Abstract
Secretion of cold-adapted α-amylase from Pseudoalteromonas haloplanktis TAB23 was studied in three Antarctic bacteria. We demonstrated that the enzyme is specifically secreted in the psychrophilic hosts even in the absence of a protein domain that has been previously reported to be necessary for α-amylase secretion in Escherichia coli. The occurrence of two different secretion pathways in different hosts is proposed.
The presence of an outer membrane in gram-negative bacteria imposes a barrier for the secretion of proteins into the extracellular medium. At least five different, widely disseminated secretion pathways have evolved in these bacteria for the translocation of soluble proteins across the outer membrane (2, 5, 10, 13, 14, 16).
In this study we have investigated the protein secretion in Antarctic bacteria by using the α-amylase from Pseudoalteromonas haloplanktis TAB23 as model enzyme. Indeed, among the secreted psychrophilic proteins studied so far (11), the cold-adapted α-amylase is one of the best characterized for both function and structure, since it is one of the very few cold-adapted enzymes whose three-dimensional structure has been solved (1). The psychrophilic α-amylase is synthesized as a preproenzyme, composed of the signal peptide (24 residues), the mature enzyme (453 amino acids, 49 kDa), and a long C-terminal propeptide (192 residues, 21 kDa) which constitutes a structurally independent domain that does not exhibit any foldase function or affect the amylase catalytic activity (8). Considering the preproenzyme structure, the export of α-amylase precursor through the inner membrane may likely occur via the Sec pathway (15), coupled to the cleavage of the leader peptide, defining the amino-terminal end of the mature enzyme. The resulting proenzyme is found in the culture supernatant as a precursor until the P. haloplanktis late exponential phase, when the action of a nonspecific extracellular protease removes the C-terminal domain, releasing the mature enzyme (8).
When the psychrophilic enzyme is produced by recombinant Escherichia coli cells, its secretion depends on the presence of the propeptide, since the truncated α-amylase, i.e., devoid of the C-terminal domain, accumulates in the E. coli periplasm (8). These results, combined with the observation that the amy propeptide can promote its own membrane spanning and accepts a foreign passenger, led the authors to conclude that this domain displays an autonomous secretion signal function, showing several features in common with a classic β-autotransporter (8).
In the present work we have extended the study of α-amylase secretion to a more physiologic environment by setting up a novel expression vector for the recombinant protein production in three cold-adapted bacteria, P. haloplanktis TAB23 (the α-amylase source strain), P. haloplanktis TAC125, and Psychrobacter sp. strain TAD1 (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this work
| Plasmid or strain | Description | Reference |
|---|---|---|
| Plasmids | ||
| pUCL | Vector deriving from the AflIII/EcoRI digestion of pUC18, fill-in reaction of the sticky ends and ligation; NdeI disruption | This work |
| Clone Q | pGEM-4z containing the T/R box | 18 |
| pFF | PUCL containing the T/R box, the promoter and termination region of the P. haloplanktis TAC125 aspC gene | This work |
| pαH12 | pUC12 derivative containing the P. haloplanktis α-amylase gene | 8 |
| pαH12wt* | pUC12 derivative containing the P. haloplanktis α-amylase gene devoid of the gene region encoding the C-terminal domain | 8 |
| pFFamy | pFF containing the pαH12 insert | This work |
| pFFamyΔCt | pFF containing the pαH12wt∗ insert | This work |
| Bacterial strains | ||
| TAD1 | Psychrobacter sp. strain TAD1 | 6 |
| TAB23 | P. haloplanktis TAB23 (formerly Alteromonas haloplanktis A23) | 9 |
| TAC125 | P. haloplanktis TAC125 | 3 |
| DH5α | E. coli [supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] | 12 |
| S17-1(λpir) | E. coli strain S17-1 (λpir) [thi pro hsd(r− m+) recA::RP4-2-Tcr::Mu Kmr::Tn7 Tpr Smr λpir] | 17 |
Construction of pFF expression vector.
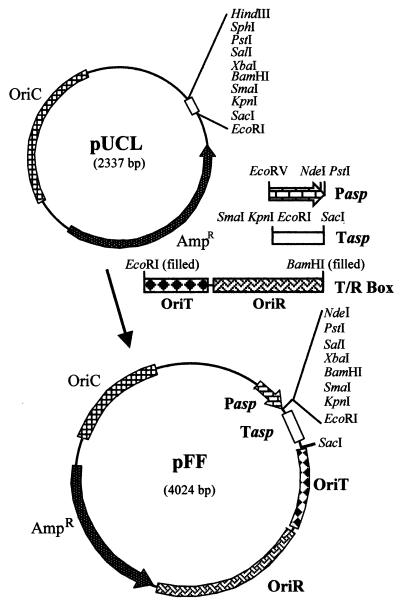
Molecular characterization of the pMtBL plasmid isolated from an Antarctic bacterium was instrumental in setting up a shuttle vector, clone Q, able to efficiently replicate either in psychrophiles or in E. coli (18). To develop an expression vector for Antarctic bacteria, the E. coli vector pUC18 was modified by destroying the unique NdeI restriction site and removing two elements, i.e., the lactose-inducible β-galactosidase promoter and the lacI gene fragment, to avoid interference with the cold-adapted promoter to be inserted, generating the pUCL vector (Fig. 1; Table 1). The T/R box, a DNA fragment containing the cold-adapted origin of replication (OriR) and the origin of conjugative transfer (OriT) derived from the pJB3 plasmid (4), was isolated from the clone Q shuttle vector (18) by double EcoRI/BamHI digestion and was cloned into pUCL EcoRI restriction site after a fill-in reaction. Transcription and translation control sequences of cold-adapted aspC gene (coding for aspartate aminotransferase from P. haloplanktis TAC125 [3]) were amplified by PCR and inserted into the above shuttle vector to generate a cold expression plasmid, the pFF construct (Fig. 1). The PhaspC promoter (Pasp) was amplified by using the primers 5′-GTTGGATCCGATATCAAAAATAGACTATG-3′ and 5′-TAATACTGCAGACATATGCTATCTCTTATC-3′, which introduces an NdeI restriction site at an optimal distance from the natural Shine-Dalgarno sequence and overlapping the aspartate aminotransferase start codon. Furthermore, the stability of the mRNAs expressed by the constitutive PhaspC promoter was increased by cloning the strong PhaspC rho-independent transcription terminator (Tasp, PCR amplified by using the primers 5′-GTTCCCGGGGTACCGAATTCATAATAGTTAACC-3′ and 5′-CCTTGAGCTCCCTGAGGTCCGG-3′) downstream of a small polylinker region.
FIG. 1.
Construction of pFF vector (see text).
Recombinant production of P. haloplanktis TAB23 α-amylase (precursor and mature forms) in three cold-adapted bacteria.
In previous work, two recombinant plasmids were constructed to produce the heterologous P. haloplanktis TAB23 α-amylase in E. coli (8). The two vectors (pαH12 and pαH12wt*) differed in the presence of the amy gene portion coding for the C-terminal propeptide of α-amylase (which was deleted and replaced by an artificial stop codon in the pαH12wt* vector). The expression of these vectors resulted in the production of the α-amylase precursor (pαH12) or the truncated native α-amylase (pαH12wt*). To construct pFFamy plasmid, the region coding for the P. haloplanktis TAB23 α-amylase was excised from pαH12 by EcoRI-XbaI double digestion and inserted into the pFF corresponding sites. The gene coding for α-amylase devoid of its C-terminal domain was obtained from the pαH12wt* vector by SalI/XbaI hydrolysis and inserted into the EcoRI/XbaI-digested pFF plasmid, generating the pFFamyΔCt vector. A fill-in reaction was necessary to make compatible SalI and EcoRI protruding ends.
The resulting plasmids, pFFamy and pFFamyΔCt (Fig. 2A), were mobilized into P. haloplanktis TAB23 (the source strain), P. haloplanktis TAC125, and Psychrobacter sp. strain TAD1 cells (both devoid of any α-amylase activity [6, 18]) by interspecific conjugation with the transformed E. coli S17-1(λpir) cells (Table 1) following the procedure previously described (18).
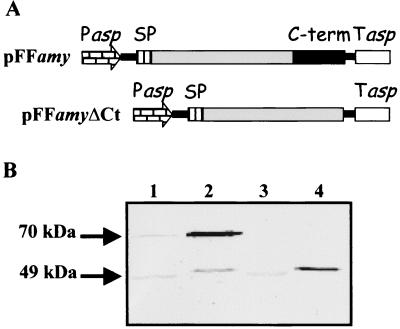
FIG. 2.
Recombinant production and cellular localization of P. haloplanktis TAB23 α-amylase (precursor and mature forms) in P. haloplanktis TAC125. (A) Schematic representation of gene constructs directing the production of the α-amylase precursor (pFFamy) and its mature form (pFFamyΔCt). Pasp, P. haloplanktis TAC125 aspC transcriptional promoter; SP, signal peptide; C-term, C-terminal propeptide. (B) Western blot analysis of cell extracts (lanes 1 and 3) and supernatants (lanes 2 and 4) of pFFamy (lanes 1 and 2) and pFFamyΔCt (lanes 3 and 4) recombinant P. haloplanktis TAC125 cells. The analyzed samples came from cultures at the same growth phase and from equal amount of cells. The immunodetection was performed by using anti-C-terminal polyclonal antiserum as described in reference 8.
Psychrophilic transconjugants were grown in liquid culture at 4°C, and samples were harvested at the mid-logarithmic phase. Culture supernatants and corresponding cell lysates were immunodetected by using anti P. haloplanktis TAB23 α-amylase antiserum (8) to evaluate production and cellular localization of the recombinant products. Western blotting analysis (Fig. 2B) of P. haloplanktis TAC125 transformed with pFFamy (lanes 1 and 2) and pFFamyΔCt (lanes 3 and 4) showed that recombinant proteins are present mainly in the culture medium (lanes 2 and 4), regardless of the presence of the C-terminal propeptide. Furthermore, a proteolytic processing, which converts the α-amylase precursor (70 kDa) into the mature enzyme (49 kDa; Fig. 2B, lanes 1 and 2), occurs even in the recombinant P. haloplanktis TAC125 cell culture. Similar results were obtained with recombinant P. haloplanktis TAB23 and Psychrobacter sp. strain TAD1 cells (data not shown).
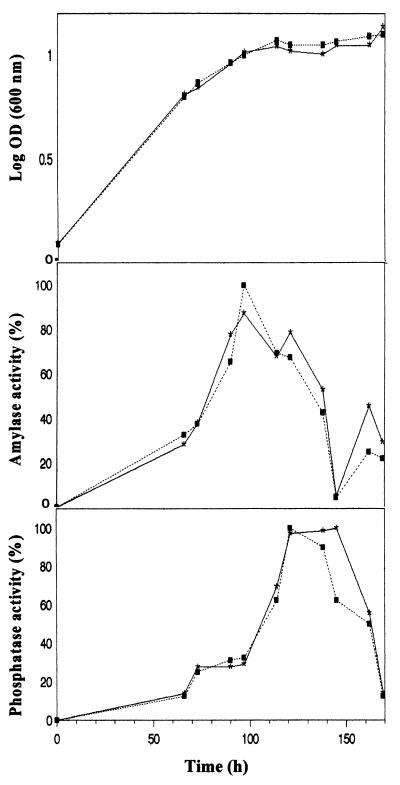
The kinetics of recombinant α-amylase production by P. haloplanktis TAC125 cells was monitored by observing the appearance of enzyme activity in the culture supernatants with the Boehringer-Roche kit AMYL as previously described (8). As shown in Fig. 3, middle panel, the extracellular targeting kinetics and the corresponding growth curves (upper panel) of the Antarctic bacterial cells, transformed with pFFamy and pFFamyΔCt vectors, were superimposable. Furthermore, the presence of the propeptide at the C terminus of the recombinant enzyme did not affect the yield of amylase secretion (Table 2). Similar results were obtained with recombinant P. haloplanktis TAB23 and Psychrobacter sp. strain TAD1 cells, although the cold α-amylase production yields for each of the three strains were slightly different (Table 2).
FIG. 3.
Secretion kinetics of recombinant precursor and mature α-amylase forms in recombinant P. haloplanktis TAC125 cells. Kinetics of bacterial growth (upper panel), recombinant α-amylase (middle panel), and alkaline phosphatase (lower panel) activities in the cell-free supernatants of P. haloplanktis TAC125 cells transformed with pFFamy (*) and pFFamyΔCt (▪) are presented. Enzyme activities are expressed as percentages of the maximal activity recorded in the cell-free supernatants. The curves were constructed from average results of three independent experiments at 4°C.
TABLE 2.
Recombinant α-amylase secretion by recombinant Antarctic bacteria at 4°C
| Vector | Maximal yielda (mg/liter) of recombinant α-amylase
|
||
|---|---|---|---|
| P. haloplanktis TAB23 | P. haloplanktis TAC125 | Psychrobacter sp. strain TAD1 | |
| pFFamy | 5.1 ± 0.1 | 4.8 ± 0.2 | 5.8 ± 0.6 |
| pFFamyΔCt | 6.2 ± 0.6 | 5.5 ± 0.5 | 6.0 ± 0.4 |
Data are average results of three independent experiments.
To demonstrate that the extracellular targeting of the recombinant α-amylase is due to a specific secretion mechanism rather than to a general leakiness of the outer membrane, the activity of a soluble periplasmic enzyme, the alkaline phosphatase, was monitored in the cell-free supernatants by using the Sigma Fast p-nitrophenyl phosphatate tablet sets. As illustrated in Fig. 3, the maximum of amylase activity in both cultures was reached about 50 h before the alkaline phosphatase release consequent to entry in stationary phase. At this time, the extracellular α-amylase activity decreased, likely due to the action of cellular proteases, whose concentration raises steadily during the prolonged cell growth (unpublished results from this laboratory). The same results were obtained with recombinant strains of P. haloplanktis TAB23 and Psychrobacter sp. strain TAD1 (data not shown).
Conclusions.
The assembly of the cold-adapted expression vector, reported in this paper, has been instrumental for the successful recombinant α-amylase production in three Antarctic bacteria (Fig. 1). This vector allows us to study the recombinant production and cellular localization of the native α-amylase and its truncated version, i.e., devoid of the C-terminal domain. In contrast to the previously reported results obtained with E. coli (8), our data demonstrate that when produced in all Antarctic bacteria tested (i) both recombinant enzymes are present mainly in the extracellular medium (Fig. 2B), (ii) the extracellular targeting of both α-amylase forms is a specific secretion rather than a general leakiness of the host outer membrane, (iii) C-terminal propeptide is not mandatory for α-amylase secretion, and (iv) the presence of the C-terminal domain does not interfere either with the secretion kinetics or with the maximal production yield (Fig. 3; Table 2).
The above results strongly suggest that the psychrophilic α-amylase carries secretion signals, besides the propeptide, that mediate the outer membrane translocation of cold-adapted bacteria, regardless of the propeptide presence at the enzyme C-terminal end. Based only on the substrate structure, the occurrence of a type II secretion pathway in these hosts can be proposed (15, 16). However, apart from the specific secretion mechanism implied, recombinant protein secretion by two distantly related bacteria (P. haloplanktis and Psychrobacter sp. strain TAD1) represents an interesting result.
The data obtained in this study, together with those reported by Feller et al. (8) for E. coli, suggest that the α-amylase from P. haloplanktis TAB23 is the only exoenzyme so far characterized that has the possibility of following two alternative secretion pathways depending on the bacterial host which is expressed. Indeed, in addition to some structural motifs likely recognized by a cold-adapted secretion machinery, this enzyme possesses an independent domain, the C-terminal propeptide, that can display its secretion helper role at least in the mesophilic E. coli.
Acknowledgments
This work was supported by grants of the European Union (program “COLDZYME,” contract ERB BIO4 CT96 0051; program “EUROCOLD,” contract ERB BIO4 CT95 0017; and program “COLDNET,” contract ERB FMRX CT97 0131), of Ministero dell'Università e della Ricerca Scientifica (Progetti di Rilevante Interesse Nazionale 1999), and of Consiglio Nazionale delle Ricerche (CNR contract 97.01138.PF49 Progetto Finalizzato “Biotecnologie”).
REFERENCES
- 1.Aghajari, N., G. Feller, C. Gerday, and R. Haser. 1998. Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level. Structure 6:1503-1516. [DOI] [PubMed] [Google Scholar]
- 2.Binet, R., S. Letoffe, J. M. Ghigo, P. Delepelaire, and C. Wandersman. 1997. Protein secretion by gram-negative bacterial ABC exporters—a review. Gene 192:7-11. [DOI] [PubMed] [Google Scholar]
- 3.Birolo, L., M. L. Tutino, B. Fontanella, C. Gerday, K. Mainolfi, S. Pascarella, G. Sannia, F. Vinci, and G. Marino. 2000. Aspartate aminotransferase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. Cloning, expression, properties, and molecular modelling. Eur. J. Biochem. 267:2790-2802. [DOI] [PubMed] [Google Scholar]
- 4.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 6.Di Fraia, R., V. Wilquet, M. A. Ciardiello, V. Carratore, A. Antignani, L. Camardella, N. Glansdorff, and G. Di Prisco. 2000. NADP+-dependent glutamate dehydrogenase in the Antarctic psychrotolerant bacterium Psychrobacter sp. TAD1. Characterization, protein and DNA sequence, and relationship to other glutamate dehydrogenases. Eur. J. Biochem. 267:121-131. [DOI] [PubMed] [Google Scholar]
- 7.Feller, G., S. D'Amico, and C. Gerday. 1999. Thermodynamic stability of a cold-active alpha-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38:4613-4619. [DOI] [PubMed] [Google Scholar]
- 8.Feller, G., S. D'Amico, A. M. Benotmane, F. Joly, J. Van Beeumen, and C. Gerday. 1998. Characterization of the C-terminal propeptide involved in bacterial wall spanning of alpha-amylase from the psychrophile Alteromonas haloplanctis. J. Biol. Chem. 273:12109-12115. [DOI] [PubMed] [Google Scholar]
- 9.Feller, G., T. Lonhienne, C. Deroanne, C. Libioulle, J. Van Beeumen, and C. Gerday. 1992. Purification, characterization, and nucleotide sequence of the thermolabile alpha-amylase from the Antarctic psychrotroph Alteromonas haloplanctis A23. J. Biol. Chem. 267:5217-5221. [PubMed] [Google Scholar]
- 10.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 11.Gerday, C., M. Aittaleb, M. Bentahir, J. P. Chessa, P. Claverie, T. Collins, S. D'Amico, J. Dumont, G. Garsoux, D. Georlette, A. Hoyoux, T. Lonhienne, M. A. Meuwis, and G. Feller. 2000. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 18:103-107. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 14.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matlack, K. E., W. Mothes, and T. A. Rapoport. 1998. Protein translocation: tunnel vision. Cell 92:381-390. [DOI] [PubMed] [Google Scholar]
- 16.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 17.Tascon, R. I., E. F. Rodriguez-Ferri, C. B. Gutierrez-Martin, I. Rodriguez-Barbosa, P. Berche, and J. A. Vazquez-Boland. 1993. Transposon mutagenesis in Actinobacillus pleuropneumoniae with a Tn10 derivative. J. Bacteriol. 175:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tutino, M. L., A. Duilio, E. Parrilli, E. Remaut, G. Sannia, and G. Marino. 2001. A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles 5:257-264. [DOI] [PubMed] [Google Scholar]