Abstract
Staphylococcus aureus isolates (n = 225) from bovine teat skin, human skin, milking equipment, and bovine milk were fingerprinted by pulsed-field gel electrophoresis (PFGE). Strains were compared to assess the role of skin and milking equipment as sources of S. aureus mastitis. PFGE of SmaI-digested genomic DNA identified 24 main types and 17 subtypes among isolates from 43 herds and discriminated between isolates from bovine teat skin and milk. Earlier, phage typing (L. K. Fox, M. Gershmann, D. D. Hancock, and C. T. Hutton, Cornell Vet. 81:183-193, 1991) had failed to discriminate between isolates from skin and milk. Skin isolates from humans belonged to the same pulsotypes as skin isolates from cows. Milking equipment harbored strains from skin as well as strains from milk. We conclude that S. aureus strains from skin and from milk can both be transmitted via the milking machine, but that skin strains are not an important source of intramammary S. aureus infections in dairy cows. A subset of 142 isolates was characterized by binary typing with DNA probes developed for typing of human S. aureus. Typeability and overall concordance with epidemiological data were lower for binary typing than for PFGE while discriminatory powers were similar. Within several PFGE types, binary typing discriminated between main types and subtypes and between isolates from different herds or sources. Thus, binary typing is not suitable as replacement for PFGE but may be useful in combination with PFGE to refine strain differentiation.
Staphylococcus aureus is an important cause of mastitis in dairy cows. Infected cows' udders are the main reservoir from which S. aureus is transmitted to other cows in the herd, and prevention of pathogen transmission from cow to cow reduces mastitis incidence (22). However, when mastitis control measures are implemented, new infections continue to occur, and eradication of S. aureus intramammary infection is difficult to achieve. Infections that originate from sources outside of the mammary gland may contribute to the infection control problem (26). Many sources of S. aureus exist, including housing materials and fodder, equipment and air, bovine skin, nonbovine animals, and humans (24). Teat skin has been suggested as an important reservoir for intramammary infection (24), while human-to-bovine transmission has also been proposed (5, 34).
Because many strains of S. aureus exist, isolates must be typed to the subspecies level to pinpoint the sources and routes of spread of the pathogen in a population (33). For decades, bacteriophage typing was the standard method for typing of S. aureus. Phage typing is still widely used today, despite a number of drawbacks (31, 37, 42). Drawbacks include limited typeability of isolates, limited technical reproducibility of results, and variable expression of epidemiological determinants, resulting in limited biological reproducibility. Pulsed-field gel electrophoresis (PFGE) is a DNA-based typing technique that has higher typeability, within-laboratory reproducibility, and discriminatory power than phage typing (2, 31). In the past decades, PFGE has replaced bacteriophage typing as the “gold standard” for typing of S. aureus (2, 4).
Although well suited for outbreak analysis, PFGE lacks between-center reproducibility because results depend on experimental conditions and because interpretation of banding patterns is open to subjective differences (4, 38). To overcome such differences, library typing systems have been developed in which any typing result has universal meaning (32). Examples of library typing systems for S. aureus are binary typing (41) and multilocus sequence typing (6). Binary typing does not require DNA sequencing equipment and is more likely than multilocus sequence typing to be available to peripheral microbiology laboratories (39). Binary typing was developed for human S. aureus. In a pilot study, it appeared to be a promising tool for typing of bovine S. aureus (43). Large-scale experiments, including comparison to epidemiological data and known typing methods (33), will be necessary to determine the usefulness of binary typing for bovine S. aureus as a routine typing technique (35).
The aim of the present study was to examine a collection of S. aureus isolates from bovine and human skin, milking machine unit liners, and bovine milk to assess the role of skin and the milking machine as reservoirs of mastitis pathogens. The collection had previously been characterized by means of phage typing (9). It is anticipated that typing by means of the current gold standard, PFGE, will reveal more-detailed information and additional insight. Secondly, the usefulness of binary typing for large-scale studies of bovine S. aureus was evaluated, based on comparison with epidemiological data and results from PFGE.
MATERIALS AND METHODS
Bacterial isolates.
S. aureus isolates had been collected by Fox and coworkers during a cross-sectional study of dairy herds in Washington State in 1987 (9). For the present study, 225 viable S. aureus isolates with known phage type were available from 43 herds, including isolates from bovine teat skin (n = 70), milkers' hands (n = 4), milking machine unit liners (n = 34), and bovine milk (n = 117). Teat skin isolates and milk isolates had been obtained from a random selection of cows in each herd. All milkers' hands had been sampled before milking of the cows, and all milking equipment had been sampled after the herd had been milked (9). Per herd, 1 to 20 isolates were available.
Phage typing.
Data on isolate origin and phage typing results were provided by Fox and colleagues (9). They used 18 phages and identified phage types by a six-digit code, where every digit represents the combined results for three phages. Per triplet of phages, eight permutations of susceptibility (+) and resistance (−) exist, with the following digits representing the results indicated: 0 = −, −, −; 1 = +, +, +; 2 = +, +, −; 3 = +, −, +; 4 = −, +, +; 5 = +, −, −; 6 = −, +, −; 7 = −, −, +. Thus, phage types that differed by one digit could differ in susceptibility to one, two, or three phages.
PFGE.
PFGE of SmaI-digested bacterial DNA of all isolates (n = 225) was performed as described before (33). DNA macrorestriction fragments were separated in a 1% SeaKem agarose gel (FMC, SanverTech, Heerhugowaard, The Netherlands) using a contour-clamped homogeneous electric field mapper (Bio-Rad, Veenendaal, The Netherlands) (43). A four-band difference between macrorestriction patterns was interpreted as a different pulsotype, with pulsotype being indicated by a capital letter. If isolates differed by up to three bands, they were classified as subtypes of a pulsotype, with subtypes being indicated by a numeral suffix (10, 36).
Binary typing.
Binary typing of a random sample of 142 isolates was performed with probes AW-1, 2, 3, 5, 6, 9, 11, 14, and 15 developed by Van Leeuwen et al. (41), using the method described for typing of bovine S. aureus (43). Binary typing of all isolates was not deemed necessary, because performance characteristics of typing techniques can be evaluated with sample collections of a hundred isolates or less (13, 27, 35). Macrorestriction fragments obtained through PFGE were Southern blotted onto Hybond N+ membranes (Amersham Life Science, Little Chalfont, Buckinghamshire, United Kingdom) and hybridized with each probe using enhanced chemiluminescence direct labeling and detection, according to the manufacturer's recommendations (Amersham). Presence or absence of a hybridization signal was scored with a one or a zero, resulting in a nine-digit binary code for each isolate. Binary codes were converted into decimal numbers represented as binary types (41).
Statistical analysis.
For isolates from bovine teat skin and milk, the association between site of isolation (teat skin versus milk) and strain (specified type versus all other types) was examined. Statistical significance of associations was tested by chi-square analysis or two-sided Fisher's exact test, as appropriate (Statistix for Windows, version 1.0; Analytical Software Co., La Jolla, Calif.). Human skin isolates were excluded from the analysis because the number of isolates was too limited to warrant the use of statistical testing. Liner isolates were excluded from analysis because liners could be contaminated with S. aureus originating from teat skin or from milk. Analyses were performed across herds. Numbers of isolates from individual herds were too small to permit significance testing within herds. Statistical significance was declared for a P of <0.05. Fisher's exact test was also used to test the association between pulsotype and binary type for isolates with pulsotype A or A.1. For other main type-subtype combinations, significance testing was not meaningful due to small numbers or lack of association between specific types.
Typeability was calculated as defined by Struelens et al. (31). To compare the discriminatory ability of techniques, Simpson's index of discrimination was calculated (13), as well as the approximate 95% confidence interval around this index (11). Nonoverlapping confidence intervals were considered indicative of statistically significant differences in discriminatory power (11).
RESULTS
Phage typing.
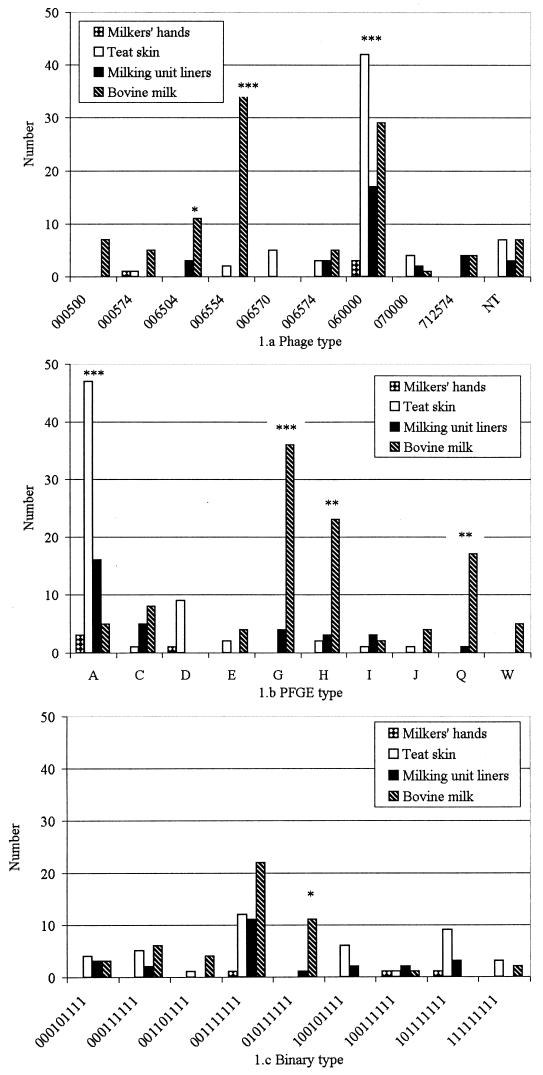
Of 225 PFGE-typed isolates, 208 were successfully typed by phage typing (Table 1). Of 21 phage types that were identified, 12 phage types were represented by four or fewer isolates. For the remaining nine phage types, the distribution over isolates from teat skin, hands, liners, and milk was determined (Fig. 1a). Phage type 0005000 was only isolated from milk, while types 000570 and 006570 were not isolated from milk at all. Phage type 060000 was significantly (but not exclusively) associated with isolation from teat skin (187 isolates included in statistical analysis; chi-square = 23.1; 1 df; P < 0.0001), and phage types 006554 and 006504 were significantly (but not exclusively) associated with isolation from milk (chi-square = 19.3, 1 df, and P < 0.0001 and chi-square = 6.99, 1 df, and P < 0.01, respectively). The majority of phage types that where represented by more than one isolate were found in multiple herds (10 of 13 or 77%).
TABLE 1.
Various typing methods for identification of S. aureus strains from bovine milk, skin, and milking equipment
| Technique | Collec- tiona | No. of typesb | Type- ability (%) | Simpson index of discriminationc (95% CI) |
|---|---|---|---|---|
| PFGE (main types only) | Full | 24 | 100 | 0.84 (0.81-0.87) |
| PFGE (main and subtypes) | Full | 24 + 17 | 100 | 0.92 (0.90-0.94) |
| PFGE (main types only) | BT | 20 | 100 | 0.80 (0.74-0.86) |
| PFGE (main and subtypes) | BT | 20 + 13 | 100 | 0.90 (0.88-0.93) |
| Phage typing | Full | 21 | 92.4 | 0.76 (0.71-0.82) |
| Phage typing | BT | 16 | 93.0 | 0.67 (0.58-0.76) |
| Binary typing | BT | 20 | 97.2 | 0.86 (0.81-0.90) |
Full, full collection of 225 samples; BT, subset of 142 samples used for binary typing.
For PFGE with distinction of subtypes, the number of main types + number of subtypes is given.
Point estimate and 95% confidence interval (CI) with calculation as described by Grundmann et al. (11) based on data from typeable isolates.
FIG. 1.
Distribution of phage types (a), PFGE types (b), and binary types (c) over sources of isolation. Only types that were represented by five or more S. aureus isolates are shown. Asterisks indicate significant differences between frequencies of isolation from teat skin and milk (∗, P ≤ 0.01; ∗∗, P ≤ 0.001; ∗∗∗, P ≤ 0.0001). When nothing is indicated, differences were not significant. NT, not typeable.
PFGE.
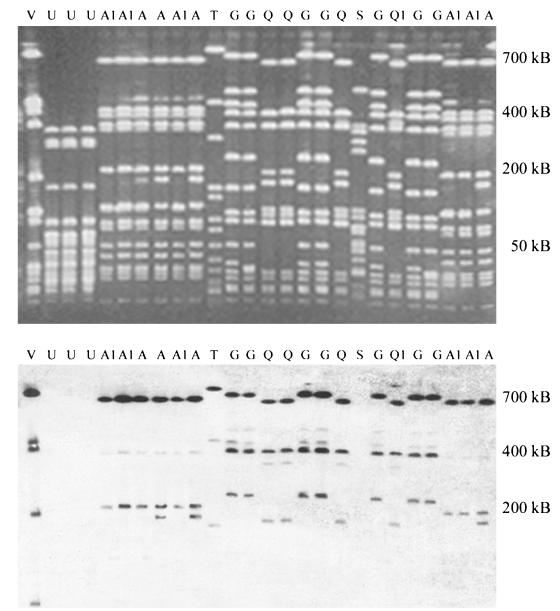
PFGE identified 24 main types and 17 subtypes (Table 1). Representative examples of PFGE banding patterns are shown in the upper panel of Fig. 2. Ten pulsotypes were represented by five or more isolates, and their distribution over teat skin, hands, liners, and milk is shown in Fig. 1b. Pulsotype D was only isolated from skin, while pulsotype W was only isolated from milk. Pulsotype A was significantly (but not exclusively) associated with isolation from teat skin (187 isolates included in analysis; chi-square = 86.2; 1 df; P < 0.0001), while types G, H, and Q were significantly (but not exclusively) associated with isolation from milk (chi-square = 26.7, 10.7, and 11.2, respectively; 1 df [for all three types]; P < 0.0001, P = 0.001, and P < 0.001, respectively). When main types and subtypes were considered separately, associations for main type A, subtype A.1, main type G, main type H, and main type Q were significant at P < 0.0001, P < 0.0001, P < 0.0001, P < 0.05, and P = 0.001, respectively. Within the subset of binary typed isolates (113 isolates included in statistical analysis), associations between site of isolation and types A, A.1, G, H, and Q were significant at P < 0.0001, P < 0.0001, P < 0.01, P < 0.01, and P < 0.001, respectively. The majority of pulsotypes that were represented by more than one isolate were found in multiple herds (9 of 13 or 69% of main types and 15 of 21 or 71% when main types and subtypes were considered separately).
FIG. 2.
Example of PFGE profiles of SmaI macrorestriction fragments (upper panel) and binary typing (lower panel) of S. aureus isolates from bovine teat skin, milking machine unit liners, and bovine milk. The lower panel shows hybridization of probe AW-14 to the macrorestriction fragments displayed in the upper panel. PFGE types are indicated with capital letters, and subtypes are indicated with a numeral suffix.
Binary typing.
Of 142 isolates that were binary typed, 138 were typeable while four isolates did not hybridize with any of the probes used. The proportion of isolates that hybridized ranged from 16.2% for probe AW-2 to 96.5% for probe AW-15, with a median of 92.3%. Representative examples of probe hybridization patterns are shown in the lower panel of Fig. 2. Binary typing identified 20 binary types (Table 1). Nine binary types were represented by five or more isolates, and their distribution over teat skin, hands, liners, and milk is shown in Fig. 1c. Binary type 9811 (code 010111111) was significantly associated with isolation from milk (113 isolates included in statistical analysis; chi-square = 10.0; 1 df; P < 0.01). This binary type was found in one herd only. No other binary types were significantly associated with milk or teat skin as the site of isolation. The majority of binary types (12 of 15 or 80%) were found in multiple herds.
Comparison of phage typing, PFGE, and binary typing.
Typeability and the discriminatory power of the techniques are summarized in Table 1. For comparison, results are given for calculations based on the full sample collection and for calculations based on the binary-typed subset. PFGE typing results are displayed for the interpretation of subtypes as belonging to the same strain as their main type and for interpretation of subtypes are displayed as separate strains. PFGE with interpretation of subtypes and main types as separate types is significantly more discriminatory than phage typing or PFGE with differentiation of main types only. PFGE with differentiation of main types only is more discriminatory than phage typing, but the difference is not significant. The discriminatory power of phage typing may have been overestimated, because some phage types differed in susceptibility to one phage only, and this is not necessarily indicative of different strains (12, 31). The discriminatory power of binary typing was similar to that of PFGE, but typeability was lower.
Several phage types and binary types were associated with multiple pulsotypes and vice versa (Table 2). For some types, close agreement between typing techniques was observed, e.g., for phage type 000574 and pulsotype E, phage type 000500 and pulsotype J, binary type 9811 and pulsotype Q, or binary type 30291 and pulsotype X. For other types, it was difficult to determine which combination of types should be considered concordant or discordant, e.g., for phage types 006504 or 006554 and pulsotypes G or H and for binary type 5715 and pulsotypes A, G, H, and I. To calculate concordance between techniques under a strict definition, the most frequent combination of a specified phage type or binary type with a specified pulsotype was considered concordant (such results are shown in boldface type in Table 2), while all other combinations were considered discordant. Based on this classification, concordance was 44% for phage typing and PFGE (100 of 225 isolates) and 38% for binary typing and PFGE (54 of 142 isolates). Under a lenient classification scheme (data italicized in Table 2), associations of a phage type or binary type with multiple pulsotypes and vice versa were considered to be in agreement for frequently occurring combinations. Under this lenient definition, concordance was 64% for phage typing and PFGE (133 of 225 isolates) and 63% for binary typing and PFGE (89 of 142 isolates).
TABLE 2.
Cross tabulation of main PFGE types and phage types or binary typesa
| Type | No. of isolates with PFGE type
|
Total no. of isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | D | E | G | H | I | J | Q | W | X | MX | ||
| Phage | |||||||||||||
| 000500 | 1 | 1 | 4 | 1 | 7 | ||||||||
| 000574 | 1 | 4 | 2 | 7 | |||||||||
| 006504 | 4 | 7 | 2 | 1 | 14 | ||||||||
| 006554 | 1 | 2 | 20 | 10 | 3 | 36 | |||||||
| 006570 | 5 | 5 | |||||||||||
| 006574 | 8 | 2 | 1 | 11 | |||||||||
| 060000 | 57 | 2 | 1 | 4 | 14 | 4 | 2 | 8 | 92 | ||||
| 070000 | 5 | 1 | 6 | ||||||||||
| 712574 | 2 | 4 | 2 | 8 | |||||||||
| MX | 3 | 3 | 3 | 1 | 4 | 3 | 1 | 2 | 2 | 22 | |||
| NT | 5 | 1 | 1 | 4 | 1 | 4 | 1 | 17 | |||||
| Total | 71 | 14 | 10 | 6 | 40 | 28 | 6 | 5 | 18 | 5 | 4 | 18 | 225 |
| Binary | |||||||||||||
| 000101111 = 1107 | 7 | 1 | 1 | 1 | 10 | ||||||||
| 000111111 = 1619 | 5 | 2 | 1 | 3 | 2 | 1 | 13 | ||||||
| 001101111 = 5203 | 3 | 1 | 5 | ||||||||||
| 001111111 = 5715 | 11 | 1 | 2 | 10 | 13 | 6 | 1 | 2 | 46 | ||||
| 010111111 = 9811 | 1 | 10 | 11 | ||||||||||
| 100101111 = 17491 | 8 | 9 | |||||||||||
| 100111111 = 18003 | 5 | 5 | |||||||||||
| 101111111 = 22099 | 13 | 13 | |||||||||||
| 111111111 = 30291 | 4 | 1 | 5 | ||||||||||
| MX | 8 | 3 | 3 | 4 | 8b | 26 | |||||||
| Total | 58 | 6 | 1 | 6 | 14 | 13 | 6 | 4 | 14 | 3 | 4 | 13 | 142 |
For phage types, results for 225 S. aureus isolates from bovine milk, skin, and milking equipment were considered. For binary types, results for 142 S. aureus isolates were considered. Data in boldface type are predominant PFGE and phage types. Data in italic type are considered to be in agreement under a lenient classification scheme. Abbreviations: MX, miscellaneous; NT, not typeable.
Concordant results were obtained for five of eight isolates from miscellaneous categories.
PFGE showed better agreement with epidemiological data than phage typing, as it divided the main phage type (060000) into multiple pulsotypes in accordance with site of isolation (pulsotype A predominantly from skin and liners and pulsotypes Q and W predominantly from milk [Fig. 1; Table 2]). Across herds, concordance between epidemiologic data and typing data was higher for PFGE than for binary typing, as significant associations between source and type were found for multiple pulsotypes but not binary types (Fig. 1). Within several pulsotypes, binary typing differentiated between herds and sources of isolation (Table 3) or main types and their associated subtypes (main types A, J, and W [Tables 3 and 4 ]). For pulsotypes that are not included in Tables 3 and 4, binary typing did not divide pulsotypes in agreement with epidemiological origin or subtype identity.
TABLE 3.
Subdivision of S. aureus pulsotypes by binary typing in agreement with herd of origin, source of isolation, or subtype identification
| Pulso- type | Herd(s) of origin | Sourcea | Binary code (type) | No. of isolates | Differentiation by binary typing |
|---|---|---|---|---|---|
| C | 7 | L | 000111111 (1619) | 2 | Herds |
| 8 | M/L | 011111111 (13907) | 3 | ||
| 15 | M | 001111111 (5715) | 1 | ||
| E | 13 | M | 000101111 (1107) | 1 | Source |
| M | 001101111 (5203) | 3 | |||
| TS | 001111111 (5715) | 2 | |||
| F | 4 | TS | 001111111 (5715) | 1 | Herds |
| 12 | M | 000101111 (1107) | 1 | ||
| 24 | M | 000111111 (1619) | 1 | ||
| J | 1 | M | 000111111 (1619) | 3 | Herds, subtypes |
| J.1 | 13 | TS | 001111111 (5715) | 1 | |
| Q | 5 | M | 010111111 (9811) | 10 | Herds |
| 19 | M | 000011111 (595) | 1 | ||
| 20 | M/L | 010101011 (9283) | 3 | ||
| W | 5, 7 | M | 000111111 (1619) | 2 | Herds, subtypes |
| W.1 | 12 | M | 000101111 (1107) | 1 |
Abbreviations: L, liner; M, milk; TS, teat skin; M/L, milk and liner.
TABLE 4.
Subdivision of S. aureus pulsotypes A and A.1 by binary typing for herds that harbored both pulsotypesa
| Binary type | No. of isolates in pulsotype:
|
Pb | |
|---|---|---|---|
| A | A.1 | ||
| 001.111.111 = 5715 | 0 | 7 | <0.01 |
| 000.111.111 = 1619 | 1 | 4 | 0.13 |
| 000.101.111 = 1107 | 1 | 4 | 0.13 |
| 100.101.111 = 17491 | 3 | 0 | 0.12 |
| 101.111.111 = 22099 | 12 | 1 | <0.001 |
| Miscellaneous | 1 | 1 | Not tested |
Results from other herds and for other subtypes of pulsotype A were excluded.
Statistical significance according to the Fisher exact test of association between pulsotype (A versus A.1) and binary type (specified type versus all other binary types listed in the table).
DISCUSSION
The first purpose of this study was to compare S. aureus isolates from skin and milk. PFGE differentiates between S. aureus strains that are predominantly isolated from healthy bovine teat skin and S. aureus strains that are predominantly isolated from milk. The association between site of isolation and strain is highly significant for the most frequently isolated pulsotypes and subtypes in this study. From these data, we conclude that teat skin is not an important reservoir for bovine intramammary infection. This is in contrast to conclusions from earlier studies that were based on phage typing of S. aureus (9) or quantitative analysis of bacteriological data (24). A discrepancy between conclusions based on PFGE typing and phage typing is easily explained. PFGE typing was more discriminatory than phage typing, and the predominant phage type was divided into multiple distinct pulsotypes in accordance with epidemiological data. Typeability and discriminatory power of phage typing for our sample collection were similar to results reported for previous studies of bovine S. aureus (1). PFGE is known to be more discriminatory than phage typing (10, 27, 37, 42). This is one of the reasons why PFGE replaced phage typing as the gold standard for S. aureus typing (2). Our study confirms that the discriminatory power of phage typing may not be sufficient to detect statistical or biological associations between strains and their sources.
The discrepancy between our results and the aforementioned quantitative analysis (24) may be due to epidemiological differences between the study populations and to differences in data interpretation. Our study deals with lactating cows, while Roberson et al. (24) studied nonlactating young stock. Roberson and his coworkers found heifers with S. aureus “on teats” (i.e., on teat skin, on the teat orifice, or in the teat canal) to be more likely to have S. aureus in milk samples at parturition than heifers without S. aureus on teats. The odds ratio was estimated at 3.34, and the statistical significance of this result was reported as P = 0.07. Because the number of intramammary infections that was observed in their study was limited, an exact test was more appropriate than the chi-square analysis reported in the paper. Reanalysis of the data supplied in the original paper by means of logistic regression and an exact test (LogXact, version 1.2; Cytel Software Corporation, Cambridge, Mass.) resulted in an estimated odds ratio of 3.33 with a P value of 0.14. Thus, the results are suggestive of a role of teat skin as reservoir for intramammary infection in heifers but are not conclusive. In the quantitative analysis, bacteria were identified to the species level but not to the strain level. In another study, Roberson et al. (25) examined the role of teat skin as a source of S. aureus infections detected at calving using phage typing. Teat skin was a possible source of infection in one of eight heifers based on phage typing results, but genotypic methods were not used.
In our study, some overlap between strains from skin and milk was observed by phenotypic and genotypic methods. This could indicate that skin may incidentally be a source of intramammary infection, as suggested by the studies discussed above (24, 25), that typing methods did not have sufficient discriminatory power to distinguish fully between skin and milk isolates, or that some samples from teat skin and milk were cross-contaminated. Care was taken to collect samples aseptically, but swabbing of the teat skin and teat orifice may result in contact of the swab with milk, especially if milk letdown occurs. Similarly, milk samples may get contaminated with bacteria from other sources. To avoid misclassification of contamination and infection, multiple milk samples are used to define infection status of the mammary gland in many studies (21). In our study, single milk samples were used. Only four pulsotype A isolates were identified among S. aureus isolates from 1,703 milk samples that were originally collected (0.2%). Even if type A isolates in milk originated from contamination of samples, this would indicate a very low level of cross-contamination.
The discriminatory power of PFGE depends in part on the interpretation of typing results. Tenover and colleagues (36) formulated guidelines for interpretation of restriction patterns. Their categorization of isolates as “indistinguishable” (no band differences), “closely related” (one to three band differences), “possibly related” (four to six band differences), or “different” (more than six band differences) was developed to classify strains from disease outbreaks. It has been applied in many other settings, e.g., to study large collections of staphylococcal isolates from multiple hospitals (42) or isolates collected over long periods of time (10). Often, the classification is simplified, and isolates that differ from a main type by one to three bands are classified as subtypes of the main type, while isolates with four or more band differences are considered different main types (10, 27). In large-scale studies of dairy herds, more-extreme classifications have been used. On the one hand, “splitters” considered any band difference indicative of a strain difference, arguing that isolates were selected from multiple herds and not from a limited outbreak (14, 16). On the other hand, “lumpers” have classified isolates as subtypes when they differed from the main type by six bands (3, 30). In our study, subtypes were usually identified in herds where the main type was also present (14 of 17 subtypes). Hence, subtypes and main types could be epidemiologically related, which would justify the interpretation of a one- to three-band difference as indicative of a subtype. However, binary typing differentiated between main types and subtypes for several PFGE types as shown in Tables 3 and 4. This would support interpretation of any band difference as indicative of a new type. The conclusion that skin isolates are different from milk isolates held true when PFGE subtypes were included with main types and when subtypes were treated as separate types.
The majority of phage types, pulsotypes, and binary types were found in multiple herds. This is in contrast with results from Korea (14) and Denmark (18), where the majority of types was unique to a herd, but in agreement with results from many other studies. Based on phage typing (18, 19), ribotyping (18, 23), multilocus enzyme electrophoresis (7, 15), random amplified polymorphic DNA typing (7), PFGE (3, 30, 43), coagulase gene typing (20), and binary typing (43), a number of strains and clones of S. aureus were found to be common to multiple herds, regions, countries, and continents. The predominance of a limited number of S. aureus strains may be the result of an increased resistance to the host immune response (20). The differences between studies depend partly on data interpretation, as discussed, and partly on the choice of typing techniques. Techniques that target different characteristics of the bacteria may result in a different outcome (35), partly because markers differ in molecular clock speed (29, 40). As a result, S. aureus isolates that appear identical when typed by one method, and hence appear common to multiple herds, may be discriminated and found to be unique to a herd when typed by a different technique. The choice of technique or combination of techniques should depend on spatial and temporal aspects of the epidemiological question at hand and on availability of laboratory facilities and expertise (31, 37). In the present study, PFGE was a satisfactory technique to demonstrate nonidentity of isolates from milk and other sources.
The number of human skin isolates in our study was limited. Isolates were obtained through swabbing of milkers' hands before milking of the herd and are therefore unlikely to be the result of hand contamination during milking. Pulsotypes of human skin isolates were akin to pulsotypes of bovine skin isolates and differed from bovine milk isolates. Phage typing and binary typing failed to discriminate between human skin isolates and isolates from other sources. In most comparative studies of human and bovine S. aureus isolates, the focus is on host specificity. Studies based on phage typing or ribotyping found S. aureus strains from human skin to be similar to those from bovine milk or skin (18, 34), but enterotoxin typing and binary typing revealed differences between human S. aureus isolates and bovine mammary isolates in situations where there was no epidemiological association between study objects (17, 43). Results of multilocus enzyme electrophoresis typing of bovine and human S. aureus isolates were originally interpreted as “consistent with the concept of host specialization” (15) and subsequently were interpreted as “indicating that many bovine isolates are more closely allied to human isolates than to other bovine isolates” (8). Our present data, though scarce, suggest that in addition to host specificity there may be organ specificity and that this organ specificity may run across lines of host specificity. Adaptation of pathogenic clones to specialized niches, e.g., the mammary gland, has also been proposed based on DNA microarray analysis of two bovine and two ovine S. aureus isolates (8).
Machine milking unit liners are important fomites for transmission of S. aureus in dairy herds, and the use of liner backflush reduces contamination with bacteria (9). Our PFGE analysis revealed that liners could be contaminated with S. aureus from teat skin and with S. aureus from milk. This implies that liners are fomites for skin flora and for intramammary infections. Although liners can be contaminated with skin and udder flora, transmission from skin flora to the mammary gland and vice versa seems rare, as indicated by the site-specific pulsotypes obtained in this study. This would be in line with the proposed hypothetical concept of organ specificity of S. aureus strains.
The second purpose of the present study was to determine whether binary typing was suited for large-scale molecular epidemiological studies of bovine S. aureus. Binary typing yielded higher typeability than phage typing but lower typeability than PFGE. The combination of PFGE with Southern blotting and subsequent binary typing that was used in this study had technical drawbacks. In this procedure, DNA concentrations could not be standardized. As a result, several isolates gave weak hybridization signals, making interpretation difficult and defeating one of the main purposes of a library typing technique, i.e., unequivocal interpretation of typing results. In addition, DNA fragments smaller than 25 kb may have been lost during electrophoresis, which could account for nontypeability of isolates. Most probe binding occurred to large macrorestriction fragments, but hybridization to smaller fragments was observed (example: strain V in the lower panel of Fig. 2). Alternatively, nontypeability may be the result of the failure of bovine strains to bind probes that were developed for typing of human S. aureus, because bovine and human strains largely belong to different S. aureus lineages.
The discriminatory power of binary typing was comparable to that of PFGE typing, in agreement with results from studies on binary typing of human S. aureus (41). Use of additional probes could further improve the discriminatory power of binary typing (41), and development of host-specific probes could contribute to improved typeability. Overall concordance with epidemiological data was poorer for binary typing than for PFGE. Concordance with epidemiological data was assessed semiquantitatively, because the quantitative analysis proposed by Struelens et al. (31) depends on knowledge of outbreak-related strains. There was no single defined outbreak in our study, and any modification of Struelens's calculation method would amount to a semiquantitative analysis similar to the results from our statistical analysis. Within several pulsotypes, binary typing identified multiple strains, and this subdivision was partly in agreement with epidemiological data such as herd of origin or site of isolation. The combined use of multiple methods is known to be more discriminatory than the use of PFGE alone (28), and it can be anticipated that other techniques will supplement or even replace PFGE as the gold standard for typing of S. aureus (6, 28, 29, 39). Ideally, library typing methods with high discriminatory power and ease of use for peripheral laboratories would be available. Depending on improved typeability and concordance with epidemiological data, binary typing of bovine S. aureus might be developed into such a method.
To summarize, PFGE showed that S. aureus isolates from bovine teat skin were different from isolates from bovine milk and similar to a small number of isolates from human skin. Milking machine unit liners may transmit strains from skin and milk. PFGE had better typeability and concordance with site of isolation than phage typing or binary typing, and higher discriminatory power than phage typing. The discriminatory power of binary typing is similar to that of PFGE. Based on typeability and concordance with epidemiological data, binary typing is not suited yet to be used as a stand-alone technique for large-scale molecular studies of bovine S. aureus, but it is a useful addition to PFGE for refinement of strain identification.
REFERENCES
- 1.Aarestrup, F. M., H. C. Wegener, and V. T. Rosdahl. 1995. Evaluation of phenotypic and genotypic methods for epidemiological typing of Staphylococcus aureus isolates from bovine mastitis in Denmark. Vet. Microbiol. 45:139-150. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., D. Hancock, F. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buzzola, F. R., L. Quelle, M. I. Gomez, M. Catalano, L. Steele-Moore, D. Berg, E. Gentilini, G. Denamiel, and D. O. Sordelli. 2001. Genotypic analysis of Staphylococcus aureus from milk of dairy cows with mastitis in Argentina. Epidemiol. Infect. 126:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aries de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Devriese, L. A., and J. Hommez. 1975. Epidemiology of methicillin-resistant Staphylococcus aureus in dairy herds. Res. Vet. Sci. 19:23-27. [PubMed] [Google Scholar]
- 6.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald, J. R., W. J. Meaney, P. J. Hartigan, C. J. Smyth, and V. Kapur. 1997. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol. Infect. 119:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, L. K., M. Gershman, D. D. Hancock, and C. T. Hutton. 1991. Fomites and reservoirs of Staphylococcus aureus causing intramammary infections as determined by phage typing: the effect of milking time hygiene practices. Cornell Vet. 81:183-193. [PubMed] [Google Scholar]
- 10.Givney, R., A. Vickery, A. Holliday, M. Pegler, and R. Benn. 1998. Evolution of an endemic methicillin-resistant Staphylococcus aureus population in an Australian hospital from 1967 to 1996. J. Clin. Microbiol. 36:552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter, P. R. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo, Y. S., L. K. Fox, W. C. Davis, G. A. Bohach, and Y. H. Park. 2001. Staphylococcus aureus associated with mammary glands of cows: genotyping to distinguish different strains among herds. Vet. Microbiol. 80:131-138. [DOI] [PubMed] [Google Scholar]
- 15.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange, C., M. Cardoso, D. Senczek, and S. Schwarz. 1999. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet. Microbiol. 67:127-141. [DOI] [PubMed] [Google Scholar]
- 17.Larsen, H. D., A. Huda, N. H. Eriksen, and N. E. Jensen. 2000. Differences between Danish bovine and human Staphylococcus aureus isolates in possession of superantigens. Vet. Microbiol. 76:153-162. [DOI] [PubMed] [Google Scholar]
- 18.Larsen, H. D., K. H. Sloth, C. Elsberg, C. Enevoldsen, L. H. Pedersen, N. H. Eriksen, F. M. Aarestrup, and N. E. Jensen. 2000. The dynamics of Staphylococcus aureus intramammary infection in nine Danish dairy herds. Vet. Microbiol. 71:89-101. [DOI] [PubMed] [Google Scholar]
- 19.Mackie, D. P., D. A. Pollock, S. P. Rodgers, and E. F. Logan. 1987. Phage typing of Staphylococcus aureus associated with subclinical bovine mastitis. J. Dairy Res. 54:1-5. [DOI] [PubMed] [Google Scholar]
- 20.Mullarky, I. K., C. Su, N. Frieze, Y. H. Park, and L. M. Sordillo. 2001. Staphylococcus aureus agr genotypes with enterotoxin production capabilities can resist neutrophil bactericidal activity. Infect. Immun. 69:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neave, F. K. 1975. Diagnosis of mastitis by bacteriological methods alone, p. 19-36. In Proceedings of the Seminar on Mastitis Control, 1975. International Dairy Federation, Brussels, Belgium.
- 22.Neave, F. K., F. H. Dodd, R. G. Kingwill, and D. R. Westgarth. 1969. Control of mastitis in the dairy herd by hygiene and management. J. Dairy Sci. 52:696-707. [DOI] [PubMed] [Google Scholar]
- 23.Rivas, A. L., R. N. Gonzalez, M. Wiedmann, J. L. Bruce, E. M. Cole, G. J. Bennett, H. F. Schulte III, D. J. Wilson, H. O. Mohammed, and C. A. Batt. 1997. Diversity of Streptococcus agalactiae and Staphylococcus aureus ribotypes recovered from New York dairy herds. Am. J. Vet. Res. 58:482-487. [PubMed] [Google Scholar]
- 24.Roberson, J. R., L. K. Fox, D. D. Hancock, J. M. Gay, and T. E. Besser. 1994. Ecology of Staphylococcus aureus isolated from various sites on dairy farms. J. Dairy Sci. 77:3354-3364. [DOI] [PubMed] [Google Scholar]
- 25.Roberson, J. R., L. K. Fox, D. D. Hancock, J. M. Gay, and T. E. Besser. 1998. Sources of intramammary infections from Staphylococcus aureus in dairy heifers at first parturition. J. Dairy Sci. 81:687-693. [DOI] [PubMed] [Google Scholar]
- 26.Saperstein, G., L. S. Hinckley, and J. E. Post. 1988. Taking the team approach to solving staphylococcal mastitis infection. Vet. Med. 83:939-947. [Google Scholar]
- 27.Shimizu, A., J. Kawano, C. Yamamoto, O. Kakutani, and M. Fujita. 1997. Comparison of pulsed-field gel electrophoresis and phage typing for discriminating poultry strains of Staphylococcus aureus. Am. J. Vet. Res. 58:1412-1416. [PubMed] [Google Scholar]
- 28.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shopsin, B., M. Gomez, M. Waddington, M. Riehman, and B. N. Kreiswirth. 2000. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 38:3453-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sordelli, D. O., F. R. Buzzola, M. I. Gomez, L. Steele-Moore, D. Berg, E. Gentilini, M. Catalano, A. J. Reitz, T. Tollersrud, G. Denamiel, P. Jeric, and J. C. Lee. 2000. Capsule expression by bovine isolates of Staphylococcus aureus from Argentina: genetic and epidemiologic analyses. J. Clin. Microbiol. 38:846-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Struelens, M., and Members of the European Study Group on Epidemiological Markers (ESGEM) of the European Society for Clinical Microbiology and Infectious Diseases (ESCMID). 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 32.Struelens, M. J., Y. De Gheldre, and A. Deplano. 1998. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect. Control Hosp. Epidemiol. 19:565-569. [DOI] [PubMed] [Google Scholar]
- 33.Struelens, M. J., A. Deplano, C. Godard, N. Maes, and E. Serruys. 1992. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swartz, R., P. J. Jooste, and J. C. Novello. 1985. Bacteriophage typing of Staphylococcus aureus strains isolated from Bloemfontein dairy herds. J. S. Afr. Vet. Assoc. 56:69-73. [PubMed] [Google Scholar]
- 35.Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hebert, B. Hill, and R. Hollis. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Belkum, A. 2000. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains: state of affairs and tomorrow' s possibilities. Microb. Drug Resist. 6:173-188. [DOI] [PubMed] [Google Scholar]
- 38.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Leeuwen, W., C. Libregts, M. Schalk, J. Veuskens, H. Verbrugh, and A. van Belkum. 2001. Binary typing of Staphylococcus aureus strains through reversed hybridization using digoxigenin-universal linkage system-labeled bacterial genomic DNA. J. Clin. Microbiol. 39:328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Leeuwen, W., A. van Belkum, B. Kreiswirth, and H. Verbrugh. 1998. Genetic diversification of methicillin-resistant Staphylococcus aureus as a function of prolonged geographic dissemination and as measured by binary typing and other genotyping methods. Res. Microbiol. 149:497-507. [DOI] [PubMed] [Google Scholar]
- 41.van Leeuwen, W., H. Verbrugh, J. van der Velden, N. van Leeuwen, M. Heck, and A. van Belkum. 1999. Validation of binary typing for Staphylococcus aureus strains. J. Clin. Microbiol. 37:664-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, J., R. Borrow, R. V. Goering, S. Egerton, A. J. Fox, and B. A. Oppenheim. 1999. Subtyping of methicillin-resistant Staphylococcus aureus isolates from the north-west of England: a comparison of standardised pulsed-field gel electrophoresis with bacteriophage typing including an inter-laboratory reproducibility study. J. Med. Microbiol. 48:297-301. [DOI] [PubMed] [Google Scholar]
- 43.Zadoks, R., W. van Leeuwen, H. Barkema, O. Sampimon, H. Verbrugh, Y. H. Schukken, and A. van Belkum. 2000. Application of pulsed-field gel electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J. Clin. Microbiol. 38:1931-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]