones, R. Striker, G. Soto, F. Jacob-Dubuisson, J. Pinkner, M. J. Wick, L. Makowski, and S. J. Hultgren. 1996. Development of pilus organelle subassemblies in vitro depends on chaperone uncapping of a beta zipper. Proc. Natl. Acad. Sci. USA 93:12890-12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavard, D., C. Lazdunski, and S. P. Howard. 1989. The acylated precursor form of the colicin A lysis protein is a natural substrate of the DegP protease. J. Bacteriol. 171:6316-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 6.Danese, P. N., and T. J. Silhavy. 1997. The σE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding systems in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 7.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. B. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates degP transcription. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 8.Dodson, K. W., F. Jacob-Dubuisson, R. T. Striker, and S. J. Hultgren. 1993. Outer membrane PapC usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc. Natl. Acad. Sci. USA 90:3670-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elzer, P. H., S. D. Hagius, G. T. Robertson, R. W. Phillips, J. V. Walker, M. B. Fatemi, F. M. Enright, and R. M. Roop II. 1996. Behaviour of a high-temperature-requirement A (HtrA) deletion mutant of Brucella abortus in goats. Res. Vet. Sci. 60: 48-50. [DOI] [PubMed] [Google Scholar]
- 10.Elzer, P. H., R. W. Phillips, G. T. Robertson, and R. M. Roop II. 1996. The HtrA stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect. Immun. 64:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Gen. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies in transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 14.Hultgren, S. J., C. H. Jones, and S. N. Normark. 1996. Bacterial adhesins and their assembly, p. 2730-2756. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 15.Hultgren, S. J., S. Normark, and S. N. Abraham. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383-415. [DOI] [PubMed] [Google Scholar]
- 16.Hung, D. L., S. D. Knight, R. M. Woods, J. S. Pinkner, and S. J. Hultgren. 1996. Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 15:3792-3805. [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 18.Jones, C. H., T. C. Bolken, K. F. Jones, G. O. Zeller, and D. E. Hruby. 2001. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes. Infect. Immun. 69:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, K. I., S. C. Park, S. H. Kang, G. W. Cheong, and C. H. Chung. 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J. Mol. Biol. 294:1363-1374. [DOI] [PubMed] [Google Scholar]
- 21.Kolmar, H. 1998. DegP or protease DO CLAN SA. In J. F. Woessner (ed.), Handbook of proteolytic enzymes, vol. CD-ROM. Academic Press, London, Great Britain.
- 22.Kolmar, H., P. R. H. Waller, and R. T. Sauer. 1996. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J. Bacteriol. 178:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. (Erratum, 417:102.) [DOI] [PubMed] [Google Scholar]
- 24.Kuehn, M. J., D. J. Ogg, J. Kihlberg, L. N. Slonim, K. Flemmer, T. Bergfors, and S. J. Hultgren. 1993. Structural basis of pilus subunit recognition by the PapD chaperone. Science 262:1234-1241. [DOI] [PubMed] [Google Scholar]
- 25.Laskowska, E., D. Kuczynska-Wisnik, J. Skorko-Glonek, and A. Taylor. 1996. Degradation by proteases Lon, Clp, and HtrA of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol. Microbiol. 22:555-571. [DOI] [PubMed] [Google Scholar]
- 26.Levchenko, I., C. K. Smith, N. P. Walsh, R. T. Sauer, and T. A. Baker. 1997. PDZ-like domains mediate binding specificity in the CLP/HSP100 family of chaperones and protease regulatory subunits. Cell 91:939-947. [DOI] [PubMed] [Google Scholar]
- 27.Li, S.-R., N. Dorrell, P. H. Everest, G. Dougan, and B. W. Wren. 1996. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect. Immun. 64:2088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipinska, B., S. Sharma, and C. Georgopoulos. 1988. Sequence analysis and regulation of the htrA gene of Escherichia coli: a σ 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 16:10053-10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipinska, B., M. Zylicz, and C. Georgopoulos. 1990. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J. Bacteriol. 172:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, C. G. 1996. Protein degradation and proteolytic modification, p. 938-954. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 31.Morrison, H. G., and R. C. Desrosiers. 1993. A PCR-based strategy for extensive mutagenesis of a target DNA sequence. BioTechniques 14:454-457. [PubMed] [Google Scholar]
- 32.Noone, D., A. Howell, and K. M. Devine. 2000. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J. Bacteriol. 182:1592-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, R. W., P. H. Elzer, G. T. Robertson, S. D. Hagius, J. V. Walker, M. B. Fatemi, F. M. Enright, and R. M. Roop II. 1997. A Brucella melitensis high-temperature-requirement A (htrA) deletion mutant is attenuated in goats and protects against abortion. Res. Vet. Sci. 63:165-167. [DOI] [PubMed] [Google Scholar]
- 36.Poquet, I., V. Saint, E. Seznec, N. Simones, A. Bolotin, and A. Gruss. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 37.Raina, S., D. Missiakas, L. Baird, S. Kumar, and C. Georgopoulos. 1993. Identification and transcriptional analysis of the Escherichia coli htrE operon which is homologous to pap and related pilin operons. J. Bacteriol. 175:5009-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, M., S. Chatfield, D. Pickard, J. Li, and A. Bacon. 2000. Comparison of abilities of Salmonella enterica serovar Typhimurium aroA aroD and aroA htrA mutants to act as live vectors. Infect. Immun. 68:6041-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouviere, P. E., A. de las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sassoon, N., J. P. Arie, and J. M. Betton. 1999. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol. 33:583-589. [DOI] [PubMed] [Google Scholar]
- 41.Sauer, F. G., K. Futterer, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 42.Seol, J. H., S. K. Woo, E. M. Jung, S. J. Yoo, C. S. Lee, K. J. Kim, K. Tanaka, A. Ichihara, D. B. Ha, and C. H. Chung. 1991. Protease Do is essential for survival of Escherichia coli at high temperatures: its identity with the htrA gene product. Biochem. Biophys. Res. Commun. 176:730-736. [DOI] [PubMed] [Google Scholar]
- 43.Skorko-Glonek, J., A. Wawrzynow, K. Krzewski, K. Kurpierz, and B. Lipinska. 1995. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene 163:47-52. [DOI] [PubMed] [Google Scholar]
- 44.Slonim, L. N., J. S. Pinkner, C. I. Branden, and S. J. Hultgren. 1992. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 11:4747-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smeds, A., P. Varmanen, and A. Palva. 1998. Molecular characterization of a stress-inducible gene from Lactobacillus helveticus. J. Bacteriol. 180:6148-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Songyang, Z., A. S. Fanning, C. Fu, J. Xu, S. M. Marfatia, A. H. Chishti, A. Crompton, A. C. Chan, J. M. Anderson, and L. C. Cantley. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275:73-77. [DOI] [PubMed] [Google Scholar]
- 47.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 49.St. Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 50.Strauch, K. L., and J. Beckwith. 1988. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc. Natl. Acad. Sci. USA 85:1576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tacket, C. O., M. B. Sztein, G. A. Losonsky, S. S. Wasserman, J. P. Nataro, R. Edelman, D. Pickard, G. Dougan, S. N. Chatfield, and M. M. Levine. 1997. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect. Immun. 65:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tacket, C. O., M. B. Sztein, S. S. Wasserman, G. Losonsky, K. L. Kotloff, T. L. Wyant, J. P. Nataro, R. Edelman, J. Perry, P. Bedford, D. Brown, S. Chatfield, G. Dougan, and M. M. Levine. 2000. Phase 2 clinical trial of attenuated Salmonella enterica serovar Typhi oral live vector vaccine CVD 908-htrA in U.S. volunteers. Infect. Immun. 68:1196-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waller, P. R., and R. T. Sauer. 1996. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol. 178:1146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, Z., C. H. Jones, D. Haslam, K. Dodson, J. Kihlberg, and S. J. Hultgren. 1995. Molecular dissection of PapD interaction with PapG reveals two chaperone binding sites. Mol. Microbiol. 16:1011-1020. [DOI] [PubMed] [Google Scholar]