Abstract
ClpP and ClpC are subunits of the Clp ATP-dependent protease, which is ubiquitous among prokaryotic and eukaryotic organisms. The role of these proteins in stress tolerance, stationary-phase adaptive responses, and virulence in many bacterial species has been demonstrated. Based on the amino acid sequences of the Bacillus subtilis clpC and clpP genes, we identified one clpC gene and two clpP genes (designated clpP1 and clpP2) in Bacillus thuringiensis. Predicted proteins ClpP1 and ClpP2 have approximately 88 and 67% amino acid sequence identity with ClpP of B. subtilis, respectively. Inactivation of clpC in B. thuringiensis impaired sporulation efficiency. The clpP1 and clpP2 mutants were both slightly susceptible to salt stress, whereas disruption of clpP2 negatively affected sporulation and abolished motility. Virulence of the clp mutants was assessed by injecting bacteria into the hemocoel of Bombyx mori larvae. The clpP1 mutant displayed attenuated virulence, which appeared to be related to its inability to grow at low temperature (25°C), suggesting an essential role for ClpP1 in tolerance of low temperature. Microscopic examination of clpP1 mutant cells grown at 25°C showed altered bacterial division, with cells remaining attached after septum formation. Analysis of lacZ transcriptional fusions showed that clpP1 was expressed at 25 and 37°C during the entire growth cycle. In contrast, clpP2 was expressed at 37°C but not at 25°C, suggesting that ClpP2 cannot compensate for the absence of ClpP1 in the clpP1 mutant cells at low temperature. Our study demonstrates that ClpP1 and ClpP2 control distinct cellular regulatory pathways in B. thuringiensis.
Living organisms have evolved complex regulatory networks to maintain cell viability under adverse environmental conditions (nutrient starvation; changes in temperature, humidity, or osmolarity; oxidative stress, host defense systems; etc.). An essential element of these networks is the induction of targeted intracellular proteolysis performed by energy-dependent proteases, such as the Clp ATP-dependent proteases (18, 49).
Proteins of the Clp ATPase family are highly conserved and ubiquitous in prokaryotes and higher organisms and are classified into two groups; proteins of the first group (ClpA, ClpB, ClpC, and ClpD), also known as the Hsp100 family, contain two distinct ATP-binding domains, while those of the second group (ClpM, ClpN, ClpX, and ClpY) contain smaller-size proteins with only one such domain (58). In Escherichia coli, ClpA or ClpX ATPase subunits associate with the ClpP proteolytic subunit to form the Clp ATP-dependent protease (21). This complex is involved in the degradation of several substrates such as endogenous regulator of programmed cell death MazE (3), stationary-phase transcriptional factor RpoS (61), and SsrA-tagged proteins (20). Clp ATPases can also function as molecular chaperones, preventing polypeptides from misfolding and aggregating (19, 21, 67, 69).
In E. coli, the majority of the clp genes are heat inducible, and their regulation depends mainly on the σ32 sigma factor (70). In Bacillus subtilis, clpP and clpC genes belong to the class III group of heat shock genes, whose expression is negatively regulated by the CtsR repressor, encoded by the first gene of the clpC operon (10, 29).
ClpP and ClpC of B. subtilis have been shown to play essential roles in growth at high temperature and stress tolerance (salt, ethanol, puromycin), presumably due to their role in the degradation of misfolded proteins generated by these types of stress (16, 30, 43, 45). Similar observations were reported for Lactococcus lactis and Listeria monocytogenes (14, 15, 55, 56).
Besides stress tolerance, ClpP and ClpC have been linked to many physiological, morphological, and developmental processes in bacteria. For instance, in B. subtilis they are required for motility, division, degradative enzyme synthesis, sporulation, and competence development (42). ClpP is required for the mycelium formation of filamentous soil bacterium Streptomyces lividans (9) as well as for the initiation of biofilm formation of surface-attached microorganism Pseudomonas fluorescens (48).
Clp proteins, including ClpX of Staphylococcus aureus (41), ClpE and ClpC ATPases of Streptococcus pneumoniae (31, 51), and ClpP, ClpC, and ClpE of Listeria monocytogenes (15, 47, 55), have been shown to play a major role in the virulence of several pathogens.
Bacillus thuringiensis, a gram-positive spore-forming bacterium well known for its entomopathogenic properties, is a member of the Bacillus cereus group, which also includes the closely related species B. cereus sensu stricto, Bacillus anthracis, and Bacillus mycoïdes (8, 25). The insecticidal activity of B. thuringiensis resides mainly in the proteinaceous crystal of δ-endotoxins produced during the stationary phase, which is lethal to susceptible insects upon ingestion (59). Many studies have shown that B. thuringiensis and B. cereus spores have a synergistic effect when fed in association with δ-endotoxins to susceptible larvae (26, 37, 57). In addition, it was shown that B. thuringiensis and B. cereus cells are highly pathogenic when injected into the hemocoel of the insect host (23, 24, 57, 62, 71). Several putative virulence factors, including phospholipases C, enterotoxins, hemolysins, immune inhibitors, and flagella, have been identified in B. thuringiensis and B. cereus, and there has been speculation that they facilitate the development of these bacteria within the host (22, 60, 71). However, none of these factors on its own has been confirmed to be essential for the pathogenic mechanisms leading to systemic septicemia.
To identify genes responsible for the virulence of B. thuringiensis in insects, we focused our interest on the clpP and clpC genes of this bacterium, since as mentioned above their role in the virulence of many pathogens has been established. Here we report that, in B. thuringiensis, there are two copies of the clpP gene, named clpP1 and clpP2. Mutants with clpC and both clpP genes inactivated were constructed and assayed for their phenotypical characteristics with respect to stress tolerance and virulence against Bombyx mori larvae. Results indicate that ClpP1 and ClpP2 control distinct cellular regulatory pathways.
MATERIALS AND METHODS
Bacterial strains, transformation, and culture conditions.
Sporogenous acrystalliferous B. thuringiensis strain 407 Cry−, belonging to serotype 1 (36), was used throughout this study. E. coli K-12 strain TG1 [Δ(lac-proAB) supE thi hsdΔ5 (F′ traD36 proA+ proB+ lacIq lacZΔM15)] (17) and E. coli K-12 strain TG1 RepA+ (33) were used as hosts for plasmid construction. E. coli strains SCS 110 [rpsL (Strr) thr leu endA thi-1 lacY galK galT ara tonA tsx dam dcm supE44 Δ(lac-proAB) (F′ traD36 proA+ proB+ lacIq lacZΔM15)] (Stratagene, La Jolla, Calif.) and ET12567 (F− dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara14 pacY1 xyl-5 leuB6 thi-1) were used to generate unmethylated plasmid DNA prior to transformation of B. thuringiensis. Conventional CaCl2 (7) or electroporation procedures (11) were used for E. coli transformation. B. thuringiensis strain 407 Cry− was transformed by electroporation as previously described (36).
E. coli and B. thuringiensis cells were routinely grown in Luria broth (LB) medium under vigorous agitation at 37 and 30°C, respectively. Antibiotic concentrations used for bacterial selection were as follows: ampicillin, 100 μg ml−1 for E. coli; spectinomycin, 100 μg ml−1 for E. coli and 300 μg ml−1 for B. thuringiensis; erythromycin, 10 μg ml−1 for B. thuringiensis.
For the phenotypical studies, different stress conditions were established as follows. Frozen glycerol stocks (exponentially growing cells; optical density at 600 nm [OD600] = 1) of the different strains were diluted 100-fold into LB medium and grown under vigorous shaking at 37°C. At an OD600 of 0.1, the culture was divided and one half was grown at 37°C whereas the other half was either shifted from 37 to 43°C or exposed to a final concentration of 6% (wt/vol) sodium chloride. Motility assays were performed on LB soft agar swarm plates (0.3% agar final concentration) followed by 16 h of incubation at 37°C. Columbia medium agar plates (Biomérieux) containing 5% sheep blood were used to evaluate the hemolytic activity of B. thuringiensis strains. Sporulation assays were performed as follows. Frozen glycerol stocks of exponentially growing cells were diluted 100-fold into sporulation-specific (HCT) medium (32) and grown for 3 days at 39°C. Serial dilutions of sporulating cells were plated at 24-h intervals before and after heat treatment (80°C; 12 min). Sporulation frequencies were established on the basis of viable-cell and spore counts.
DNA manipulations.
Plasmid DNA was extracted from E. coli by standard alkaline lysis with QIAprep spin columns (Qiagen). Chromosomal DNA was extracted from B. thuringiensis cells harvested in mid-log phase as described previously (44). Restriction enzymes and T4 DNA ligase were used as recommended by the manufacturer (New England Biolabs). Oligonucleotide primers (Table 1) were synthesized by Genset (Paris, France). PCRs were performed in a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer). Amplified DNA fragments were purified with the QIAquick PCR purification kit (Qiagen) and separated on 0.7% agarose gels after digestion. Digested DNA fragments were eluted from agarose electrophoresis gels with a centrifugal filter device (Ultrafree-DA; Amicon Laboratories).
TABLE 1.
Primer sequences used in this study
| Name | Nucleotide sequencea (5′-3′) | Restriction site |
|---|---|---|
| SH1 | GAAGAATTCGTTTGGCTCGTAATTTTAAAAAATATCAATTC | EcoRI |
| SH2 | GGAGGATCCGAAATCGCCTCCTACTTGCGCTTAGTTTTTTTC | BamHI |
| SH3 | GAAGAATTCGTATAGACAAACTAAGAGGGCTACGAGATAGCC | EcoRI |
| SH4 | AAGAAGCTTTCTTGTTCCTTCCATTGAAGCAACCACTGTTG | HindIII |
| Sp1 | CGCGGATCCGGATCTTCACCTAGATCC | BamHI |
| Sp2 | CCGGAATTCGTTACAAATTGTTTCACTAAATTAAAG | EcoRI |
| Pi2 | CCCAAGCTTGCTGTCATGAAGTTGTCGCGG | HindIII |
| Pi3 | CGCGGATCCGGTATGGCTGCATCTATGGGTGC | BamHI |
| Pi4 | CCCAAGCTTGGTTGACCTGTGCGGTCAGC | HindIII |
| P2.i3 | CGCGGATCCAATAGCCCCGGCGGTTCAACG | BamHI |
| P2.i4 | CCCAAGCTTGCCCTTGTGCACCACCAAGC | HindIII |
| PclpP1 | AAACTGCAGCCCTAGAAACAAAGTCGAAATC | PstI |
| PclpP2 | CGCGGATCCCGTAAGCGCGTTCTCCACG | BamHI |
| pP2.1 | AAACTGCAGCACATCAAAAGTTCCCGCACC | PstI |
| pP2.5 | CGCGGATCCGAGATCTTCCCCATCCCATTTG | BamHI |
| P8 | GCTGTCTTAGAACCGAGCGGAG | |
| P9 | CGTTTAGTGAATTGCGAAACAGC | |
| clpP2.1 | CGCTCGTATATAAAATAGCTAG | |
| clpP2.2 | CGGTTGCCCTGTTTTTTCTGC | |
| clpP2.3 | CCAGACGTGCAAACGCTATGC | |
| clpP2.6 | CTTGTCCAATCTTCTGAAGCTGC |
Restriction sites are underlined.
Plasmid and mutant strain constructions.
Plasmids pGhost9 (38) and pRN5101, a pE194 derivative (65), conferring resistance to erythromycin in gram-positive hosts and to ampicillin in E. coli, were used for gene deletion and gene disruptions, respectively. These vectors contain a thermosensitive origin of replication that prevents them from replicating in gram-positive hosts at nonpermissive temperatures (≥37°C).
Deletion of the clpC gene was carried out as follows. A 996-bp EcoRI/BamHI DNA fragment and a 1,010-bp EcoRI/HindIII DNA fragment, corresponding to the DNA chromosomal regions located immediately upstream and downstream from the clpC gene, respectively, were generated by PCR using B. thuringiensis strain 407 Cry− chromosomal DNA as a template and oligonucleotide pairs SH1-SH2 and SH3-SH4, respectively (Table 1). The primers were designed from the sequence of the clpC operon identified in the B. anthracis incomplete genome sequence. A 1,134-bp BamHI/EcoRI DNA fragment carrying the S. aureus spectinomycin resistance gene spc (46) was amplified with primer pair Sp1-Sp2 (Table 1) and B. subtilis strain QB4756 chromosomal DNA (45) as a template. The amplified DNA fragments were digested by the appropriate restriction enzymes and cloned between the internal HindIII and EcoRI sites of vector pGhost9 to give plasmid pGhost9ΔclpC::spc, which requires E. coli strain TG1 RepA+ to replicate (33). The resulting plasmid was introduced into B. thuringiensis strain 407 Cry− by electroporation, and the deletion of the clpC gene was obtained by a double-crossover event as described previously (34). The complete deletion of the gene was verified by PCR using additional oligonucleotides located further upstream and downstream from the original fragments. The corresponding mutant strain was designated 407 Cry− ΔclpC.
The chromosomal B. thuringiensis clpP1 and clpP2 genes were disrupted by insertion, through homologous recombination, of derivatives of plasmid pRN5101. Constructions were performed as follows. DNA fragments corresponding to internal regions of the clpP1 and clpP2 genes (203 and 205 bp, respectively) were generated by PCR using primer pairs Pi3-Pi4 and P2.i3-P2.i4 (Table 1) and cloned between the BamHI/HindIII sites of pRN5101. The resulting plasmids were introduced into B. thuringiensis strain 407 Cry− by electroporation. Transformants were grown at 30°C and then transferred to a nonpermissive temperature (39°C) and finally plated onto LB agar plates supplemented with erythromycin (10 μg ml−1) at 39°C. Integration of the recombinant plasmid was confirmed by PCR and Southern hybridization. The corresponding insertional mutant strains were designated 407 Cry− ΔclpP1 and 407 Cry− ΔclpP2 and were found to be highly stable during growth at the nonpermissive temperature.
Construction of clpP1′-lacZ and clpP2′-lacZ transcriptional fusions.
A clpP1′-lacZ transcriptional fusion was constructed by cloning a 395-bp BamHI/PstI DNA fragment, corresponding to the chromosomal DNA region upstream from the clpP1 gene, between the BamHI and PstI sites of plasmid pHT304-18′Z (2) to give plasmid pHT304ΩclpP1′Z. This DNA fragment was generated by PCR using synthetic oligonucleotides PclpP1 and PclpP2 (Table 1).
A clpP2′-lacZ transcriptional fusion was constructed by amplifying a 326-bp BamHI/PstI DNA fragment by PCR using synthetic primers pP2.1 and pP2.5 (Table 1), designed on the available B. cereus nucleotide sequence. This fragment contains 227 bp upstream from the extracytoplasmic function (ECF) RNA polymerase σ factor gene as well as the first 100 bp of the coding sequence. The fragment was cloned between the BamHI and PstI restriction sites of the pHT304-18′Z vector to give plasmid pHT304ΩclpP2′Z.
The recombinant plasmids were introduced into strain 407 Cry− by electroporation to give strains 407 Cry− [pHT304ΩclpP1′Z] and 407 Cry− [pHT304ΩclpP2′Z].
β-Galactosidase activity.
Colonies expressing lacZ fusions were detected on media containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg ml−1) and erythromycin (10 μg ml−1). Cells were grown in LB medium devoid of antibiotics at 25 and 37°C with vigorous shaking and assayed for β-galactosidase-specific activities as described previously (44).
PCR amplification and sequencing of clpP1 and clpP2 genes.
Four pairs of synthetic oligonucleotides (p1 [P8-Pi2], p2 [P9-Pi3], p3 [clpP2.1-clpP2.2], and p4 [clpP2.3-clpP2.6]) (Table 1) were designed from the sequence of B. anthracis genomic regions encompassing the clpP1 and clpP2 genes. The p1, p2, p3, and p4 oligonucleotide pairs were used to amplify four overlapping fragments, corresponding to the clpP1 and clpP2 regions, from positions −640 to +900 and −470 to +392 with respect to the ATG start codon and the TAA terminal codons of clpP1 and clpP2, respectively. PCRs were carried out in a reaction volume of 100 μl containing 200 μM deoxynucleoside triphosphates, 2.5 mM MgSO4, 50 pmol of each primer, 0.5 μg of B. thuringiensis strain 407 Cry− chromosomal DNA, and 0.5 U of Pwo DNA polymerase (Roche Boehringer). Purified PCR products were sequenced on both strands by Genome Express (Paris, France)
Database comparisons and sequence analysis.
Alignment and sequence comparisons with the GenBank database were performed by using the National Center for Biotechnology Information BLAST2 (4) network server with the default parameter values provided. B. subtilis sequences were from Subtilist (http://genolist.pasteur.fr/SubtiList/), and B. cereus sequences were from http://ergo.integratedgenomics.com/B_cereus.html. Unfinished B. anthracis genome sequences were kindly provided by The Institute for Genomic Research website (http://www.tigr.org).
Insects and in vivo experimental infections.
Eggs of B. mori strain nistari provided by the Institut National de la Recherche Agronomique (Unité Nationale Séricicole, Lyon, France) were incubated at 25°C. The resulting larvae were reared on a commercially available artificial diet (Fukui and Co., Ltd., Yokohama, Japan).
Pathogenicity assays of B. thuringiensis vegetative cells were carried out as follows. Cells of wild-type and mutant strains of B. thuringiensis were grown in LB medium devoid of antibiotics at 30°C and with shaking. Bacterial concentrations were monitored by optical density measurement at 600 nm and verified by plating dilutions onto LB agar plates. Different dilutions of exponentially growing B. thuringiensis cells were inoculated into groups of 30 B. mori larvae (10 μl larva−1). The control group was injected with sterile water. B. mori larvae were on the first day of the fourth instar and weighed about 150 to 200 mg. B. thuringiensis cell suspensions were injected through the intersegmental membrane between the fourth and the fifth abdominal legs of the larva with a 1-ml Terumo syringe and a microapplicator (Buckard type LV 65). Inoculated larvae were incubated individually in plastic containers at 25°C. Mortality was recorded daily over a 3-day period.
In vitro experimental infections.
In vitro experimental infections were performed in insect hemolymph pooled from fifth-instar B. mori larvae. The surfaces of the larvae were cleaned with 70% ethanol and dried. Cell-free hemolymph was collected by capillary action in a sterile, ice-cooled Eppendorf tube by cutting off the abdominal legs of the larvae. Cell-free hemolymph was prepared by centrifugation at 16,000 × g for 2 min. Twenty microliters of a dilution of a B. thuringiensis culture were added to 200 μl of hemocoel, and the mixture was incubated at 28 or 37°C for 3 h. During this period, 20-μl aliquots were withdrawn at 30-min intervals and spread on LB plates for viable-cell counts. Experiments were repeated three times for each test.
Statistical analysis.
Mortality data were analyzed by calculating 50% lethal doses (LD50s) with the Log-Probit program (12; M. Raymond, G. Prato, and D. Ratsira, PROBIT analysis of mortality assays displaying quantal response, license no. L93019-Avenix [Praxeme, Montpellier, France], 34680 St. George d'Orques, France, 1993).
Nucleotide sequence accession numbers.
The nucleotide sequences of clpP1 and clpP2 from B. thuringiensis reported in this paper have been submitted to the GenBank database under accession no. AF454757 and AF454758, respectively.
RESULTS
Nucleotide and amino acid sequence analysis of B. thuringiensis clpP genes.
The B. subtilis ClpP protein sequence (43) was used to screen the incomplete B. anthracis genome sequence. This revealed the presence of two highly similar gene products, having approximately 88 and 66% identity with ClpP of B. subtilis. Primer pairs were designed from the B. anthracis sequence and used to amplify the corresponding DNA regions from B. thuringiensis strain 407 Cry− (see Materials and Methods). The nucleotide sequences of the PCR products were determined on both strands, and the B. thuringiensis DNA regions encompassing both open reading frames (ORFs) were entirely reconstituted. Analysis of the nucleotide sequence of the first DNA fragment (2,121 bp) revealed an ORF starting with an ATG codon and predicted to encode a protein of 194 amino acid residues having 88% identity with ClpP of B. subtilis (Fig. 1). The ATG codon is preceded by a typical ribosome binding site sequence (GGAGG) at an appropriate distance, and a potential promoter sequence with significant similarity to σA-type −35 and −10 promoter recognition sequences (TTGACCN17TATTAT) was found 35 bp upstream from the start codon. A CtsR consensus sequence (A/GGTCAAANANA/GGTCAAA), reported in the promoter regions of several clp genes of various bacterial species (10), in the reverse orientation (5′-GGTCAATAAAGGTCAAA-3′) was identified 66 bp upstream from the translational start site, overlapping the potential −35 sequence. A likely rho-independent transcription terminator stem-loop sequence (GAAGCGCCCTATAATTGGGCGCTTC, ΔG = −103.7 kJ mol−1) followed by a poly(T) stretch is located 135 bp after the TAA stop codon. This ORF appeared to be organized as a single transcriptional unit and was designated clpP1.
FIG. 1.
Alignment of the B. thuringiensis (Bth) ClpP1 and ClpP2 amino acid sequences with that of B. subtilis ClpP. The B. subtilis (Bsu) ClpP sequence is as described by Msadek and colleagues (43). Numbers indicate positions in the amino acid sequence. Identical residues are shaded. The conserved catalytic Ser-98, His-123, and Asp-172 residues are underlined.
The second DNA fragment contained two ORFs that appear to be organized as an operon. The second ORF, beginning with an ATG codon, encodes a 193-amino-acid protein displaying 67% identity with ClpP of B. subtilis (Fig. 1). This ORF is therefore referred to as clpP2. The fact that the stop codon of the upstream ORF and the ATG codon of the clpP2 ORF are separated by only 21 bases and the absence of a potential stem-and-loop structure in the intergenic region suggest that the two ORFs are organized as an operon. The clpP1 and clpP2 genes are organized similarly in the B. cereus strain ATCC 14579 and B. anthracis genomes. The deduced amino acid sequence encoded by the first ORF of the B. cereus clpP2 operon displayed similarities with an RNA polymerase ECF-type sigma factor from Bacillus halodurans (63). No CtsR box could be identified in the upstream region of the clpP2 operon in B. cereus. The high degree of identity over the entire length of the aligned sequences (B. thuringiensis clpP1, B. thuringiensis clpP2, and B. subtilis clpP; Fig. 1) indicates that the B. thuringiensis ClpP proteins are members of the ClpP family of proteolytic subunits (40). The serine-98 residue, the histidine-123 residue, and the aspartate-172 residue, which constitute the catalytic triad of the serine protease (39, 40, 66), are conserved (Fig. 1).
Since a CtsR consensus sequence was found upstream of clpP1 ORF, we searched for a ctsR-like gene in B. cereus and B. anthracis genomes. This led to the identification, in the genomes of both species, of an ortholog of the ctsR gene organized as in B. subtilis, with ctsR and clpC as the first and last genes of a four-gene operon.
Phenotypical features of the mutant strains.
To address the question of whether the ClpP1, ClpP2, and ClpC proteins are required for stress tolerance, mutations inactivating the corresponding genes were generated and the mutants were assessed for altered phenotypes with respect to high temperature (43°C) and salt concentration (6% NaCl) (see Materials and Methods).
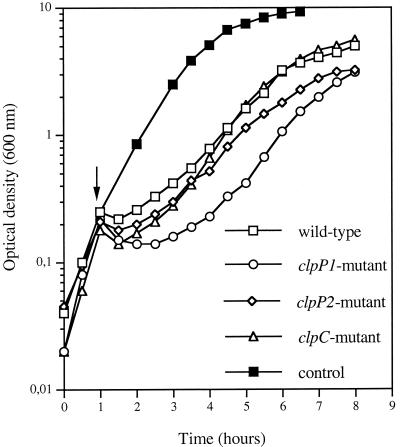
Regarding stress tolerance, all mutants displayed growth rates when grown at 37°C that were the same as those when they were shifted from 37 to 43°C, the upper limit for growth of B. thuringiensis 407 Cry− (data not shown). However, diminished growth rates of the ΔclpP1 and ΔclpP2 strains were observed in the presence of 6% NaCl, with a larger lag phase for the ΔclpP1 mutant strain (Fig. 2). These results indicate a possible role for the B. thuringiensis clpP2 gene in salt tolerance.
FIG. 2.
Growth curves of B. thuringiensis strains in LB at 37°C after the addition of 6% (wt/vol) NaCl. Arrow, OD at which the cultures were exposed to the salt stress. The control indicates the growth curve of the wild type at 37°C in LB devoid of NaCl.
As shown in Fig. 3, the ΔclpP2 cells were severely impaired in motility whereas all the other mutants were as motile as the parental strain. Further confirmation of the nonmotile phenotype of ΔclpP2 cells was obtained by microscopic examination, indicating that ClpP2 is required for motility. Sporulation assays were assessed by establishing sporulation frequencies after 24 h of incubation in HCT medium. As shown in Table 2, the sporulation frequencies of the ΔclpC and the ΔclpP2 mutants were diminished approximately 103- and 105-fold, respectively, whereas that of the ΔclpP1 mutant was lowered only 1.4-fold. These results suggest that ClpP2 and ClpC play an important role in the sporulation pathway whereas ClpP1 does not.
FIG. 3.
The clpP2 gene is required for motility. The B. thuringiensis (Bt) strains were incubated on LB soft agar as described in Materials and Methods.
TABLE 2.
Effects of clpP and clpC mutations on sporulation
| B. thuringiensis strain | Sporulation frequency (CFU ml−1) after 24 h incubation in HCT, plating:
|
% Sporulation | |
|---|---|---|---|
| Before heat treatment | After heat treatment | ||
| 407 Cry− | 2.88 × 108 | 2.5 × 108 | 86 |
| 407 Cry− ΔclpC | 1.72 × 108 | 1.4 × 105 | 0.072 |
| 407 Cry− ΔclpP1 | 2.84 × 108 | 1.71 × 108 | 60.2 |
| 407 Cry− ΔclpP2 | 4 × 107 | 300 | 0.00075 |
B. thuringiensis genes encoding extracellular virulence factors (including phospholipases C PlcA and PlcB, enterotoxins Hbl and Nhe, hemolysins and proteases) are positively regulated by the PlcR pleiotropic regulator (1, 35). Inactivation of the plcR gene prevents the hemolytic activity of B. thuringiensis and B. cereus cells (57). All the mutant strains (ΔclpP1, ΔclpP2, and ΔclpC strains) were fully hemolytic on sheep erythrocytes (data not shown), suggesting that the PlcR regulon is not affected in the clp mutant backgrounds.
Virulence of B. thuringiensis clp mutants in insects.
The virulence of the different B. thuringiensis clp mutants was assessed by injecting exponentially growing cells into the hemocoel of fourth-instar B. mori larvae. This lepidopteran species is a useful model because the larvae are highly susceptible to the parental-strain cells (LD50 < 5 vegetative cells/larva; Table 3). As shown in Table 3, LD50s of the ΔclpC and ΔclpP2 mutants were not significantly different from that of the 407 Cry− strain, suggesting that the ClpP2 and ClpC proteins are not required for expression of virulence. In contrast, the virulence of the clpP1-deficient mutant was severely attenuated since no dead larvae were obtained from the insects infected with 100 cells of the mutant strain.
TABLE 3.
Effects of clpP and clpC mutations on the virulence of B. thuringiensis in B. mori larvae
| Strain | LD50a (CFU/injected larva) |
|---|---|
| 407 Cry− | 4.79 (3.28-6.12) |
| 407 Cry− ΔclpC | 5.27 (3.57-6.95) |
| 407 Cry− ΔclpP1 | >100b |
| 407 Cry− ΔclpP2 | 7.094 (3.56-9.91) |
LD50s were calculated 1 day postinfection by Probit analysis (Raymond et al., license no. L93019-Avenix). Values in parentheses are 95% confidence intervals (12).
Injection of 100 cells of a clpP-deficient mutant causes 0% mortality.
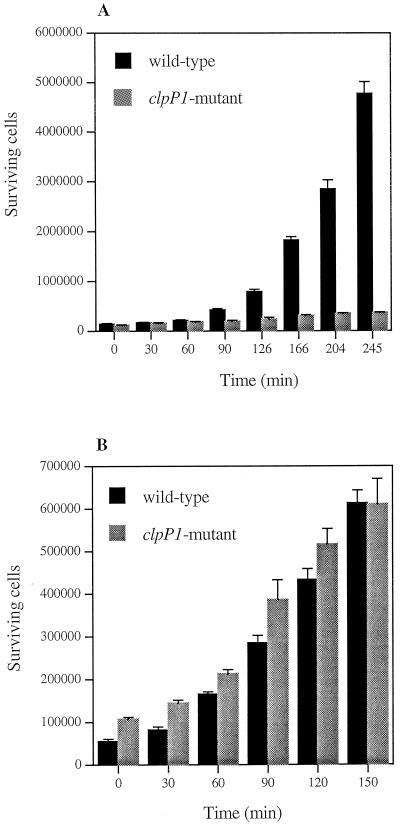
To determine whether the avirulent phenotype of the ΔclpP1 strain was due to the loss of specific pathogenic properties or to the inability of the mutant to survive within the host, we tested bacterial survival in vitro over a 3-h period of incubation in B. mori hemolymph (see Materials and Methods). The test was performed at 28°C without shaking to mimic the in vivo temperature and partially anaerobic conditions. The ΔclpP1 mutant was unable to grow in the hemolymph in contrast to the wild type (Fig. 4A). These results suggest that the reduced virulence of the clpP1-deficient mutant was due to the restricted in vivo development of the strain.
FIG. 4.
Surviving B. thuringiensis wild-type and ΔclpP1 cells after incubation with hemolymph from B. mori larvae. Two hundred microliters of hemolymph and 20 μl of a dilution containing about 105 B. thuringiensis viable cells were mixed. The reaction mixture was incubated without shaking at 28 (A) and 37°C (B). Aliquots were withdrawn at the time points indicated, and the surviving bacteria were counted. Experiments were repeated three times. Error bars, standard errors of the means.
ClpP1 is required for growth at low temperature.
To investigate the altered growth of the ΔclpP1 strain, we first assumed that the partially anaerobic conditions encountered in the insect hemolymph could prevent the growth of the mutant strain. However, both wild-type and mutant strains grew similarly at 37°C in anaerobiosis either in LB (data not shown) or in hemolymph in vitro (Fig. 4B), suggesting that anaerobiosis is not the reason that the ΔclpP1 mutant is unable to grow in vivo.
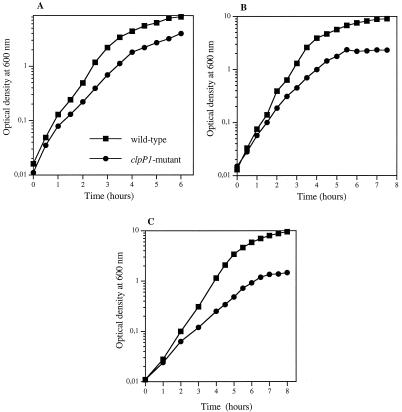
At 30°C, the ΔclpP1 mutant gave small-size colonies, suggesting that this strain presented a sensitivity to low growth temperatures. To test this hypothesis, we compared the growth rates of the ΔclpP1 mutant and the wild-type strains in LB medium at 30, 28, and 25°C, the last temperature being that routinely used for in vivo infection tests. As shown in Fig. 5, the ability of ΔclpP1 mutant cells to grow decreased at lower growth temperatures. At 25°C, the cell concentration did not exceed that corresponding to an OD600 of 1.5 while the OD600 for the wild type reached a value of 10 (Fig. 5C). Furthermore, microscopic examination revealed a filamentous phenotype for the mutant cells during growth at 25°C, which was not observed for the wild-type cells. This suggests that ClpP1 is essentially required for cell separation at low temperature.
FIG. 5.
Growth curves of B. thuringiensis wild-type and clpP1 mutant strains in LB medium at 30 (A), 28 (B), and 25°C (C).
To determine whether low temperature is the only factor that might affect in vivo septicemia due to the ΔclpP1 strain, we assessed the virulence of the mutant strain at 37°C. A dose of 20 cells of the strain was inoculated into groups of 30 B. mori larvae. Nonentomopathogenic bacterium B. subtilis 168 was used as a negative control. By 24 h of incubation of infected insects at 37°C, we recorded the same mortality values for both wild-type and ΔclpP1 mutant strains (90 to 95% mortality). The B. subtilis control used did not induce any lethality even at a dose of 100 cells, indicating that the mortality was not due to the relatively high temperature of incubation. This suggests that the loss of virulence of the ΔclpP1 strain resulted from its reduced growth at low temperatures such as those present in the insect host.
Analysis of clpP1 and clpP2 expression in B. thuringiensis during growth at low temperatures.
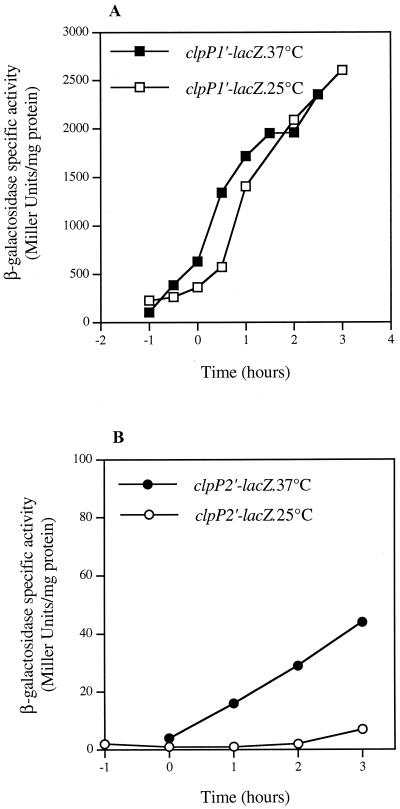
The expression of the clpP genes at different temperatures was analyzed by using transcriptional fusions with the lacZ gene. Plasmid pHT304ΩclpP1′Z was introduced into B. thuringiensis strain 407 Cry−, and the β-galactosidase activity of the recombinant strain was monitored during growth in LB medium at 37 and 25°C (Fig. 6A). clpP1′-lacZ was expressed during the exponential growth phase: about 200 to 300 U of β-galactosidase mg−1 was produced before the onset of stationary phase (time zero) at each temperature. As reported for the B. subtilis clpP gene (43), induction of clpP1 expression is observed upon entry into stationary phase. In cells grown at 25 and 37°C, the transcription patterns were similar. The significant level of clpP1 expression during the exponential growth phase at low temperature is in agreement with its requirement for low-temperature growth of B. thuringiensis.
FIG. 6.
Expression of B. thuringiensis clpP genes in strain 407 Cry−. (A) Expression of clpP1′-lacZ at 37 and 25°C. (B) Expression of clpP2′-lacZ at 37 and 25°C. Cells were grown in LB medium. Time zero indicates the onset of the stationary phase.
A transcriptional fusion between a DNA fragment carrying the 370 bp directly upstream from clpP2 and lacZ was not significantly expressed in B. thuringiensis (data not shown), in agreement with the proposed operon structure of the gene. We therefore analyzed clpP2 expression by fusing the region upstream from the first gene of the operon to the lacZ reporter gene (plasmid pHT304ΩclpP2′Z) (Fig. 6B). A relatively weak clpP2 operon-directed β-galactosidase activity was noted at 37°C, and no β-galactosidase production (<10 U mg−1) was detected during growth at 25°C.
DISCUSSION
This is the first study reporting an analysis of clp genes in the entomopathogenic bacterium B. thuringiensis and their involvement in stress tolerance, stationary-phase responses, and virulence.
We have shown that, in contrast to B. subtilis, B. thuringiensis contains two copies of the clpP gene (clpP1 and clpP2). The clpP1 gene appears to belong to a monocistronic transcription unit, whereas clpP2 is organized as an operon with a gene encoding an ECF RNA polymerase sigma factor homologue. The B. thuringiensis ClpP1 and ClpP2 proteins were typical of the highly conserved family of ClpP proteolytic subunits (40). Most bacteria (e.g., E. coli, B. subtilis, and Yersinia enterocolitica) possess a single clpP gene; however, some have recently been shown to contain more than one copy. For instance, Mycobacterium tuberculosis has two ClpP proteins (28) and cyanobacteria Synechocystis sp. strain PCC6803 (27) and Synechococcus sp. strain PCC7942 (6) possess three distinct ClpP isoenzymes. S. lividans has been recently reported to possess a clpP multigenic family with four clpP representatives (9, 64). Since B. thuringiensis and B. subtilis are closely related species, it was unexpected to find two clpP copies in B. thuringiensis.
In B. subtilis, transcriptional repressor CtsR negatively regulates the expression of class III heat shock genes (clpP, clpE, and clpC operon) by binding to a directly repeated heptanucleotide operator sequence (A/GGTCAAA NAN A/GGTCAAA) which has been identified upstream of many bacterial clp genes (10). A gene encoding a CtsR homologue exists in the clpC operon in the genomes of B. cereus (http://ergo.integratedgenomics.com/B_cereus.html) and B. anthracis (http://www.tigr.org), and a CtsR consensus sequence was found in the promoter region of clpP1 in B. thuringiensis but not in that of the clpP2 operon. This suggests that B. thuringiensis clpP1 is negatively controlled by CtsR.
The role of ClpP and ClpC in the stress survival of several gram-positive bacteria, including B. subtilis, L. monocytogenes, L. lactis, Synechococcus, and S. pneumoniae, has previously been reported (5, 54). In B. subtilis and L. monocytogenes, ClpP and ClpC were shown to be required for growth at high temperature and stress tolerance (exposure to salt, ethanol, or puromycin) (15, 16, 30, 56). Furthermore, B. subtilis Clp proteins were found to play central roles in many cellular developmental processes such as cell division, motility, degradative enzyme synthesis, sporulation, and genetic competence (43, 45). To study the involvement of ClpP1, ClpP2, and ClpC proteins of B. thuringiensis in stress tolerance and stationary-phase adaptive responses, we constructed mutants by inactivating the corresponding genes. Our results showed that, in contrast to what was found for B. subtilis, inactivation of clpC did not lead to pleiotropic effects. The clpC-deficient mutant displayed only a sporulation defect. Moreover, none of the mutant strains was sensitive to the different forms of stress imposed (high temperature and elevated salt concentrations) except for a slight sensitivity to high osmolarity for the clpP1 mutant. The ΔclpP2 mutant was the most affected since it was both nonmotile and deficient in sporulation. These results pointed out the involvement of B. thuringiensis ClpP1 in salt tolerance, of ClpP2 in both sporulation and motility, and of ClpC in sporulation. Our results also emphasize the functional diversity of the ClpP and ClpC proteins among prokaryotic organisms since the phenotypical features of the mutants vary strongly among bacteria.
Clp proteases have been shown to play a major role in the virulence of many bacterial pathogens including L. monocytogenes, Y. enterocolitica, and Salmonella enterica serovar Typhimurium (15, 50, 55, 68). We therefore assessed the pathogenicity of ΔclpC, ΔclpP1, and ΔclpP2 mutants against B. mori larvae and found that the virulence was markedly decreased for clpP1-deficient bacteria. The in vitro growth measurement data and the in vivo infection experiments performed at 37°C demonstrated that the avirulent phenotype of the clpP1 mutant strain was due to a lower growth rate at temperatures less than 30°C. The importance of ClpP1 in tolerance of low temperature might reflect a possible mechanism allowing the bacterium to adapt to the physical conditions encountered in the insect host since invertebrates are not able to regulate their internal temperature. Furthermore, the loss of ClpP1 impaired cell separation after septum formation during the exponential growth phase at 25°C, indicating an involvement of ClpP1 in normal cell division at low temperatures. The characteristics of the B. thuringiensis ΔclpP1 mutant with regard to the effect of exposure to low temperature are reminiscent of the ClpP1− and ClpB− phenotypes in Synechococcus sp. strain PCC7942. In this organism, ClpP1 and ClpB were shown to play critical roles during acclimation to low temperature (52, 53). Indeed, cyanobacterial clpP1 and clpB mutant cells were arrested during cell division at 25°C and lost viability after prolonged cold treatment. In addition, as for the B. thuringiensis ΔclpP1 mutant, the loss of ClpB in Synechococcus sp. strain PCC7942 led to altered bacterial division, with the cells remaining attached after septum formation. On the other hand, the inactivation of cyanobacterial clpP1 caused the formation of severely elongated cells, a morphological phenotype previously reported for the B. subtilis clpP mutant (6, 43).
The phenotypical changes resulting from the disruption of the B. thuringiensis clpP1 gene prompted the investigation of clpP1 expression at different growth temperatures. Analysis of clpP1′-lacZ expression showed that clpP1 is expressed at both temperatures tested (25 and 37°C). This result suggests that the ClpP1 protein is present in the B. thuringiensis cells during vegetative growth at low temperature, in agreement with its requirement for growth. An increase in ClpP1 content of Synechococcus wild-type cultures shifted from 37 to 25°C has been reported (53). It has been suggested that one likely function for the cyanobacterial ClpP1 protein would be to maintain the correct extracellular protein environment through its proteolytic activity in the cold. Indeed, as for high temperature, the abrupt shift to low temperature is considered to be a form of thermal stress that causes extensive protein denaturation and subsequent aggregation, whose magnitudes depend on the range of the temperature shift (13). B. thuringiensis ClpP1 may play the same role as that suggested for Synechococcus sp. ClpP1.
The finding that clpP2 was not expressed at 25°C suggests that the loss of ClpP1 in the ΔclpP1 mutant is not compensated by ClpP2 at this temperature. This is consistent with the presence of ClpP1 being crucial for the cell's tolerance of low temperature.
A major finding of this work is that the two related ClpP proteins of B. thuringiensis (ClpP1 and ClpP2) control distinct cellular regulatory pathways. While ClpP1 seems to be essential for normal cell division at low temperature, ClpP2 is required for motility and sporulation but is unnecessary for low-temperature growth. The apparent functional difference between ClpP1 and ClpP2 might explain the significance of the duplication of clpP genes in B. thuringiensis.
Acknowledgments
This work was supported by the Institut National de la Recherche Agronomique (AIP Microbiologie), the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires of the Ministère de la Recherche, and the Institut Pasteur. Sinda Fedhila was supported by a grant from the Tunisian government and by a grant from INRA (Département Santé des Plantes et Environnement).
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Økstad, A. B. Kolstø, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse, H., and D. Lereclus. 1994. Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J. Bacteriol. 176:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. (Erratum, 93:9991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, and W. Miller. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chastanet, A., M. Prudhomme, J.-P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, A. K., J. Schelin, and J. Porankiewicz. 1998. Inactivation of the clpP1 gene for the proteolytic subunit of the ATP-dependent Clp protease in the cyanobacterium Synechococcus limits growth and light acclimation. Plant Mol. Biol. 37:791-801. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. N., A. C. Y. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daffonchio, D., A. Cherif, and S. Borin. 2000. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group.” Appl. Environ. Microbiol. 66:5460-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Crecy-Lagard, V., P. Servant-Moisson, J. Viala, C. Grandvalet, and P. Mazodier. 1999. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32:505-517. [DOI] [PubMed] [Google Scholar]
- 10.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 11.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finney, D. J. 1971. Probit analysis. Cambridge University Press, Cambridge, United Kingdom.
- 13.Franks, F. 1995. Protein destabilization at low temperatures. Adv. Protein Chem. 46:105-139. [DOI] [PubMed] [Google Scholar]
- 14.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 15.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 16.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, T. J. 1984. Ph.D thesis. University of Cambridge, Cambridge, United Kingdom.
- 18.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 19.Gottesman, S., M. R. Maurizi, and S. Wickner. 1997. Regulatory subunits of energy-dependent proteases. Cell 91:435-438. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 22.Guttmann, D. M., and D. J. Ellar. 2000. Phenotypic and genotypic comparisons of 23 strains from the Bacillus cereus complex for a selection of known and putative B. thuringiensis virulence factors. FEMS Microbiol. Lett. 188:7-13. [DOI] [PubMed] [Google Scholar]
- 23.Heierson, A., I. Sidén, A. Kivaisi, and H. G. Boman. 1986. Bacteriophage-resistant mutants of Bacillus thuringiensis with decreased virulence in pupae of Hyalophora cecropia. J. Bacteriol. 167:18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heimpel, A. M. 1955. Investigations of the mode of action of strains of Bacillus cereus Fr. and Fr. pathogenic for the larch sawfly, Pristiphora erichsonii (Htg). Can. J. Zool. 33:311-326. [DOI] [PubMed] [Google Scholar]
- 25.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. E., and W. H. McGaughey. 1996. Contribution of Bacillus thuringiensis spores to toxicity of purified Cry proteins towards Indianmeal moth larvae. Curr. Microbiol. 33:54-59. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 28.Knipfer, N., A. Seth, S. G. Roudiak, and T. E. Shrader. 1999. Species variation in ATP-dependent protein degradation: protease profiles differ between mycobacteria and protease functions differ between Mycobacterium smegmatis and Escherichia coli. Gene 231:95-104. [DOI] [PubMed] [Google Scholar]
- 29.Kruger, E., and M. Hecker. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 180:6681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruger, E., U. Volker, and M. Hecker. 1994. Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance. J. Bacteriol. 176:3360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 32.Lecadet, M. M., M. O. Blondel, and J. Ribier. 1980. Generalized transduction in Bacillus thuringiensis var. berliner 1715, using bacteriophage CP54 Ber. J. Gen. Microbiol. 121:203-212. [DOI] [PubMed] [Google Scholar]
- 33.Leenhouts, K. J., B. Tolner, S. Bron, J. Kok, G. Venema, and J. F. Seegers. 1991. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26:55-66. [DOI] [PubMed] [Google Scholar]
- 34.Lereclus, D., H. Agaisse, M. Gominet, and J. Chaufaux. 1995. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Bio/Technology 13:67-71. [DOI] [PubMed] [Google Scholar]
- 35.Lereclus, D., H. Agaisse, M. Gominet, S. Salamitou, and V. Sanchis. 1996. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 178:2749-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lereclus, D., O. Arantes, J. Chaufaux, and M.-M. Lecadet. 1989. Transformation and expression of a cloned ∂-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 60:211-218. [DOI] [PubMed] [Google Scholar]
- 37.Li, R. S., P. Jarrett, and H. D. Burges. 1987. Importance of spores, crystals, and ∂-endotoxins in the pathogenicity of different varieties of Bacillus thuringiensis in Galleria mellonella and Pieris brassicae. J. Invertebr. Pathol. 50:277-284. [Google Scholar]
- 38.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maurizi, M. R., W. P. Clark, Y. Katayama, S. Rudikoff, J. Pumphrey, B. Bowers, and S. Gottesman. 1990. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265:12536-12545. [PubMed] [Google Scholar]
- 40.Maurizi, M. R., W. P. Clark, S. H. Kim, and S. Gottesman. 1990. ClpP represents a unique family of serine proteases. J. Biol. Chem. 265:12546-12552. [PubMed] [Google Scholar]
- 41.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 42.Msadek, T. 1999. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 7:201-207. [DOI] [PubMed] [Google Scholar]
- 43.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 44.Msadek, T., F. Kunst, D. Henner, A. Klier, G. Rapoport, and R. Dedonder. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172:824-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Msadek, T., F. Kunst, and G. Rapoport. 1994. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc. Natl. Acad. Sci. USA 91:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy, E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3") (9). Mol. Gen. Genet. 200:33-39. [DOI] [PubMed] [Google Scholar]
- 47.Nair, S., C. Frehel, L. Nguyen, V. Escuyer, and P. Berche. 1999. ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 31:185-196. [DOI] [PubMed] [Google Scholar]
- 48.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 49.Parsell, D. A., and S. Lindquist. 1993. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27:437-496. [DOI] [PubMed] [Google Scholar]
- 50.Pederson, K. J., S. Carlson, and D. E. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 51.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porankiewicz, J., and A. K. Clarke. 1997. Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 179:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porankiewicz, J., J. Schelin, and A. K. Clarke. 1998. The ATP-dependent Clp protease is essential for acclimation to UV-B and low temperature in the cyanobacterium Synechococcus. Mol. Microbiol. 29:275-283. [DOI] [PubMed] [Google Scholar]
- 54.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 55.Rouquette, C., C. de Chastellier, S. Nair, and P. Berche. 1998. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27:1235-1245. [DOI] [PubMed] [Google Scholar]
- 56.Rouquette, C., M. T. Ripio, E. Pellegrini, J. M. Bolla, R. I. Tascon, J. A. Vazquez-Boland, and P. Berche. 1996. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21:977-987. [DOI] [PubMed] [Google Scholar]
- 57.Salamitou, S., F. Ramisse, M. Brehelin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology. 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 58.Schirmer, E. C., J. R. Glover, M. A. Singer, and S. Lindquist. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289-296. [PubMed] [Google Scholar]
- 59.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schnepf, H. E., and H. R. Witheley. 1985. Protein toxins of Bacillus spp., p. 209-216. In J. Hoch and P. Setlow (ed.), Molecular biology of microbial differentiation. American Society for Microbiology, Washington, D.C.
- 61.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σS) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stephens, J. M. 1952. Disease in codling moth larvae produced by several strains of Bacillus cereus. Can. J. Zool. 30:30-40. [Google Scholar]
- 63.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viala, J., G. Rapoport, and P. Mazodier. 2000. The clpP multigenic family in Streptomyces lividans: conditional expression of the clpP3 clpP4 operon is controlled by PopR, a novel transcriptional activator. Mol. Microbiol. 38:602-612. [DOI] [PubMed] [Google Scholar]
- 65.Villafane, R., D. H. Bechhofer, C. S. Narayanan, and D. Dubnau. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, J., J. A. Hartling, and J. M. Flanagan. 1997. The structure of ClpP at 2.3 [angst]A resolution suggests a model for ATP-dependent proteolysis. Cell 91:447-456. [DOI] [PubMed] [Google Scholar]
- 67.Wawrzynow, A., B. Banecki, and M. Zylicz. 1996. The Clp ATPases define a novel class of molecular chaperones. Mol. Microbiol. 21:895-899. [DOI] [PubMed] [Google Scholar]
- 68.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112-123. [DOI] [PubMed] [Google Scholar]
- 69.Wickner, S., and M. R. Maurizi. 1999. Here's the hook: similar substrate binding sites in the chaperone domains of Clp and Lon. Proc. Natl. Acad. Sci. USA 96:8318-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yura, T., K. Nakahigashi, and M. Kanemori. 1996. Transcriptional regulation of stress-inducible genes in procaryotes. EXS 77:165-181. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, M.-Y., A. Lövgren, M. G. Low, and R. Landén. 1993. Characterization of an avirulent pleiotropic mutant of the insect pathogen Bacillus thuringiensis: reduced expression of flagellin and phospholipases. Infect. Immun. 61:4947-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]