Abstract
Acyl homoserine lactone (acyl-HSL)-mediated gene regulation has been shown to influence biofilm formation in one Burkholderia cepacia cystic fibrosis isolate, but it is not known whether this relationship is a consistent feature of the several genomic species that make up the B. cepacia complex (BCC). We screened strains belonging to genomovars I to V of the BCC for biofilm formation on an abiotic surface and for acyl-HSL synthesis. We determined that organisms from each of these genomovars were capable of biofilm formation. Similarly, acyl-HSL was synthesized by organisms from each of genomovars I to V, with most isolates producing octanoyl-HSL in greatest abundance. When biofilms were grown in Luria broth, acyl-HSL synthesis and biofilm formation appeared to be associated, but these phenotypes were independent when the biofilms were grown in basal salts containing citrate. Genomovar V strains synthesized the greatest quantities of acyl-HSL, and genomovar II and III-A strains elaborated the most abundant biofilms. Quorum sensing may play a role in BCC pathogenesis, but it may not regulate biofilm formation under all growth conditions.
Burkholderia cepacia has been recognized as a problematic opportunistic pathogen, particularly among cystic fibrosis (CF) and chronic granulomatous disease patients. Accurate identification of this organism can be problematic as its taxonomy continues to evolve. The B. cepacia complex (BCC) is a group of phenotypically related but genotypically distinct organisms. The BCC is divided into at least nine closely related genomic species or genomovars. Genomovars II, IV, V, VII, VIII, and IX have been named Burkholderia multivorans, Burkholderia stabilis, Burkholderia vietnamiensis, Burkholderia ambifaria, Burkholderia anthina, and Burkholderia pyrrocinia, respectively. Genomovars I, III, and VI have not been formally named, pending the availability of differential diagnostic tests, but genomovar I is understood to be B. cepacia.
Interest in acyl homoserine lactone (acyl-HSL)-mediated quorum sensing and bacterial biofilm formation has increased in recent years. Quorum sensing regulates the expression of virulence factors in several organisms, including the CF pathogen, Pseudomonas aeruginosa (29, 30, 34). Biofilm formation by P. aeruginosa has been recognized as an important clinical problem due to the intrinsically high level of antibiotic resistance of bacteria growing in the biofilm (2, 3, 26). Maturation of a P. aeruginosa biofilm requires synthesis of the quorum sensing signal, 3-oxo-dodecanoyl HSL (3OC12-HSL), indicating that quorum sensing is necessary for biofilm formation (7). Quorum sensing and biofilm formation have also been observed with B. cepacia (10, 13, 17, 24). BCC organisms are problematic since some can be highly virulent and highly transmissible in CF patients (11, 14, 18). In addition, treatment is often complicated by the high degree of antibiotic resistance exhibited by these organisms, and this resistance may be enhanced in biofilms. The purpose of these studies was to determine if the properties of quorum sensing and biofilm formation were widespread among the BCC. Isolates were tested for their ability to form biofilms in a microtiter plate assay and for their ability to synthesize acyl-HSLs in stationary phase culture. These data were analyzed to determine if a correlation existed between acyl-HSL-mediated quorum sensing and biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth.
Bacterial strains used are listed in Table 1. Most of these organisms have been designated as part of the B. cepacia experimental strain panel by the International B. cepacia Working Group (22). Nonpanel organisms were added to increase representation of other genetically distinct strains within each of the genomovars, as determined by random amplified polymorphic DNA (RAPD) fingerprinting (21). Genomovar IV had the lowest genetic variability among isolates, with just three different RAPD groups occurring among the six strains tested. All isolates are deposited in the Canadian B. cepacia Complex Research and Referral Repository, Vancouver, British Columbia. Growth curves were determined by incubation in Luria broth (LB) (pH 7.0) at 37°C, 250 rpm. Aliquots were removed at specific time intervals and diluted in phosphate-buffered saline (pH 7.4) if necessary, and the optical density was measured at 600 nm.
TABLE 1.
Bacterial strains used in this study
| Bacterial strain | Presence of BCESMa | Source, locationb | Referencec |
|---|---|---|---|
| Genomovar I | |||
| C9139 | − | CF, Canada | This study |
| ATCC 25416 | − | Onion, U.S. | 22 |
| ATCC 17759 | − | Soil, Trinidad | 22 |
| ATCC 10856 | − | Onion | ATCC |
| CEP509 | − | CF, Australia | 22 |
| LMG 17997 | − | UTI, Sweden | 22 |
| Genomovar II (B. multivorans) | |||
| C5274 | − | CF, Canada | This study |
| C5393 | − | CF, Canada | 22 |
| 249-2 | − | Laboratory, U.S. | 22 |
| ATCC 17616 | − | Soil, U.S. | 22 |
| C1576 | − | CF-e, U.K. | 22 |
| JTC | − | CGD, U.S. | 22 |
| LMG 13010 | − | CF, Belgium | 22 |
| C1962 | − | Clinic, U.K. | 22 |
| CF-A1-1 | − | CF-e, U.K. | 22 |
| Genomovar III-A | |||
| C1257 | + | CF, Canada | This study |
| C4455 | + | CF, Canada | This study |
| C5424 | + | CF-e, Canada | 22 |
| C6433 | + | CF-e, Canada | 22 |
| J2315 | + | CF-e, U.K. | 22 |
| BC7 | + | CF-e, Canada | 22 |
| K56-2 | + | CF-e, Canada | 22 |
| Genomovar III-B | |||
| PC184 | + | CF-e, U.S. | 22 |
| C1394 | + | CF-e, U.K. | 22 |
| J415 | − | CF, U.K. | 22 |
| CEP511 | + | CF-e, Australia | 22 |
| ATCC 17765 | + | UTI, U.K. | 22 |
| Genomovar IV (B. stabilis) | |||
| C7322 | − | CF, Canada | 22 |
| ATCC 35254 | − | Disinfectant, U.S. | 4 |
| LMG 07000 | − | Clinical, Sweden | BCCM/LMG |
| LMG 14294 | − | CF, Belgium | 22 |
| LMG 14086 | − | Respirator, U.K. | 22 |
| LMG 18888 | − | Clinical, Belgium | 22 |
| Genomovar V (B. vietnamiensis) | |||
| C2822 | − | CF, Canada | This study |
| C9178 | − | CF, Canada | This study |
| PC259 | − | CF, U.S. | 22 |
| LMG 10929 | − | Rice, Vietnam | 22 |
| FC441 | − | CGD, Canada | 22 |
| LMG 16232 | − | CF, Sweden | 22 |
| G4 | − | WTF holding pond, U.S. | 25 |
Presence (+) or absence (−) of B. cepacia epidemic strain marker (23).
CF, infection of a CF patient; CF-e, strain that has spread epidemically among patients with CF; CGD, infection of a chronic granulomatous disease patient; UTI, urinary tract infection; WTF, waste treatment facility; U.S., United States; U.K., United Kingdom
ATCC, American Type Culture Collection, Manassas, Va.; BCCM/LMG, Belgian Co-ordinated Collections of Micro-organisms—Bacteria Collection, Ghent, Belgium.
Growth of bacterial biofilms.
Biofilm formation was tested by determining the ability of BCC strains to adhere to the wells of 96-well polypropylene microtiter dishes (28). BCC strains were spotted on LB agar and grown overnight at 37°C. Organisms were inoculated into media (100 μl/well) in 96-well polypropylene microtiter plates (Costar # 3790; Corning Incorporated Life Sciences, Acton, Mass.) using a pin-inoculation device. Three types of media were employed: supplemented LB (SLB) consisted of LB with 0.5% (wt/vol) Casamino Acids (CAA); supplemented basal salts medium with citrate (SBSM-citrate) consisted of basal salts medium (BSM) with 20 mM citrate and 0.5% (wt/vol) CAA; and SBSM-glucose consisted of BSM with 20 mM glucose and 0.5% (wt/vol) CAA. Microtiter plates with fitted lids were incubated at 37°C for 48 h in a closed, humidified plastic container. The plates were then washed to remove planktonic organisms, and stained for 15 min with 125 μl of 1% (wt/vol) crystal violet. After thorough washing with water, 200 μl of 95% (vol/vol) ethanol was added to the wells to release the stain. The extent of biofilm staining was determined by measuring the absorbance of the resulting solution at 590 nm.
Identification of acyl-HSLs.
Acyl-HSLs were detected using a previously described radiotracer procedure (32). BCC strains were grown in 5 ml of BSM containing 20 mM glycerol at 37°C with shaking (12). To label acyl-HSLs, 5.0 μCi of l-[1-14C]methionine was added to cultures in early stationary phase and incubated for 30 min. Radiolabeled acyl-HSLs were recovered by two extractions of the culture fluid with equal volumes of ethyl acetate containing 0.1 ml of glacial acetic acid per liter. After drying, the sample residue was dissolved in 20% methanol and fractionated by C18 reverse-phase high performance liquid chromatography (HPLC) as previously described (29). Aqueous Counting Scintillant (Amersham Pharmacia Biotech, Piscataway, N.J.) was added to each of 70 1-ml fractions and counted with a scintillation detector. To identify acyl-HSLs, the retention times of peaks of radioactivity were compared to retention times of synthetic acyl-HSL standards (Quorum Sciences Inc., Coralville, Iowa).
Southern hybridizations.
Southern hybridizations were performed using digoxigenin (DIG)-dUTP-labeled probes (Roche Molecular Biochemicals). Methods were as per the manufacturer's protocols using total DNA isolated from BCC organisms. Total DNA was recovered by incubating cells on ice with lysozyme and then lysing them with sodium dodecyl sulfate. DNA was purified from cell debris by phenol extraction followed by ethanol precipitation. Total DNA (2 μg) was digested with XhoI (New England Biolabs) and then separated on an 0.8% Tris-acetate-EDTA-buffered agarose gel. DIG-dUTP-labeled cepI and bviI gene probes were generated by PCR from the B. cepacia K56-2 chromosome and cloned B. vietnamiensis G4 bviI gene, respectively (6, 17). Southern transfer and detection of homologous sequences, under low-stringency conditions with labeled probes, were performed using a DIG detection kit (Roche Molecular Biochemicals). The low-stringency conditions were attained by incubating membranes and labeled probe overnight at 53°C and by carrying out the two 15-min stringency washes with 0.75× SSC (0.11 M NaCl plus 0.011 M sodium citrate)-0.1% sodium dodecyl sulfate at 53°C.
Statistical analysis methods.
Descriptive statistics were used to compare the frequencies with which biofilm formation (defined as A590 of ⩾0.100) and acyl-HSL synthesis (defined as an incorporation of radiolabel greater than 100 cpm/30 min) occurred together in a population. Biofilm formations in each of the three media were considered to be different populations; thus, three separate comparisons of biofilm formation and acyl-HSL synthesis were made. The G value for goodness of fit was calculated for each population using 2 × 2 tables of observed and expected frequencies (33). Williams' correction (for sample sizes where n was <200) was applied to each G value to obtain the adjusted G value (Gadj) (33).
RESULTS
BCC isolates form biofilms on polypropylene.
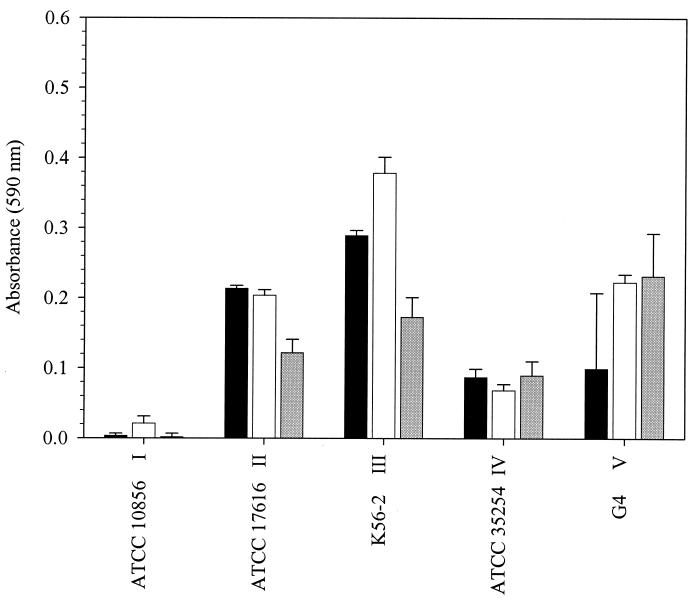
The biofilm formation assay was based on the one described by O'Toole and Kolter (see Materials and Methods) (28). Preliminary assays using 96-well polypropylene microtiter dishes and one BCC isolate from each of genomovars I to V indicated that attached growth was possible for these organisms. Biofilm formation in SLB (Fig. 1) was assessed after 24, 48, and 72 h of growth at 37°C. After 48 h, substantial attached growth was observed for three of the five test strains. Neither the genomovar I test strains nor the genomovar IV test strains formed substantial biofilms, but the amount of attached growth at 48 h was the maximum observed for the genomovar I strain and was not significantly different from the maximum growth observed for the genomovar IV strain. Similar results were obtained when the test strains were grown in SBSM-citrate (data not shown). Because measurable attached growth at or near the maximum observed had occurred in the test strains by 48 h, all subsequent biofilm assays were carried out at 37°C for 48 h.
FIG. 1.
Biofilm formation by B. cepacia complex strains at different incubation times. Representative strains from each of genomovars I to V were grown in 96-well polypropylene microtiter dishes in SLB. Strain and genomovar designations appear on the x axis. Biofilm formation was assessed at 24 h (black bars), 48 h (white bars), and 72 h (grey bars) as noted in Materials and Methods. Each bar represents the average for three replicates, and vertical lines represent standard errors.
Genomovar II and III isolates demonstrate the greatest capacity for biofilm formation.
Our approach employed different media to provide organisms with conditions favorable for biofilm formation but also to detect any significant differences among isolates. To that end we used a rich medium (SLB) and minimal media (SBSM-citrate and SBSM-glucose) supplemented with CAA. Carbon sources were chosen based on preliminary studies indicating that minimal medium containing citrate allowed the best biofilm formation among test isolates from all five genomovars (data not shown). Glucose was chosen as an alternative carbon source since growth in this medium allowed differences in biofilm formation by test isolates from the five genomovars to be observed (data not shown).
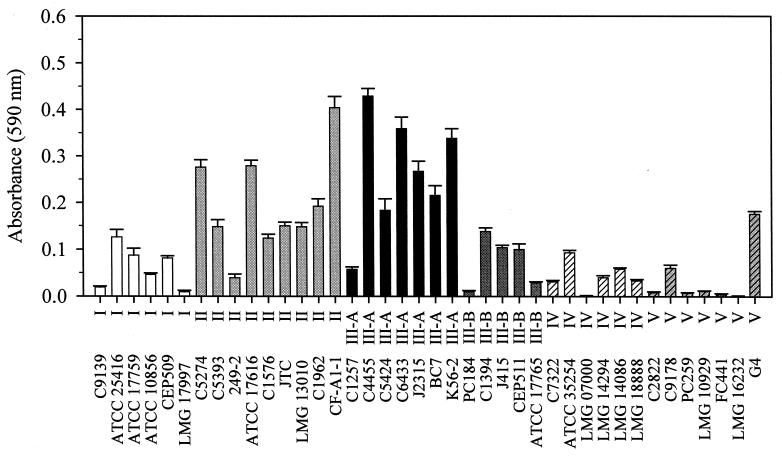
The biofilm-forming abilities of strains from genomovars I to V, grown in SLB, are shown in Fig. 2. Based on quantitation of biofilm growth by crystal violet staining, genomovar II and III-A isolates were capable of greater biofilm formation in polypropylene microtiter dishes than genomovars I, III-B, IV, and V, when grown in SLB. Similar results were obtained when organisms were grown in SBSM-citrate and SBSM-glucose (data not shown).
FIG. 2.
Biofilm formation on polypropylene by B. cepacia complex strains. Strains from genomovars I to V were tested for their ability to form biofilms in 96-well polypropylene microtiter dishes in SLB. Strain and genomovar designations appear on the x axis. Shaded bars denote genomovars: genomovar I, white; genomovar II, light grey; genomovar III-A, black; genomovar III-B, dark grey; genomovar IV, white, hatched; genomovar V, light grey, hatched. Biofilm formation after 48 h was measured as described in Materials and Methods. Each assay included at least eight replicates per organism. Bars represent averages from at least three assays. Vertical lines denote standard errors.
The separation of genomovar III isolates into two subgroups, A and B, is based on differences at the recA locus. The recA gene product is essential for repair and recombination of DNA, and sequence polymorphisms of the gene have made it a useful bacterial systematics tool (20). Genomovar III-A strains formed more biofilm than III-B strains under the conditions used. To test a diverse range of strains within the genomovars, organisms included in the screen were from different genetic lineages as determined by RAPD analysis (21). All genomovar III isolates except for J415 (III-B) carried the B. cepacia epidemic strain marker (BCESM), which is present in several transmissible strains infecting multiple CF patients at different treatment centers (Table 1) (23). BCESM DNA, which encodes the putative transcriptional regulator, EsmR, was present both in isolates that formed biofilms and those that did not. Therefore, it is unlikely that EsmR has a specific function in biofilm formation in BCC isolates.
To determine the impact of growth rate on biofilm formation, we assessed the growth rates of several isolates from each of the genomovars (Table 2). Of the 40 isolates, 30 had doubling times of approximately 1 to 2 h when grown with shaking in LB; most of these were from genomovars I, II, III, and V. Among strains with doubling times shorter than 2 h, there was considerable variation in the amount of biofilm formation observed. Strains with doubling times longer than 2 h also exhibited variable biofilm formation. The genomovar III-A strains CEP565 and BC7 were observed to have good biofilm formation with doubling times close to 3 h, whereas the four genomovar IV strains with longer doubling times formed much less biofilm. While the growth rate certainly influences biofilm formation, these data suggest that additional factors are involved and that the observed differences in biofilm formation are likely not simply the result of different growth rates.
TABLE 2.
Acyl-HSL synthesis by B. cepacia complex isolates
| Isolate | Doubling timea (h) | Most abundant acyl-HSLb | Acyl-HSL radiolabele (cpm/30 min) | ceplf-(kb) | bvilf-(kb) |
|---|---|---|---|---|---|
| Genomovar I | |||||
| C9139 | 1.1 | C8-HSL | 6,370 | 2.2 | — |
| ATCC 25416 | 0.9 | C8-HSL | 8,480 | 2.2 | — |
| ATCC 17759 | 1.0 | C8-HSL | 17,329 | 2.2 | — |
| ATCC 10856 | 1.0 | C8-HSL | 7,193 | 2.2 | — |
| CEP509 | 1.0 | C8-HSL | 685 | 2.2 | — |
| LMG 17997 | 1.2 | ND | ND | 2.2 | — |
| Genomovar II | |||||
| C5274 | 1.1 | ND | ND | — | — |
| C5393 | 1.7 | — | 0 | — | — |
| 249-2 | 1.3 | — | 0 | — | — |
| ATCC 17616 | 1.1 | C8-HSL | 134 | — | — |
| C1576 | 1.8 | C8-HSL | 744 | — | — |
| JTC | 1.4 | — | 0 | — | — |
| LMG 13010 | 1.4 | — | 0 | — | — |
| C1962 | 1.1 | C8-HSL | 148 | — | — |
| CF-A1-1 | 1.5 | — | 0 | — | — |
| Genomovar III-A | |||||
| C1257 | 1.2 | C8-HSL | 12,894 | 6.7 | — |
| C4455 | 1.2 | F32c/C8-HSL | 343/253 | 8.0 | — |
| C5424 | 1.7 | — | 0 | >12.2 | — |
| C6433 | 1.2 | — | 0 | 6.7 | — |
| J2315 | 2.8 | — | 0 | >12.2 | — |
| BC7 | 3.0 | C8-HSL | 909 | >12.2 | — |
| K56-2 | 1.7 | C8-HSL | 301 | >12.2 | — |
| Genomovar III-B | |||||
| PC184 | 2.4 | C8-HSL | 345 | 6.7 | — |
| C1394 | 1.9 | C8-HSL | 1,964 | 7.1 | — |
| J415 | 2.4 | C8-HSL | 5,182 | 11.0 | — |
| CEP511 | 1.4 | — | ND | 6.7 | — |
| ATCC 17765 | 1.6 | C4-/3OC6-HSLd | 1,510 | 6.7 | — |
| Genomovar IV | |||||
| C7322 | 3.7 | ND | ND | 7.1 | — |
| ATCC 35254 | 2.8 | C8-HSL | 8,830 | 7.1 | — |
| LMG 07000 | 1.7 | C8-HSL | 8,272 | 7.7 | — |
| LMG 14294 | 1.8 | C8-HSL | 1,455 | 8.0 | — |
| LMG 14086 | 3.5 | C8-HSL | 160 | 7.7 | — |
| LMG 18888 | 2.1 | — | 0 | 7.7 | — |
| Genomovar V | |||||
| C2822 | 1.3 | C8-HSL | 17,931 | 5.5 | 2.8/6.7 |
| C9178 | 1.3 | C8-HSL | 26,519 | 5.5 | 2.8/6.7 |
| PC259 | 1.5 | C8-HSL | 2,592 | >12.2 | 2.8/6.7 |
| LMG 10929 | 1.3 | C10-HSL | 71,206 | >12.2 | 2.8/6.7 |
| FC441 | 1.3 | C8-HSL | 24,033 | >12.2 | 2.8/6.7 |
| LMG 16232 | 1.2 | C8-HSL | 175 | >12.2 | 2.8/6.7 |
| G4 | 2.0 | C10-HSL | 56,852 | >12.2 | 4.1/6.7 |
Doubling time in LB at 37°C, 250 rpm.
ND, not determined; —, no acyl-HSL detected.
Unidentified 14C-labeled species occurring in HPLC fraction 42.
C4-HSL and 3OC6-HSL eluted in the same fractions under the HPLC conditions used.
Amount of radiolabel detected in most abundant acyl-HSL in 5 ml of 14C-labeled culture.
Sizes of bands containing the specific gene sequence detected by Southern hybridization of gene probe to XhoI-digested chromosomal DNA; —, no hybridization.
Acyl-HSL synthesis by B. cepacia complex isolates.
Acyl-HSL synthesis was assessed during planktonic growth in minimal medium using a previously described radiotracer procedure (32). Table 2 shows the amount of radioisotope incorporated into the most abundant acyl-HSL synthesized by the BCC isolates assayed. Acyl-HSLs were identified by comparing the mobilities of radiolabeled peaks with the mobilities of synthetic acyl-HSLs. Most of the BCC isolates producing acyl-HSLs synthesized C8-HSL in highest abundance. Exceptions to this were a genomovar III-A isolate, C4455, a genomovar III-B isolate, ATCC 17765, and two genomovar V isolates, LMG 10929 and G4. C4455 synthesized an unidentified species that eluted in fraction 32 under the HPLC conditions used. This species eluted between C6-HSL and C8-HSL and may resemble a previously reported unidentified species that migrates between C6-HSL and C8-HSL on C18 reversed-phase thin layer chromatography plates (19, 36). One organism, ATCC 17765 (genomovar III-B), produced C4-HSL and/or 3OC6-HSL, which elute together under the HPLC conditions used. The genomovar V strains, B. vietnamiensis G4 and LMG 10929, synthesized C10-HSL in greatest abundance. Synthesis of acyl-HSLs by the trichloroethylene degrader, G4, has been reported previously (6).
All genomovar I and V isolates tested incorporated radiolabel into acyl-HSLs, as shown in Table 2. Incorporation of radiolabel into acyl-HSL by strains from genomovars II, III, and IV was varied, with several strains failing to synthesize detectable levels of acyl-HSLs. Genomovar V strains synthesized the most acyl-HSL, as demonstrated by the level of incorporation of radiolabel. The amount of acyl-HSL synthesized can be summarized as follows: no or low detectable radiolabel incorporated into acyl-HSL, <2,000 cpm incorporated in 30 min; moderate, between 2,000 and 10,000 cpm incorporated into acyl-HSL; and high, greater than 10,000 cpm incorporated into acyl-HSL. With the exception of one genomovar I strain (ATCC 17759) and one genomovar III strain (C1257), just the genomovar V strains were observed to synthesize acyl-HSLs in large amounts. Some strains from genomovars II, III, and IV failed to incorporate detectable amounts of radiolabel into acyl-HSL. It should be noted that acyl-HSL production by one isolate, C5393, was previously detected using an Agrobacterium tumefaciens reporter strain (19). This organism did not incorporate radiolabel into acyl-HSL under the conditions described here. This may have been because a larger volume of culture was assayed previously, or it may reflect the sensitivity of the reporter to the acyl-HSL detected. Alternatively, it may reflect the ability of the organism to take up or incorporate the radioisotope. Our data indicate that if acyl-HSLs were synthesized by C5393, the amounts were small enough to be undetectable in the radiolabeling assay. For comparison, acyl-HSL synthesis by P. aeruginosa resulted in greater than 100,000 cpm being incorporated into 3OC12-HSL in 10 min (32).
Since acyl-HSL synthesis was not detected for many BCC isolates, we investigated if the cepI or bviI autoinducer synthase genes were present in these organisms. Total DNA from each isolate was digested with XhoI, electrophoretically separated, transferred to a nylon membrane, and hybridized with specific gene probes for the cepI and bviI autoinducer synthases (6, 17, 19). The results are shown in Table 2. The bviI gene was detected only in genomovar V strains, in agreement with previously published results (19). All genomovar V strains exhibited the same bviI hybridization pattern except for the C10-HSL producer, G4. The cepI gene was detected in all isolates except for genomovar II organisms. This is also consistent with published data (19). The cepI hybridization patterns for all genomovar I isolates were identical. In contrast, the cepI hybridization patterns for genomovar III isolates were quite heterogeneous. Among the seven III-A organisms, four different band sizes were detected; among the five III-B organisms, three different band sizes were detected. These banding patterns did not correlate with the RAPD designations of the organisms, since different banding patterns were observed among the strains of the same RAPD type and similar banding patterns were observed for strains from different RAPD groups. Three different hybridization patterns were observed among the six genomovar IV isolates. This suggests that the genomovar I, III, IV, and V isolates all have the genetic potential for acyl-HSL synthesis and that the autoinducer synthase genes may reside in different chromosomal locations. Yao et al. identified a member of the luxI family of autoinducer synthases, bmuI, in B. multivorans ATCC17616 (36). It is possible that the genomovar II isolates may contain bmuI sequences which are different enough from cepI and bviI to be undetectable by those gene probes.
Correlation of biofilm formation with acyl-HSL synthesis.
We assumed that with BCC isolates, as with P. aeruginosa, biofilm formation would be a process that was regulated in part by quorum sensing. Huber et al. showed that biofilm formation by a BCC genomovar III CF isolate is under control of the cep system (13). We therefore compared biofilm formation with acyl-HSL synthesis using a two-way table test of independence (33). To determine the frequencies of occurrence of acyl-HSL synthesis and biofilm formation, acyl-HSL synthesis was defined as any incorporation of radiolabel greater than twofold over background (>100 cpm), and biofilm formation was defined as an A590 value of ⩾0.100. This value was chosen because it resulted in a measured A590 value greater than twofold that of background staining of the wells of the microtiter plate. The calculated G values for biofilms grown in the different media were Gadj = 4.400 for SLB and Gadj = 2.273 for SBSM-citrate. A G value could not be calculated for SBSM-glucose since no data points fell within the “no biofilm and no acyl-HSL” section of the two-way table (frequencies are shown in Table 3). The calculated G value for SLB (Gadj = 4.400) was greater than the χ20.05 value for 1 degree of freedom (χ20.05[1] = 3.841); therefore, the null hypothesis that biofilm formation was independent of acyl-HSL synthesis was rejected. However, the calculated G value for SBSM-citrate (Gadj = 2.273) was less than the χ20.05 value for 1 degree of freedom, indicating that biofilm formation was independent of acyl-HSL synthesis. These conflicting outcomes make it difficult to reconcile the biofilm formation and acyl-HSL data. As shown in Table 3, in the cases of all three media, from 28 to 47% of strains synthesized acyl-HSLs and formed biofilms; likewise, 28 to 47% of strains synthesized acyl-HSLs but failed to form biofilms. Most (75%) of the strains tested fell into these two categories. The frequency of strains forming biofilms without synthesizing acyl-HSL was 19 to 25%, and only a few strains neither formed biofilms nor synthesized acyl-HSLs. Together, the data suggest that acyl-HSL synthesis is widespread among the BCC but not all strains with the genetic capacity for acyl-HSL synthesis make acyl-HSLs in culture. In addition, biofilm formation occurred in strains from each of the genomovars, but the greatest amount of biofilm formation was observed in genomovars II and III-A. These two phenotypes are widespread among the BCC but do not appear to be linked under all growth conditions.
TABLE 3.
Frequencies of association of biofilm formation with acyl-HSL synthesis
| Growth medium | No. (%) of isolatesa
|
|||
|---|---|---|---|---|
| Bfm and AHL | Bfm, no AHL | No Bfm, AHL | No Bfm, no AHL | |
| SLB | 10 (28) | 7 (19) | 17 (47) | 2 (6) |
| SBSM-citrate | 17 (47) | 8 (22) | 10 (28) | 1 (3) |
| SBSM-glucose | 10 (28) | 9 (25) | 17 (47) | 0 (0) |
Associated phenotypes shown as numbers of isolates and, in parentheses, percentage of total strains tested for both phenotypes. Bfm, biofilm; AHL, acyl-HSL synthesis.
DISCUSSION
The association of bacterial biofilms with a disease state in the human host has brought increased attention to the study of this communal stage of microbial development. Bacterial biofilms have been studied for decades, but recognition of their importance in opportunistic infection has come more recently (for a recent review, see reference 27). Of particular interest in the study of CF pulmonary infections are recent reports demonstrating the ability of the CF pathogen P. aeruginosa to form biofilms and the involvement of quorum sensing or cell density-dependent regulation of gene expression in this process (7, 32). In addition, the cep quorum sensing system of a B. cepacia CF isolate has been shown to play a role in biofilm formation in one BCC strain (13). Other studies have also indicated that acyl-HSL synthesis is widespread among members of the BCC (10, 19).
We report that among representative isolates from genomovars I to V of the BCC, the greatest amount of biofilm formation was observed for genomovar II and III isolates. Although members of all five genomovars have been found in CF patients, genomovars II and III are most commonly isolated. Organisms from both of these groups have been responsible for several outbreaks in CF clinics, with evidence of patient-to-patient spread (11, 20). This suggests that like P. aeruginosa, BCC genomovar II and III strains may exist in a biofilm state in the CF lung, and this may give them an advantage by providing protection from host antimicrobial defenses as well as increased resistance to antibiotic therapy. Our findings are consistent with recent studies demonstrating that in both in vitro and in vivo models, BCC strains from genomovars II and III were more invasive than strains belonging to genomovars I, IV, and V (5, 15). Of 12 invasive strains from the study by Cieri et al., 11 formed biofilms in at least one media type, compared to 3 of 7 of the noninvasive strains (5). It is possible that biofilm formation contributes to the virulence of these organisms. To our surprise, within the genomovar III strains, the recA type A organisms were more competent to grow as biofilms than the recA type B organisms. The recA gene is involved in DNA recombination and is used as a taxonomic marker because of its high degree of nucleotide sequence conservation. We do not yet understand the significance of the increase in biofilm formation for genomovar III-A strains over that for genomovar III-B strains, but it would be informative to study a larger number of strains from these two subgenomovar groups to determine if this relationship is true for a larger population.
The results of our survey of acyl-HSLs synthesized by BCC isolates was in agreement with previous reports that most of these organisms synthesize C8-HSL (10, 19, 36). Exceptions included a genomovar III isolate that produced C4-HSL and/or 3OC6-HSL and two genomovar V isolates that synthesized C10-HSL in greatest abundance. We also detected an unidentified species synthesized by another genomovar III strain. Under the HPLC conditions used, this molecule eluted between C6-HSL and C8-HSL. Other investigators have also reported unidentified molecules that eluted between C6-HSL and C8-HSL for both a genomovar II and genomovar VII isolate (19, 36). It remains to be determined if these species are acyl-HSLs.
We note that the amount of incorporation of radiolabel into acyl-HSL varied from none or low levels (<2,000 cpm incorporated per 30 min) to high levels of incorporation (>10,000 cpm incorporated per 30 min). We observed that the highest production of acyl-HSL occurred in the genomovar V strains and that several isolates from genomovars II, III, and IV synthesized low or no acyl-HSL under the conditions tested. Genomovar I and V strains were also notable in that every isolate tested produced acyl-HSL. This was not the case for genomovars II, III, and IV. We detected low or no acyl-HSL synthesis in genomovar II strains and in most genomovar III strains. Genomovar IV isolates produced low to moderate amounts of acyl-HSL, with one strain producing none. It is difficult to compare acyl-HSL synthesis rates between strains when they are determined by the radiotracer method because of potential differences in methionine uptake and incorporation into the acyl-HSL precursor S-adenosylmethionine. In two BCC strains, however, >100-fold differences in 14C incorporation into acyl-HSLs confirmed the differences previously observed in the concentration of acyl-HSLs in cell-free culture fluids of strains G4 and K56-2 (6, 17). This suggests that the differences in incorporation detected in this study may reflect true differences in acyl-HSL production in at least some cases. It is interesting that a significant proportion of BCC strains (22 of 37) produced only low levels of acyl-HSL or no acyl-HSL. In all cases, except for the genomovar II isolates, at least one autoinducer synthase gene was identified, indicating that these organisms have the genetic potential for acyl-HSL synthesis. Recently it was reported that one genomovar II organism, B. multivorans ATCC 17616, readily gave rise to mutants that produced high levels of acyl-HSL and that it contained a novel acyl-HSL synthase gene, bmuI (36). It is likely that other genomovar II strains contain bmuI and that most BCC organisms have the ability to synthesize acyl-HSLs. There may be specific environmental cues that cause the levels of acyl-HSL synthesis to increase or decrease, and the growth conditions used here have not allowed this additional level of regulation to be observed. For Vibrio fischeri and P. aeruginosa, hierarchies of control exist that additionally regulate acyl-HSL production (1, 8, 9, 35); this may also be the case for B. cepacia. Alternatively, the observed differences in acyl-HSL synthesis may be the result of genetic rearrangement, since it is known that BCC organisms have complex genomes containing multiple chromosomes and insertion sequences (16). It is also possible that expression of some quorum sensing-controlled genes requires only a low level of acyl-HSL synthesis while others may require high levels of the signal molecule: different target genes are activated at different 3OC12-HSL concentrations for P. aeruginosa (31).
For all isolates tested, the amount of biofilm formation and the amount of radiolabeled primary acyl-HSL were compared. A test for independence using a two-way table of independence determined that biofilm formation in SLB was not independent of acyl-HSL synthesis but that biofilm formation in SBSM-citrate was independent of acyl-HSL synthesis. A recently published study demonstrated that the cep quorum sensing system controlled biofilm formation and swarming motility in a genomovar III isolate (13). The results of this study, while intriguing, do not provide a clear-cut demonstration of correlation between biofilm formation and acyl-HSL synthesis in the BCC. It is possible that biofilm formation does not require a functional quorum sensing system under all conditions. It may be that BCC isolates produce different levels of acyl-HSL when grown in a biofilm than they do when grown planktonically. We are currently evaluating a method that may allow rapid screening of acyl-HSL synthesis by biofilm-grown cultures. We would like to determine if the level of acyl-HSL synthesis by some BCC isolates is different for planktonic versus biofilm-grown cells, as has been demonstrated with P. aeruginosa (32). It is possible that a consistent relationship between biofilm formation and acyl-HSL synthesis might be detected under those conditions. Although we were unable to demonstrate a clear correlation or lack of correlation between quorum sensing and biofilm formation, acyl-HSL production likely plays a role in ecology and pathogenesis in the BCC. Further studies will be required to identify phenotypic traits under control of this regulatory system.
Acknowledgments
This work was supported by grants from the Canadian Cystic Fibrosis Foundation, BC Lung Association, and Canadian Bacterial Diseases Network (to D.P.S.).
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anwar, H., M. Dasgupta, K. Lam, and J. W. Costerton. 1989. Tobramycin resistance of mucoid Pseudomonas aeruginosa biofilm grown under iron limitation. J. Antimicrob. Chemother. 24:647-655. [DOI] [PubMed] [Google Scholar]
- 3.Anwar, H., J. L. Strap, and J. W. Costerton. 1992. Establishment of aging biofilms: possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob. Agents Chemother. 36:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkelman, R. L., S. Lewin, J. R. Allen, R. L. Anderson, L. D. Budnick, S. Shapiro, S. M. Friedman, P. Nicholas, R. S. Holzman, and R. W. Haley. 1981. Pseudobacteremia attributed to contamination of povidone-iodine with Pseudomonas cepacia. Ann. Intern. Med. 95:32-36. [DOI] [PubMed] [Google Scholar]
- 5.Cieri, M. V., N. Mayer-Hamblett, A. Griffith, and J. L. Burns. 2002. Correlation between an in vitro invasion assay and a murine model of Burkholderia cepacia lung infection. Infect. Immun. 70:1081-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conway, B. D., and E. P. Greenberg. 2002. Quorum sensing signals and quorum sensing genes in Burkholderia vietnamiensis. J. Bacteriol. 184:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 8.De Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlap, P. V., and E. P. Greenberg. 1988. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J. Bacteriol. 170:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotschlich, A., B. Huber, O. Geisenberger, A. Togl, A. Steidle, K. Riedel, P. Hill, B. Tummler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is widespread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 11.Govan, J. R., P. H. Brown, and J. Maddison. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 12.Hareland, W., R. L. Crawford, P. J. Chapman, and S. Dagley. 1975. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J. Bacteriol. 121:272-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 14.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis; an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 15.Keig, P. M., E. Ingham, P. A. R. Vandamme, and K. G. Kerr. 2002. Differential invasion of respiratory epithelial cells by members of the Burkholderia cepacia complex. Clin. Microbiol. Infect. 8:47-49. [DOI] [PubMed] [Google Scholar]
- 16.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 17.Lewenza, S., B. Conway, E. P. Greenberg, and P. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LiPuma, J. J., S. E. Dasen, D. W. Nielson, R. C. Stern, and T. L. Stull. 1990. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet 336:527-532. [DOI] [PubMed] [Google Scholar]
- 19.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by random amplified polymorphic DNA (RAPD) fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenney, D., K. E. Brown, and D. G. Allison. 1995. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J. Bacteriol. 177:6989-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, M. J. K., S. O. Montgomery, E. J. O'Neill, and P. H. Pritchard. 1986. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl. Environ. Microbiol. 52:383-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nickel, J. C., I. Ruseska, J. B. Wright, and J. W. Costerton. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary tract catheter. Antimicrob. Agents Chemother. 27:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 28.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 29.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for the expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seed, P. C., L. Passador, and B. H. Iglewski. 1995. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 33.Sokal, R. R., and F. J. Rohlf. 1987. Introduction to biostatistics, 2nd ed. W. H. Freeman and Company, New York, N.Y.
- 34.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao, F., H. Zhou, and T. G. Lessie. 2002. Characterization of N-acyl homoserine lactone overproducing mutants of Burkholderia multivorans ATCC 17616. FEMS Microbiol. Lett. 206:201-207. [DOI] [PubMed] [Google Scholar]